Abstract
Studies of copy-number variation and linkage disequilibrium (LD) have typically excluded complex regions of the genome that are rich in duplications and prone to rearrangement. In an attempt to assess the heritability and LD of copy-number polymorphisms (CNPs) in duplication-rich regions of the genome, we profiled copy-number variation in 130 putative “rearrangement hotspot regions” among 269 individuals of European, Yoruba, Chinese, and Japanese ancestry analyzed by the International HapMap Consortium. Eighty-four hotspot regions, corresponding to 257 bacterial artificial chromosome (BAC) probes, showed evidence of copy-number differences. Despite a predisposing genetic architecture, no polymorphism was ever observed in the remaining 46 “rearrangement hotspots,” and we suggest these represent excellent candidate sites for pathogenic rearrangements. We used a combination of BAC-based and high-density customized oligonucleotide arrays to resolve the molecular basis of structural rearrangements. For common variants (frequency >10%), we observed a distinct bias against copy-number losses, suggesting that deletions are subject to purifying selection. Heritability estimates did not differ significantly from 1.0 among the majority (30 of 34) of loci analyzed, consistent with normal Mendelian inheritance. Some of the CNPs in duplication-rich regions showed strong LD with nearby single-nucleotide polymorphisms (SNPs) and were observed to segregate on ancestral SNP haplotypes. However, LD with the best available SNP markers was weaker than has been reported for deletion polymorphisms in less complex regions of the genome. These observations may be accounted for by a low density of SNP data in duplicated regions, challenges in mapping and typing the CNPs, and the possibility that CNPs in these regions have rearranged on multiple haplotype backgrounds. Our results underscore the need for complete maps of genetic variation in duplication-rich regions of the genome.
Variation in the human genome occurs on multiple levels, from the SNP to larger events involving contiguous blocks of DNA sequence that vary in copy number between individuals. Although the technological development of SNP detection and genotyping methods has progressed significantly in the past decade, the ability to detect copy-number variants (CNVs) on a genomewide scale has emerged only recently. Current array-based methods typically detect CNVs ⩾40 kb in size, and variation at this level of resolution has been shown to occur frequently in the human population.1–3 On the basis of a report published elsewhere, it has been estimated that any two individuals differ by >11 CNVs that are >100 kb.2 At a finer level of resolution, a recent analysis comparing a single individual with the reference human genome identified 297 intermediate-sized structural variants (ISVs) in the 8–200-kb range (77 events >40 kb).4 Structural variation is therefore an important subject for study, not only to understand the full spectrum of human genetic variation, but also to assess the significance of such variation in disease-association studies.
Several consistent themes have emerged from recently published studies of copy-number polymorphisms (CNPs), CNVs with a frequency >1%. Of primary importance to understanding the relationship between genotype and phenotype is the fact that CNPs are frequently found in genic regions. This association is exemplified by studies of toxin sensitivity and variation in the copy number of members of the glutathione S-transferase gene family GSTT1 and GSTM1.5 Also, CNPs and ISVs have been found, by multiple genomewide approaches,1–4 to be enriched in regions of intrachromosomal segmental duplication, and many deletions have been shown to be flanked by pairs of paralogous sequences in a direct orientation.6 These findings indicate that genes found in regions of segmental duplication are more likely to vary in copy number in the human population. The majority of CNP studies to date, however, have used a panel of unrelated individuals, and questions about the heritability of CNPs have been left unaddressed. More importantly, it is unknown whether CNPs are in linkage disequilibrium (LD) with nearby single-nucleotide variation. Previous studies of unique regions of the genome suggest that nearby SNPs may serve as markers for deletion polymorphisms,6,7 but the association of CNPs with SNPs in duplication-rich regions—which are more likely to have undergone multiple rearrangements—has not been addressed. Furthermore, the LD properties of structural polymorphisms involving gains of copies are almost completely unknown.
In this study, we present an analysis of CNPs within the sample populations used by the International HapMap Project8 and assess somatic variation among diverse tissue sources. By using array-based comparative genomic hybridization (array CGH) targeted to regions of segmental duplication, we focus our efforts on assessing variation in complex regions that are prone to rearrangement.3 A combination of both BAC-based and high-density oligonucleotide arrays allowed for an extremely detailed view and illuminated the molecular basis of a subset of CNPs. We assessed copy-number variation in all four HapMap population samples, a total of 269 individuals. Using DNA samples and available SNP data from the International HapMap Project, we analyzed patterns of SNP density, heritability, and LD at sites of CNP in duplication-rich regions of the genome.
Methods
Samples
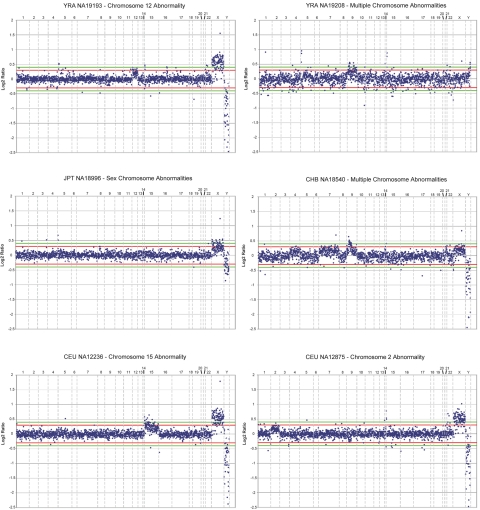
The samples profiled in this study were those used in the International HapMap Project.8,9 Hybridizations were performed on DNA from all 269 individuals sampled by the HapMap Consortium; these consisted of 90 individuals (30 trios) of European ancestry sampled in Utah (CEU); 90 individuals (30 trios) of Yoruba ancestry, sampled in Ibadan, Nigeria (YRI); 45 unrelated individuals of Han Chinese ancestry, sampled in Beijing (CHB); and 44 unrelated individuals of Japanese ancestry, sampled in Tokyo (JPT). DNA samples were obtained from Coriell Cell Repositories. A few profiles showed chromosomal aneuploidy, suggestive of cell-line artifacts; therefore, the data for those chromosomes were not considered. The following samples were affected: JPT NA18996, YRI NA19208, YRI NA19193, CHB NA18540, CEU NA12236, and CEU NA12875 (fig. 1). The reference DNA used for all hybridizations was a single male of Czechoslovakian descent, Coriell ID GM15724, which is a well-characterized sample used in a previous array CGH study.3 In the present study, a CNV was classified as a CNP if altered copy number was observed in >1% of the 269 individuals sampled. We refer to “altered copy-number frequency” (ACNF) instead of “minor-allele frequency,” because measurements of copy number are on diploid samples, and, in most cases, the actual allele structure of the variant has not been resolved at the molecular level.
Figure 1. .
Array CGH profiles of aneuploid samples. Six samples profiled in this study produced array profiles indicative of an aneuploidy event that most likely occurred during the proliferation of the cell-line source of these samples. If the profile for an entire chromosome appeared affected, those data points were removed from further analysis. In most cases, only a single chromosome was affected. The red and green horizontal lines on each plot indicate the ±0.3 and ±0.5 log2 ratio thresholds, respectively, and are a visual aid for determining the extent of duplication or deletion, since our SD cutoff method typically results in a threshold in the ±0.2–0.4 range.
BAC-Based Array CGH
Array hybridizations were performed as described by Snijders et al.,10 with use of the segmental duplication array.3,10 The segmental duplication array consists of 2,007 BACs, spotted in triplicate, that were targeted to 130 complex regions of the genome and flanked by intrachromosomal segmental duplications. All 269 individuals were hybridized, with dye-swap replicate experiments, to the segmental duplication array with use of a single reference individual for comparison.3 A locus was considered a CNV if the log ratio of fluorescence measurements for the individuals assayed exceeded twice the SD of the autosomal clones in both dye-swapped experiments. To account for asymmetry in some hybridization data, presumably due to differences in labeling efficiency between DNAs obtained from outside sources and our reference DNA extracted in-house, we developed a statistical method to identify variants in an asymmetric distribution. In brief, the total distribution of autosomal log2 ratios was divided into two groups, with the average autosomal log2 ratio as the division point. The SD was then determined for the above-average and below-average groups, after mirroring the data to simulate a symmetric distribution within each subgroup. The variant threshold for gains was then calculated as 2 SDs of the above-average group added to the mean, and the threshold for losses became 2 SDs of the below-average group subtracted from the mean. Comparison of the results of this method with those that did not account for asymmetry showed that the asymmetric method reduced the number of false-positive results, when compared with the oligonucleotide array data used for validation purposes (data not shown). Generally, hybridizations were considered good quality if they had an SD <0.2 for autosomal clones; otherwise, they were repeated. In a small subset of cases, repeated hybridizations also resulted in higher SDs, likely because of starting-DNA quality. Of the 538 hybridization profiles used in this analysis, which comprise 269 dye-swap pairs, only 6 profiles (samples CHB NA18633, CEU NA10847, CEU NA10851, CEU NA12707, CEU NA12740, and CEU NA12864) exceed an autosomal SD of 0.2. For each locus, the reported ACNF represents the percentage of unrelated individuals assayed (i.e., with exclusion of offspring from the CEU and YRI trios) who were scored as possessing a copy-number variation at that locus. Since our standard reference individual is male, to avoid difficulties in identifying variant clones on the X and Y chromosomes in sex-mismatched hybridizations, only male-male hybridizations were used to score variants on the sex chromosomes. A complete list of all BACs present on the array and the frequency of copy-number variation of each within the populations tested is shown in the tab-delimited ASCII files of data set 1 (online only).
Somatic Variation
A total of 30 self-versus-self hybridizations were performed on a panel of tissues from four individuals obtained from the Cooperative Human Tissue Network (CHTN), with use of the identical protocol that was used for the HapMap population sample hybridizations. A total of 7 or 8 tissues were profiled from each individual with splenic genomic DNA as the reference DNA, since it was abundantly available, high quality, and obtainable from all donors.
Oligonucleotide-Based Array CGH
A custom oligonucleotide array (NimbleGen Systems) was designed that consisted of 385,000 isothermal probes (45–70 bp) that covered the identical regions represented on the segmental duplication array, with an overall mean probe density of one probe per 733 bp. Probes were selected in regions devoid of high-copy repeats but within the unique and duplicated sequences that comprise the BACs on the segmental duplication array. DNA from nine individuals (YRI NA18517, YRI NA18507, YRI NA18502, YRI NA19240, YRI NA19129, CHB NA18555, JPT NA18992, CEU NA12156, and CEU NA12878), representing individuals from each of the four HapMap population samples, were then hybridized to the oligonucleotide array. The variants detected by BAC-array analysis were then directly compared with the oligonucleotide array profiles by converting the results from all oligonucleotide probes overlapping a BAC into a single statistic. The oligonucleotide array data was scored such that the duplication and deletion thresholds were computed as 2 SDs beyond the mean log2 ratio for all autosomal oligonucleotides reporting in that hybridization. For each BAC, the number of oligonucleotide probes that reported a loss was subtracted from the number of oligonucleotide probes that reported a gain and then was divided by the total number of probes overlapping the original BAC probe, which resulted in a simple scoring ratio. Ratios >0.1 or <−0.1 were scored as gains or losses, respectively. To assess the sensitivity and specificity of these criteria, we examined X chromosome loci in sex-mismatched hybridizations; this analysis indicated a false-negative rate of 5% and a false-positive rate of <0.2%, indicating it is a sensitive and specific metric for confirming copy-number changes.
Heritability
CNVs were classified as discrete or continuously variable by visual inspection of a plot of the log2 ratios from replicate dye-swap hybridizations. For discrete CNVs (defined as those in which the underlying signal intensity ratio could be visually clustered into two, three, or four well-separated copy-number classes by inspection of scatter plots of the replicate log2 hybridization values), we treated each of these clusters as a genotype, omitting genotype calls for any samples for which assignment was ambiguous or for which the dye-swap replicates were not concordant (SD >0.2) (see the tab-delimited ASCII file of data set 2 [online only]). We assessed whether the resulting genotypes were in Hardy-Weinberg equilibrium (HWE) and analyzed all trios for deviations from Mendelian inheritance. “Narrow-sense” heritability estimates,11 h2, obtained by estimating the regression coefficient (slope) of offspring values against midparental values (the mean value for both parents within a trio), are shown in table 1. Measurements of h2 close to 1.0 suggest that the copy number is stably inherited, independent of measurement noise or precision.
Table 1. .
Heritability of CNPs with a Continuous Distribution[Note]
| Population and Clone | Chromosome and hg16 Coordinatesa |
Dye-Swap R2 | ACNF | Narrow-Sense Heritability (h2±SE) |
| YRI: | ||||
| CTD-2046J21 | 1: 103532647–103647985 | .90 | .138 | 1.06 ± .21 |
| RP11-585N15 | 1: 16304321–16391174 | .66 | .249 | .97 ± .18 |
| CTD-2589H19b | 5: 662684–864137 | .87 | .424 | .86 ± .27 |
| RP11-837K1b | 5: 693297–873247 | .75 | .416 | .65 ± .25 |
| RP11-812N8b | 5: 779850–879258 | .69 | .313 | .90 ± .38 |
| RP11-262L1 | 7: 45058286–45214464 | .77 | .191 | .49 ± .55c |
| RP11-384C2 | 7: 142717297–142869087 | .62 | .141 | .83 ± .16 |
| RP11-45N9 | 7: 143297685–143451563 | .87 | .481 | .92 ± .24 |
| CTD-2142K23 | 8: 7238603–7341931 | .86 | .238 | .69 ± .19 |
| RP11-774P7 | 8: 7917017–8067760 | .92 | .328 | .73 ± .23 |
| RP11-138C5 | 15: 19199775–19364096 | .51 | .488 | 1.11 ± .23 |
| RP11-117M14 | 15: 19804700–19971720 | .71 | .401 | 1.20 ± .21 |
| RP11-351D6 | 17: 34930509–35010273 | .69 | .267 | 1.16 ± .19 |
| RP11-142H6 | 19: 8669454–8825625 | .87 | .463 | .61 ± .20 |
| RP11-775G6 | 22: 17102889–17244196 | .58 | .417 | 1.06 ± .27 |
| RP11-379N11 | 22: 19757625–19940794 | .80 | .402 | 1.20 ± .19 |
| CTD-2506I16 | 22: 20014749–20220783 | .61 | .246 | .83 ± .26 |
| CEU: | ||||
| CTD-2046J21 | 1: 103532647–103647985 | .69 | .138 | .92 ± .19 |
| RP11-1112O10 | 3: 196744968–196880879 | .72 | .186 | 1.20 ± .22 |
| CTD-2108J17 | 3: 196950243–197121995 | .70 | .183 | .84 ± .25 |
| CTD-2589H19b | 5: 662684–864137 | .62 | .424 | .32 ± .21c |
| RP11-837K1b | 5: 693297–873247 | .57 | .587 | .71 ± .21 |
| RP11-812N8b | 5: 779850–879258 | .50 | .691 | .15 ± .35c |
| RP11-24O14 | 5: 69417315–69562055 | .73 | .199 | .80 ± .19 |
| RP11-188C21 | 7: 101763594–101920490 | .59 | .117 | .50 ± .27c |
| CTD-3088N11b | 8: 7767399–7916838 | .83 | .282 | 1.08 ± .16 |
| RP11-774P7b | 8: 7917017–8067760 | .83 | .328 | .99 ± .19 |
| RP11-110H22 | 8: 86762305–86913434 | .72 | .111 | .95 ± .15 |
| CTD-2387G7 | 10: 48395333–48482422 | .79 | .062 | 1.02 ± .14 |
| RP11-138C5 | 15: 19199775–19364096 | .63 | .488 | .58 ± .19 |
| RP11-142H6 | 19: 8669454–8825625 | .82 | .463 | 1.05 ± .21 |
| CTD-3048O14 | 22: 16933331–17071291 | .51 | .172 | .90 ± .23 |
| RP11-775G6 | 22: 17102889–17244196 | .75 | .417 | .78 ± .23 |
| RP11-379N11 | 22: 19757625–19940794 | .73 | .404 | 1.10 ± .18 |
Note.— A subset of CNPs with continuously distributed copy-number measurements was tested for narrow-sense heritability, estimated by the slope of the regression line fitting offspring copy-number measurements to midparental (mean of the parents) copy-number measurements. Of the 34 analyzed CNPs, 30 (88%) demonstrated significant heritability in the CEU and YRI subpopulations. ACNF indicates the frequency at which this variant was found among all HapMap sample populations. The coefficient of determination (R2) was calculated from the dye-swap replicate data points of the BAC array hybridization data and is an indicator of reproducibility. Sites with R2<0.5 were removed from further analysis. Three further loci were also analyzed and showed narrow-sense heritability values <0 (data not shown). All three corresponded to the IGH and IGL gene clusters, which are known sites of somatic variation.2
Based on the hg16 reference sequence.
Overlapping BAC clones were analyzed independently.
BAC does not show significant heritability.
LD
To assess the LD of discretely varying CGH measurements with SNPs, we used an approach used elsewhere to analyze discretely varying copy-number measurements obtained by quantitative PCR.6 In brief, we recoded the discrete CNP genotype as a SNP genotype (“+/+” = AA, “+/−” = AT, and “−/−” = TT) and combined this with SNP genotype data from Phase I HapMap.9 We used SNP genotype data from all SNPs in a region extending 200 kb beyond the edges of the BAC probe, which was based on the hg16 reference sequence. We used the Haploview program12 to phase CNP and SNP genotypes and to calculate R2.
To assess the correlation of continuously varying CGH measurements with SNPs, we obtained SNP genotype data from all SNPs from Phase I HapMap that spanned a region 200 kb from both edges of the CGH probe, transformed these SNP genotypes into integers (e.g., AA=0, AC=1, and CC=2), and calculated the coefficient of determination (R2) for each of these SNP genotypes with the CNP measurements. To assess the significance of these correlation measurements, we performed a permutation test in which the CGH measurements were permuted across the trios in a population sample (maintaining the relationships within each trio) and again compared with the same SNP genotypes in that region. We considered a correlation significant (P<.05) if it was observed in <5% of these simulations.
Genome Build and Physical Coordinates
All genome physical coordinates referred to in this work are on the hg16 (build 34) coordinate system.
Results
HapMap CNPs
Using a BAC array targeted to regions of intrachromosomal segmental duplication,3 we analyzed 269 DNA samples corresponding to 209 unrelated individuals and 60 parents-child trios,9 by array CGH against a single reference individual. Of the samples, 263 passed quality assessment criteria (see the “Methods” section). From this set, we identified 384 CNV BACs, of which 127 (∼33%) were observed only once, and 257 (∼67%) were observed in more than one individual (fig. 2 and data set 1 [online only]). Of these variants, 103 have not been reported elsewhere (table 2). When adjacent clones, mapping within 250 kb of each other, are merged, these variant BACs represent 222 CNV regions. The average multi-BAC CNV region spanned 436 kb, with a range of 145 kb to 1.4 Mb. Several multi-BAC CNV regions showed evidence of heterogeneity, suggestive of additional genomic complexity. For example, a variant region from chromosome 22 consisted of four BAC clones (RP11-105A23, RP11-157B2, RP11-1143M16, and RP11-229C18) within a span of 605 kb. The four clones in this region were observed as a copy-number loss in the CEU subpopulation, three of the four were observed as a loss in the CHB population, and two of the four were observed as a loss in the JPT and YRI populations.
Figure 2. .
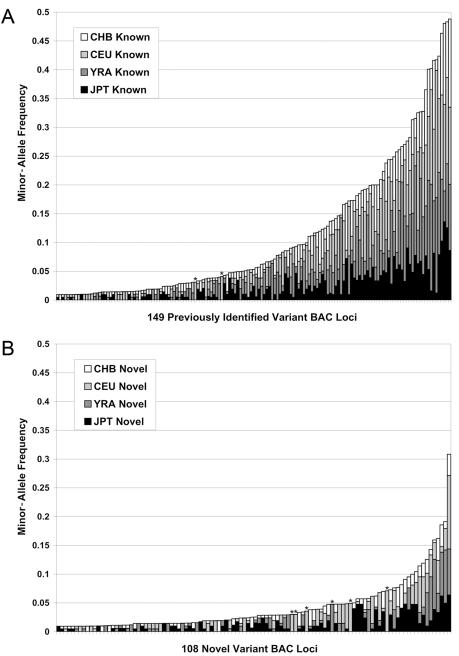
Frequency spectrum of novel and previously identified segmental duplication–array variants. The frequencies of a total of 257 autosomal BAC variants observed in multiple individuals are shown, 149 previously described variants1–3,13 (A), and 108 novel variants (B). Sites are coded by HapMap sample subpopulation. Note that only 11 novel variants were observed with an ACNF >10%, indicating that the majority of large, common CNPs in duplicated regions have likely been described. No common variants (ACNF >10%) were population specific, suggesting that these CNVs either predate the dispersal of the assayed populations or are recurrent events (population-specific variants >2.5% ACNF are indicated with an asterisk [*]). Only variants observed in two or more individuals are shown.
Table 2. .
Summary of Autosomal Variant BACs by Array CGH[Note]
| No. of Variant BACs |
No. of Variants |
|||||||||
| HapMap Subgroup | No. of Samples | Total | With Gain | With Loss | With Gain and Loss |
Corroborated | Novela | Percentage of Corroboration | Singleton Filtering Improvement (%) |
No. of Corroborated Singletons |
| CHB with singletons | 45 | 206 | 82 | 104 | 20 | 120 | 86 | 58 | … | 36 |
| CHB without singletons | 45 | 122 | 60 | 42 | 20 | 84 | 38 | 69 | 11 | … |
| CEU with singletons | 60 | 224 | 70 | 118 | 36 | 139 | 85 | 62 | … | 38 |
| CEU without singletons | 60 | 142 | 65 | 41 | 36 | 101 | 42 | 71 | 9 | … |
| JPT with singletons | 45 | 201 | 77 | 97 | 27 | 121 | 80 | 60 | … | 28 |
| JPT without singletons | 45 | 138 | 50 | 61 | 27 | 93 | 45 | 67 | 7 | … |
| YRI with singletons | 60 | 186 | 59 | 99 | 28 | 115 | 71 | 62 | … | 26 |
| YRI without singletons | 60 | 128 | 42 | 58 | 28 | 89 | 39 | 70 | 8 | … |
| Nonredundant total with singletons | 210 | 384 | 118 | 195 | 71 | 194 | 190 | 51 | … | 40 |
| Nonredundant total without singletons | 210 | 257 | 63 | 123 | 71 | 154 | 103 | 60 | 9 | … |
Note.— BACs with variant log2 ratios in both dye-swap replicated experiments were compared with previously published data sets of CNP and with our results from an oligonucleotide array targeted to the identical regions as the BAC-based segmental duplication array. In general, filtering out the lowest frequency variants—the singletons (i.e., those observed in only a single individual)—substantially increased the corroboration with other data sets1–3,13 as well as our own additional oligonucleotide-array experiments.
Novel within the subgroup but not necessarily novel with respect to all subgroups, except in the nonredundant category.
We classified a variant as a putative CNP if it was observed in two or more unrelated individuals. In total, of the BAC variants observed in multiple individuals, 154 (60%) of 257 were confirmed by previous studies (fig. 3) or by our own experimental validation with use of an oligonucleotide array in a small subset (n=9) of the original 209 individuals (see below). Among the 194 validated sites (including sites observed only once but corroborated by other sources), we observed 66 gains, 66 losses, and another 62 BACs as both gains and losses, with respect to the reference DNA sample. The underlying data as well as a UCSC browser version comparing these sites can be found at the Eicherlab Human Structural Variation Database.
Figure 3. .
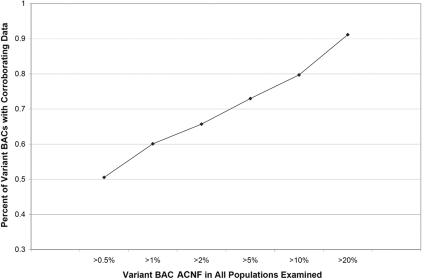
CNP ACNF and corroboration with other data sources. When stratified by frequency, the variants more commonly seen in the HapMap population samples were more often supported by other sources of CNP data, such as previous reports,1–3,13 and by our own comparison with oligonucleotide-array data. This is partly because of the nature of rare variants, in that they are less likely to be seen by other studies of small populations, but also implies that a proportion may be false-positive results and should be considered with more caution until sequence data are generated to fully document the variant in question.
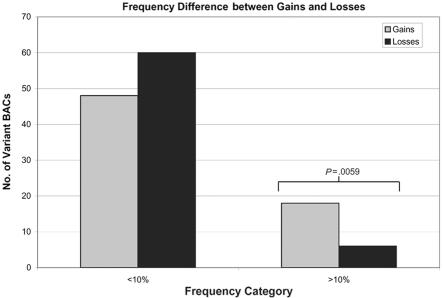
Population specificity of the CNPs was limited and correlated inversely with the frequency with which ACNF was observed. As expected, the most common variant sites (ACNF >10%; see the “Methods” section for definition) were observed across multiple populations of the HapMap set. Of the 105 variants with ACNF >5%, only 1 was confined to a single population. Some low-frequency CNPs were apparently population specific, with 9 of 63 sites showing population specificity in the 2%–5% frequency category (fig. 2). The ACNF spectrum differs strikingly between gains and losses. Dividing CNPs into two bins of ACNF either ⩾10% or <10%, we observed a significant difference between the two groups (P<.006 [Fisher’s exact test]), with common gain CNPs (ACNF >10%) outnumbering loss CNPs >3:1 (fig. 4). In contrast, deletions appear to predominate among low-frequency CNPs and singletons. Of the 127 singleton observations, for example, 72 (57%) were losses, and the remaining 55 (43%) were gains, indicating a slight bias for deletions.
Figure 4. .
Frequency differences between gain and loss CNPs. Of the total set of CNPs that had been corroborated by additional experiments or previously published reports (n=194), the gain (n=66) and loss (n=66) CNPs were divided into bins on the basis of ACNF with a threshold of 10%. A significant reduction in the number of sequence-loss CNPs was observed in the bin of common variants. This bias against common copy-number losses suggests that deletions are subject to purifying selection. Similar observations have been reported for smaller deletion variants (median size ∼7.0 kb).14
Fine-Scale Validation with Use of an Oligonucleotide Array
To validate and further investigate the CNPs detected using the segmental duplication array, we designed a custom high-density oligonucleotide array (NimbleGen Systems) for the same regions represented by BACs on the segmental duplication array (spanning 283.6 Mb, or 9.2%, of the human genome). Nine samples from the HapMap collection were analyzed with the custom oligonucleotide array with use of the same reference DNA that was used for the BAC array hybridizations. The array data were scored with a system based on the overall ratio of oligonucleotides above and below a threshold based on the SD of all autosomal oligonucleotides on the array (see the “Methods” section). These variant sites were then compared with those obtained using the BAC array data.
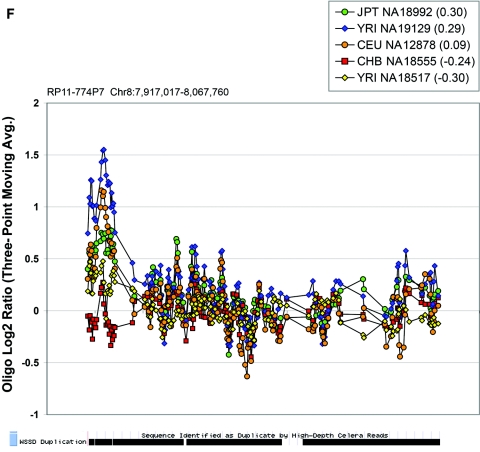
In general, the correspondence of BAC and oligonucleotide array data was reasonable, with 66% (136 of 207) of the CNVs identified by both platforms. After exclusion of some sites with insufficient oligonucleotide coverage, the remaining 63 variants identified by our BAC array were compared with published CNP data sets, and 64% (40 of 63) of these sites shared >40 kb overlap with one or more independent studies, indicating that the lack of validation of these sites is likely a false-negative result from the oligonucleotide-array experiments. Thus, 85% (176 of 207) of the CNVs identified with the segmental duplication array had supporting data from either our oligonucleotide-array experiments and/or reports published elsewhere. Of the remaining 15% (31 of 207) of sites without validation, 4% (8 of 207) were represented by <30 oligonucleotide probes and likely did not have sufficient density to report a variant, leaving 11% (23 of 207) sites as potential segmental duplication array false-positive results. Because 48% (11 of 23) of these potential false-positive results were found in a single individual of the nine profiled, we believe the false-positive results were limited to a small subset of BAC-array hybridizations. We also noted examples in which BAC-array variants had a high correspondence between dye-swap replicate hybridizations and demonstrated heritability in the HapMap trios, but no variant was detected using the oligonucleotide array (RP11-188C21 and RP11-774P7). These likely represent false-negative results of the oligonucleotide-array data or of oligonucleotide-array variant-scoring method, as opposed to false-positive results with use of the BAC array.
Detailed analysis of sites that disagree between the two platforms (potential false-negative results from the oligonucleotide array) suggests these resulted from a combination of reduced probe density, reduced magnitude of the underlying copy-number change, and an inherent bias in oligonucleotide placement against segmental duplications compared with unique sequence. First, the mean BAC log2 ratio of sites validated by the oligonucleotide array was 0.493; the mean of sites that did not validate was 0.452 (an 8.3% difference), indicating that sites that produce a lower amplitude log2 variation are closer to the threshold of detection. Second, sites that agreed between both platforms had an average oligonucleotide probe density of one probe per 769 bp, whereas sites that did not agree had an average density of one probe per 806 bp, a 4.6% reduction. Third, we examined the distribution of oligonucleotide array probes in relation to segmental duplications. Numerous studies have found a significant association between structural variation and segmental duplications,1–4,6 and our own observations with use of oligonucleotide arrays demonstrate that many sites of structural variation coincide precisely with the location of segmental duplications (figs. 5 and 6B–6H). Comparison of the density of oligonucleotides in unique sequence versus in segmental duplications showed that the oligonucleotide array we used contained a reduced probe coverage in segmental duplications, compared with unique sequence. Unique sequences showed a significantly higher probe density, containing, on average, one probe every 716 bp, compared with one probe every 814 bp in segmental duplications of >90% identity, falling to one probe every 860 bp in segmental duplications of >99% identity. Thus, the oligonucleotide array was biased against the detection of CNPs that coincided with sites of segmental duplication.
Figure 5. .
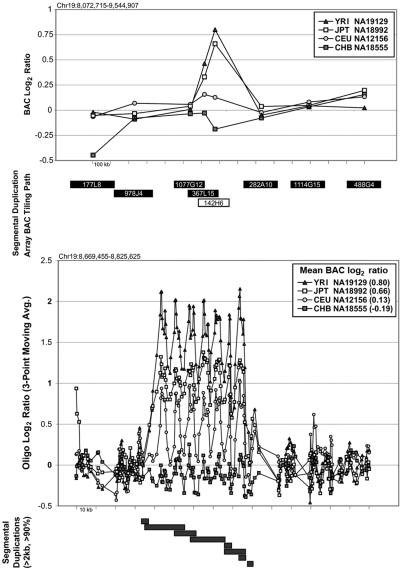
Fine structure variation of a common CNP. The CNP initially observed in BAC RP11-142H6 was observed as a common variant found in all subgroups of the HapMap population samples (ACNF 0.46). BAC-based array CGH results (top panel) for eight BAC clones spanning ∼1.5 Mb of chromosome 19 show continuous variability. Fine-tiling oligonucleotide array CGH profiles of the same individuals confirms subtle copy-number differences over the 156-kb interval encompassed by RP11-142H6. Note the close correlation between the log2 hybridization data from BAC-based array CGH and oligonucleotide microarray data. The recurrent pattern of log2 ratio data corresponds to a tandem organization of a 60-kb region composed of intrachromosomal segmental duplication (gray bars). Avg. = average.
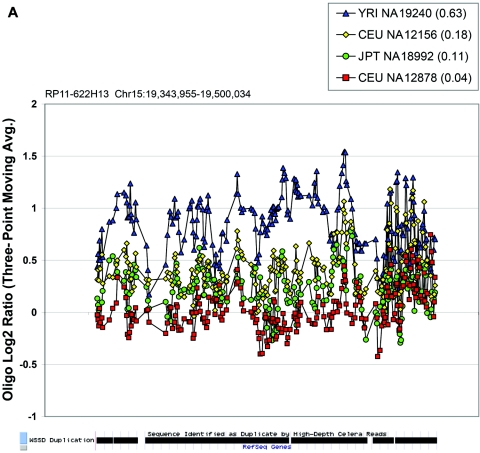
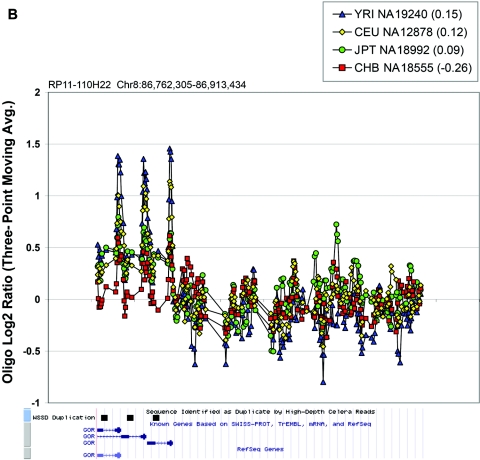
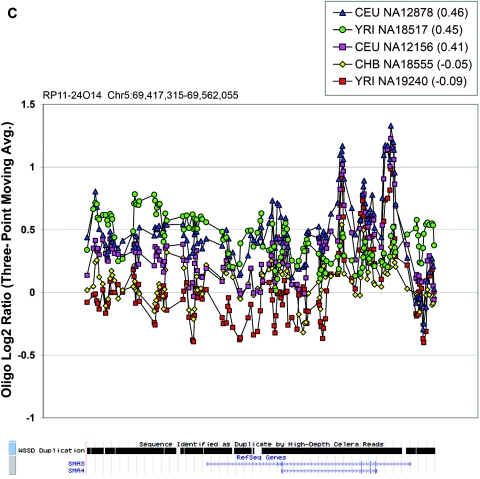
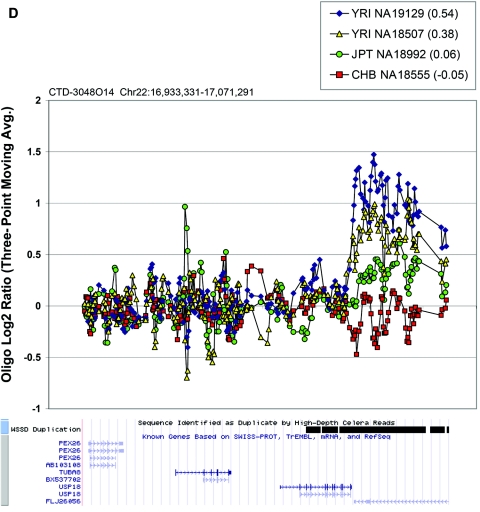
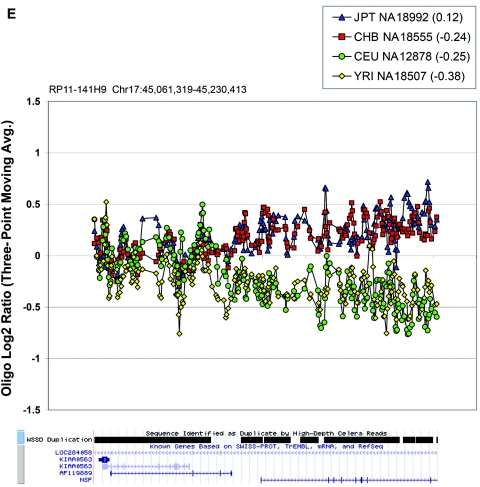
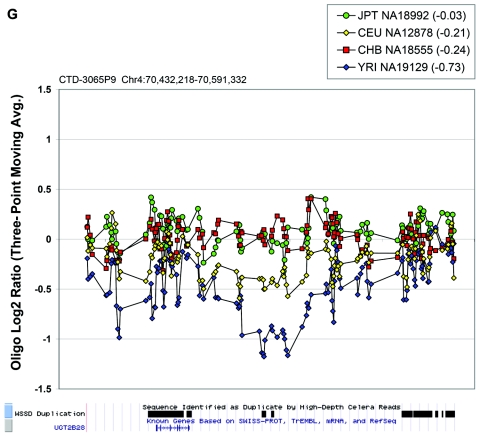
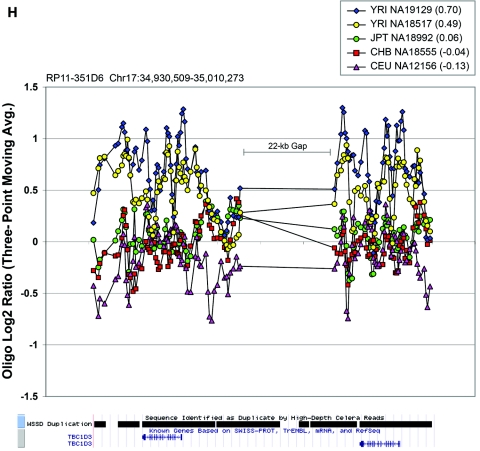
Figure 6. .
Oligonucleotide array data underlying CNPs detected via BAC array. Each plot displays the oligonucleotide hybridization data for a subset of samples tested. The BAC ID and build 34 interval for that BAC are indicated at the top left corner of each plot. The average log2 ratio determined by BAC array CGH is listed in parentheses within the graph key. The tracks depicting segmental duplications detected using a whole-genome depth of coverage statistic (WSSD) and known genes, extracted from the UCSC Genome Browser, are shown below each plot. Note that a three-point moving average of the oligonucleotide data is plotted. Avg. = average.
In the majority of cases, the oligonucleotide-array data not only confirm but also further refine the location of the CNV (fig. 6A–6H). More importantly, these additional data provide confirmatory evidence that even subtle differences in BAC-array log2 relative hybridization signals reflect real variants. The power of the high-resolution oligonucleotide array to reveal structure and additional information regarding the allelic state is illustrated in figure 5. In this example, the BAC-array data identified a candidate BAC (RP11-142H6) as a CNV within the population. Comparison of the BAC-array data with the oligonucleotide-array data clearly refined the breakpoints of the region responsible for the copy-number difference. In addition, it also revealed the presence of at least four different copy-number levels among the nine individuals assayed, something that had not been evident from the BAC array CGH data. Retrospective analysis showed an excellent correlation between the BAC-array and oligonucleotide-array log2 relative hybridization intensities for these individuals. Analysis of the underlying sequence revealed a complex architecture comprising multiple tandemly arranged segmental duplications of 21 kb (98.5% sequence identity), 11 kb (97.5% sequence identity), and 1.7 kb (91% sequence identity), suggesting variability in copy number of these segmental duplications as the likely basis for the subtle difference in log2 relative hybridization intensity ratio. Examination of the oligonucleotide-array data indicated that, at the available resolution (∼1 kb), the majority of CNPs exhibited breakpoints that were indistinguishable among different individuals.
Somatic Variation Analysis
Elsewhere, we analyzed self-versus-self comparisons of lymphoblastoid cell lines from the same passage as well as from different passages, to estimate both false-positive rates and rearrangements that may have emerged during culture. Although several apparent cell-culture artifacts were identified (fig. 1), our previous studies suggested that our false-positive rate was low (<0.08%). We wished to extend this analysis to address the frequency of normal somatic variation in such studies. We assessed the rate of somatic copy-number variation among tissues from the same individual. We performed a total of 30 self-versus-self hybridization experiments, using a panel of tissues (heart, lung, liver, testes, ovary, pituitary, cerebellum, cerebrum, medulla, occipital lobe, and pons) from four unrelated individuals against spleen DNA as our reference (table 3). Overall, the apparent level of somatic CNP was extremely low, with a median value of 1 CNV, compared with 12–15 for interindividual HapMap experiments. However, one sample in particular, the lung sample from CHTN-32364, showed 37 CNV gains, which accounted for nearly half (49%) of all CNVs detected among all somatic tissues examined. This particular experiment was repeated four times, with similar results. Although the cause of death in this individual was myocardial infarction, not lung cancer, it may be noteworthy that a subset of the CNP gains in the lung tissue sample involve BAC clones that overlap several oncogenes and genes implicated in cancer. If this sample is eliminated from the rate calculation, of the 1,871 autosomal BACs that were assessed for variation among 29 tissues, a total of 39 CNPs were detected. At this gross level of resolution—whole-tissue DNA preparation—this translates to an estimated rate of 0.00072 somatic CNVs per clone. Since our previous analysis of self-versus-self hybridizations had indicated that our analysis thresholds yield a false-positive rate of ∼0.08%,3 these data are very similar to the margin of error for array CGH experiments. We recognize, however, that there is extensive heterogeneity of cell types among the different tissues. A much more detailed study is now required to assess individual variability between specific cell types.
Table 3. .
Variation in Somatic Tissues[Note]
| No. of Copy-Number Variations (Gain, Loss) in |
|||||||||||||
| Sample | Sex | Age (years) |
Heart | Liver | Lung | Testis | Ovary | Pituitary | Cerebellum | Cerebrum | Medulla | Occipital Lobe | Pons |
| CHTN-32505 | Male | 64 | 0, 0 | 2, 0 | 0, 0 | 0, 0 | … | NA | 0, 0 | NA | 1, 1 | 0, 0 | 1, 4 |
| CHTN-32364 | Female | 55 | 0, 1 | 0, 1 | 37, 0 | … | 1, 0 | NA | 1, 0 | NA | 1, 0 | 0, 2 | 2, 0 |
| CHTN-32176 | Male | 42 | 0, 0 | 0, 1 | 2, 1 | 0, 0 | … | 2, 0 | NA | NA | 2, 1 | 0, 6 | NA |
| CHTN-31871 | Male | 89 | 2, 0 | 2, 0 | 1, 0 | 1, 0 | … | NA | 0, 0 | 1, 0 | NA | NA | 0, 0 |
Note.— Self-versus-self hybridizations, with use of splenic DNA from an individual as a reference against all other tissues examined, demonstrated a very low level of variance, with an average of 0.00072 CNVs per clone. Since our previous analysis of self-versus-self hybridizations had indicated that our analysis thresholds yield a false-positive rate of ∼0.08%,3 these data lie within the margin of error for array CGH experiments. Note that the excess of variants in the CHTN-32364 lung tissue sample were excluded from rate calculations and that they include several regions associated with oncogenesis. NA = tissue not available.
Heritability Analysis: Discrete versus Continuous CNPs
All genetic variation is discrete at the nucleotide level; nonetheless, assays for typing variants involve measurements (such as fluorescence measurements) that are less discretely distributed than the underlying variants because of measurement noise. For SNP assays, such measurements still typically cluster into discrete groups that allow an individual to be assigned a discrete genotype (such as AA, AT, or TT). For BAC array CGH data, whereas measurements for most probes showed an almost-continuous pattern of variation (fig. 7A), we found that copy-number measurements for some probes clustered into similarly discrete groups (fig. 7B). We refer to these as “discretely distributed” and “continuously distributed” copy-number measurements, respectively. Continuously distributed patterns of variation might be expected to arise when there are multiple or multiallelic variants (such as VNTRs) corresponding to the region covered by a single BAC clone, since such variants could involve changes in DNA content (fluorescence ratios) that are small relative to the resolving power of BAC-array-hybridization measurements.
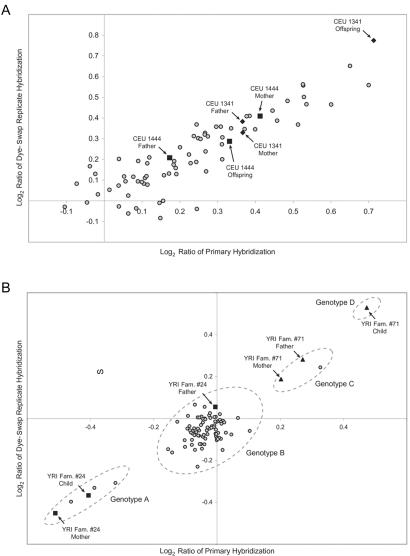
Figure 7. .
Continuous versus discrete CNPs. A comparison of log2 relative hybridization intensity ratio values between primary and replicate experiment for CEU and YRI pedigree samples distinguished continuously (A) and discretely (B) variable CNPs. A discrete CNP was defined as one in which two, three, or four distinct, well-separated genotype classes could be distinguished. Note that, unlike for SNPs, the genotype of the offspring (YRI family 71) can be a novel log2 ratio category, presumably because of the additive effect of the inheritance of two alleles with high copy number. More-common scenarios, such as a parent and child sharing a genotype (YRI family 24), were also observed. For continuous distributions, the additive effect seen for discrete sites was also observed (CEU family 1341), as well as the case in which the offspring log2 ratio value appeared to be intermediate beween the parental values (CEU family 1444). Therefore, each CNP can have a complex inheritance pattern within any given set of trios.
In assessing the heritability and LD of CNPs, we sought to understand both types of patterns of copy-number variation. We therefore developed approaches for analyzing both patterns of variation. For discretely distributed copy-number measurements, we visually clustered the copy-number measurements into copy-number classes, considered each discrete cluster as a discrete genotype, and analyzed these variants much as SNPs are analyzed. If there were more than four clusters or if clusters could not be discerned, CNPs were classified as “continuous.” For continuously distributed copy-number measurements, we treated the copy-number measurement as a quantitative trait and assessed the heritability and LD of this trait. We acknowledge that this approach may underestimate the extent of LD around CNPs with continuously distributed measurements, since CGH measurements integrate data from variation across 150 kb, which is several times larger than the scale of LD. However, this approach is likely to be sound when changes in copy number under a BAC are all due to variation at the same site within that BAC. Earlier studies of CNPs1,2 relied on a binary classification of a CNP as “present” or “absent” in each individual; it became clear that such approaches did not adequately track the segregation of CNPs through pedigrees. For example, because an individual could have more than one copy of a particular variant, CNPs can produce a genotype in an offspring, because of an additive effect, that was not seen in the parental generation. This phenomenon was observed for both discrete and continuous distributions (fig. 7).
Heritability of Discretely Distributed Copy-Number Measurements
When the copy-number measurements at a CNP clustered into two, three, or four discrete genotype classes, we encoded the variation into corresponding discrete genotypes (fig. 7). We first assessed whether the resulting genotype frequencies were in HWE. For 7 of the 8 YRI CNPs, the genotypes were in HWE (table 4). The locus that failed HWE showed a multiallelic pattern of variation, with apparent low-frequency variations that represented gains and losses of material relative to the prevailing, common copy-number class (fig. 7B); we subsequently analyzed the copy-number gain and copy-number loss variants separately (table 4), with each of the two variants conforming to HWE on its own. We further analyzed the trios for deviations from Mendelian inheritance and found that 7 of the 8 YRI loci were consistent with Mendelian inheritance. Finally, we analyzed the variants for stable transmission rates. Variants at all the YRI loci appeared to transmit with a frequency not significantly different from 0.5. Array CGH measurements for these loci appear to reflect an underlying, stably inherited genetic polymorphism.
Table 4. .
Heritability of YRI CNPs with a Discrete Distribution[Note]
| No. with Genotype |
Altered Copy-Number |
||||||||
| Clone | Chromosome and hg16 Coordinatesa |
+/+ | +/− | −/− | HWE | MIb | Transmissions (Total) | Frequency | Dye-Swap R2 |
| RP11-97F19 | 2: 89779383-89954029 | 78 | 5 | 1 | + | 0 | 3 (4) | .146 | .523 |
| CTD-3065P9 | 4: 70432218-70591332 | 28 | 36 | 11 | + | 0 | 11 (25) | .316 | .552 |
| RP11-177L24: | 10: 46859266-47011568 | 4 | 79 | 5 | … | 0 | .053 | .771 | |
| RP11-177L24 (CNP gain) | 0 | 4 | 84 | + | 1 | 1 (2) | |||
| RP11-177L24 (CNP loss) | 83 | 5 | 0 | + | 0 | 2 (3) | |||
| RP11-958F14 | 11: 55221653-55393983 | 82 | 7 | 0 | + | 0 | 3 (4) | .019 | .664 |
| RP11-1068E21 | 15: 28321608-28499440 | 60 | 22 | 3 | + | 0 | 9 (19) | .038 | .783 |
| RP11-261P7 | 15: 28207785-28330872 | 63 | 20 | 3 | + | 0 | 8 (18) | .200 | .671 |
| RP11-141H9 | 17: 45061319-45230413 | 30 | 36 | 13 | + | 0 | 15 (38) | .276 | .882 |
| RP11-1143M16 | 22: 24050173-24204080 | 86 | 3 | 0 | + | 0 | 1 (2) | .050 | .632 |
Note.— A subset of CNPs was analyzed in the YRI trios who presented a distribution of log2 ratios in which distinct genotype classes could be identified (see fig. 7). These “discrete” sites were tested for HWE, Mendelian inconsistencies, and rates of minor-allele transmission, to assess the heritability of a CNP event. The coefficient of determination of dye-swap replicate values (R2) is an indicator of reproducibility in the data, and sites with R2<0.5 were removed from further analysis. The CNV defined by BAC RP11-177L24 initially failed HWE and showed a distribution consistent with multiallelic variation (fig. 7B); the gain and loss variants at this site were henceforth analyzed separately and conformed to Hardy-Weinberg expectations on their own. ACNF indicates the frequency at which this variant was found among all HapMap sample populations.
Based on the hg16 reference sequence.
MI = Mendelian inconsistencies.
Heritability of Continuously Distributed Copy-Number Measurements
To analyze continuously distributed copy-number measurements, we treated the copy-number measurement as a quantitative trait. Narrow-sense heritability was estimated through regression of offspring copy-number measurements against the mean of their parents’ copy-number measurements. The heritability (h2) is given by the slope of the resulting regression line. With use of this methodology, a high level of heritability was observed from the majority of loci examined (table 1). Of 34 CNP probes examined, 30 yielded h2 measurements that were significantly greater than zero, and 32 yielded h2 measurements that were not significantly different from 1.0. Three further CNP probes that showed the lowest heritability measurements mapped inside immunoglobulin loci, which would be expected to have undergone somatic rearrangements in the lymphoblastoid cells from which DNA was obtained. The SEs for these heritability measurements were generally large, reflecting the fact that a relatively small number of trios were informative for any particular variant.
LD between CNPs and SNPs
An important question regarding structural variation is whether CNPs result from ancestral mutations and segregate on ancestral haplotypes or whether CNPs reflect multiple mutational events that have occurred on different haplotype backgrounds. If most CNPs are ancestral mutations and are in LD with SNPs, then SNPs may be used as markers for CNPs in genetic association studies; if CNPs do not have good SNP markers, then assessing the association of CNPs with phenotypes would require assays that type copy number explicitly. Whereas deletion polymorphisms in unique regions of the human genome appear to result from ancestral mutations and to segregate on ancestral haplotypes,6,7,15 little is known about the LD properties of deletions or duplications in repeat-rich, structurally complex regions of the genome such as those analyzed here. To begin to address this question, we assessed whether BAC-array–derived copy-number measurements showed evidence of LD with SNPs near the genomic locations of the BAC probes. We examined 42 CNP loci, including 8 with discretely distributed and 34 with continuously distributed copy-number measurements. The best available SNP markers tend to be found close to the breakpoints of deletion polymorphisms.6 Analysis of LD around CNPs ascertained by BAC arrays is complicated by the fact that, in most cases, the breakpoints of the structural variants are not known: the CNP could reside entirely within the BAC or could extend beyond the BAC on either or both sides. To search for SNPs that could potentially be used as markers for CNPs, we searched a 0.5-Mb genomic region centered on the genomic coordinates of the BAC probe.
We first assessed the LD between copy-number measurements and SNPs near eight BAC probes for which copy-number measurements showed discrete patterns of variation. Four of these CNPs showed significant LD (LOD >3.0) with nearby SNPs, although only two had perfect (R2=1.0) SNP proxies. The ability of SNPs to serve as surrogates for copy-number variation, therefore, appeared to be reduced when compared with less complex regions of the genome,6,7 although this assessment is complicated by the fact that only two of these variants were common (ACNF >10%), and rare variants typically are less likely to have proxies among the common SNPs typed by HapMap than are common variants.
We then assessed the correlation between copy-number measurements and SNPs near 34 BAC probes for which copy-number measurements showed continuous patterns of variation. The significance of these correlations was assessed by a permutation test in which the copy-number measurements were permuted across the 30 trios in a way that preserved parent-offspring relationships; a potential SNP marker was considered significant (P<.05) if its correlation to the copy-number measurements exceeded the 95th percentile (across 100 simulations) of the maximum correlation observed in each simulation. The resulting significance threshold varied from BAC to BAC because of local variation in SNP density, SNP frequency, and SNP distribution with respect to the BAC; however, the threshold typically fell within a range of 0.15–0.18 (fig. 8). A potential marker SNP was qualified as “significant” (and was included in table 5) if the correlation to the actual CNP measurements exceeded this threshold for that locus.
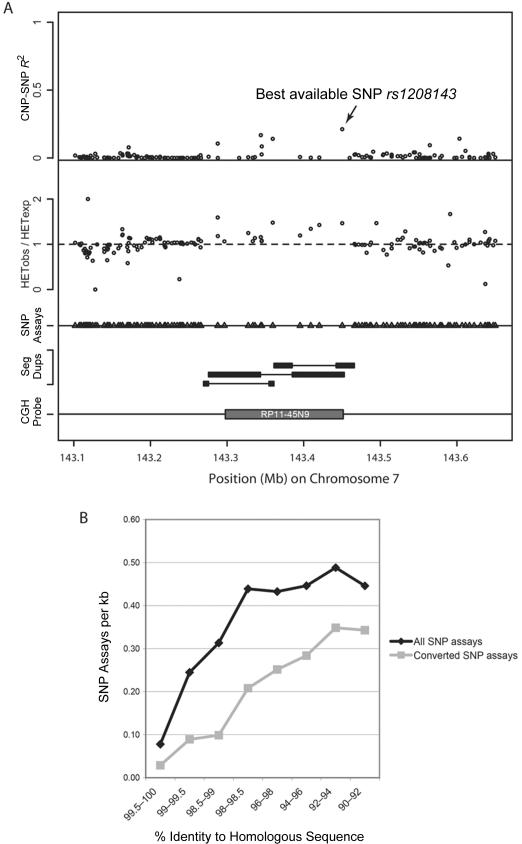
Figure 8. .
LD at a continuous site of CNP variation. A, Multiple tracks (top to bottom) depicting the distribution of R2 for SNPs within a 200-kb region flanking the CNP variant, the ratio of observed:expected heterozygosity (HETobs/HETexp), the location of SNP assays (triangles), the position of segmental duplications (Seg Dups) (gray bars, with pairwise relationships indicated by joining lines), and the position of the BAC array CGH probe (dark gray box). The best available SNP, rs1208143, was selected on the basis of a permutation test of all SNPs within an interval encompassing 200 kb proximal and distal of the BAC coordinates. Note the reduced SNP density in the BAC interval, along with increased heterozygosity, likely because of the presence of segmental duplications.20 B, The number of SNP assays per kilobase, plotted with respect to the percentage of similarity of segmental duplications. The trend clearly indicates a reduction in the number of SNP assays, with increasing homology in duplicated space. This trend implies that the regions most prone to complex rearrangement are less likely to have dense SNP coverage and thus are less apt to associate with a nearby SNP. In addition, methods for the identification of CNPs based on the analysis of SNP data have reduced power to detect structural variations in regions of segmental duplication.6,7,14
Table 5. .
LD at Continuous CNP Sites[Note]
| Population and Clone |
Chromosome and Coordinates | ACNFa | Dye-Swap R2 |
Associated HapMap SNP |
Chromosome and Position | SNP R2 | R2SNP:R2dye-swap |
| YRI: | |||||||
| RP11-585N15 | 1: 16304321–163911174 | .249 | .66 | rs2796146 | 1: 16497921 | .26 | .39 |
| CTD-2046J21 | 1: 103532647–103647985 | .133 | .90 | rs11185321 | 1: 103779317 | .20 | .22 |
| CTD-2589H19b | 5: 662684–864137 | .492 | .88 | rs508016 | 5: 931263 | .51 | .59 |
| RP11-837K1b | 5: 693297–873247 | .317 | .75 | rs508016 | 5: 931263 | .58 | .78 |
| RP11-812N8b | 5: 779850–879258 | .304 | .69 | rs508016 | 5: 931263 | .46 | .66 |
| RP11-262L1 | 7: 45058286–45214464 | .191 | .77 | ||||
| RP11-384C2 | 7: 142717297–142869087 | .141 | .62 | rs10249881 | 7: 142516332 | .25 | .40 |
| RP11-45N9 | 7: 143297685–143451563 | .186 | .87 | rs1208143 | 7: 143450282 | .21 | .24 |
| CTD-2142K23 | 8: 7238603–7341931 | .200 | .86 | ||||
| RP11-774P7 | 8: 7917017–8067760 | .305 | .92 | rs3860876 | 8: 7755676 | .20 | .22 |
| RP11-138C5 | 15: 19199775–19364096 | .400 | .51 | rs2203858 | 15: 19186415 | .29 | .57 |
| RP11-117M14 | 15: 19804700–19971720 | .458 | .71 | ||||
| RP11-351D6 | 17: 34930509–35010273 | .467 | .69 | ||||
| RP11-142H6 | 19: 8669454–8825625 | .367 | .86 | ||||
| RP11-775G6 | 22: 17102889–17244196 | .417 | .58 | rs5992185 | 22: 17008532 | .20 | .34 |
| RP11-379N11 | 22: 19757625–19940794 | .417 | .80 | ||||
| CTD-2506I16 | 22: 20014749–20220783 | .246 | .61 | rs2930770 | 22: 20161914 | .52 | .86 |
| CEU: | |||||||
| CTD-2046J21 | 1: 103532647–103647985 | .233 | .69 | rs1161064 | 1: 103658457 | .46 | .67 |
| RP11-1112O10b | 3: 196744968–196880879 | .150 | .72 | rs7633103 | 3: 196799287 | .60 | .83 |
| CTD-2108J17b | 3: 196950243–197121995 | .183 | .7 | rs7633103 | 3: 196799287 | .46 | .66 |
| CTD-2589H19b | 5: 662684–864137 | .491 | .62 | rs508016 | 5: 931263 | .49 | .79 |
| RP11-837K1b | 5: 693297–873247 | .587 | .57 | rs508016 | 5: 931263 | .73 | 1.29 |
| RP11-812N8b | 5: 779850–879258 | .691 | .50 | rs508016 | 5: 931263 | .67 | 1.35 |
| RP11-24O14 | 5: 69417315–69562055 | .418 | .73 | ||||
| RP11-188C21 | 7: 101763594–101920490 | .186 | .59 | ||||
| CTD-3088N11b | 8: 7767399–7916838 | .276 | .83 | rs2698913 | 8: 7808534 | .19 | .23 |
| RP11-774P7b | 8: 7917017–8067760 | .333 | .83 | rs2740621 | 8: 7814659 | .24 | .29 |
| RP11-110H22 | 8: 86762305–86913434 | .138 | .72 | ||||
| CTD-2387G7 | 10: 48395333–48482422 | .062 | .79 | rs11594866 | 10: 4844171749 | .25 | .32 |
| RP11-138C5 | 15: 19199775–19364096 | .475 | .63 | rs2203858 | 15: 19186415 | .24 | .38 |
| RP11-142H6 | 19: 8669454–8825625 | .492 | .82 | ||||
| CTD-3048O14 | 22: 16933331–17071291 | .300 | .51 | ||||
| RP11-775G6 | 22: 17102889–17244196 | .417 | .75 | ||||
| RP11-379N11 | 22: 19757625–19940794 | .404 | .73 |
Note.— Best available SNPs were tested for significance by permutation test (see the “Methods” section). Blank rows indicate that no significantly associated SNP was identified. The ratio R2SNP:R2dye-swap expresses the fraction of reproducible variation in copy-number measurements that is captured by the associated SNP marker.
ACNF is within each respective population.
Overlapping or adjacent BAC clones tested independently.
Of the 34 CNPs, 21 had nearby SNPs that accounted for a statistically significant fraction of the variation in copy-number measurements. In some cases, nearby BAC probes showed LD with the same explanatory SNPs (table 5), suggesting that they report the same underlying variant; copy-number measurement at two CNPs that were evaluated in both the CEU and YRI population samples were linked to the same SNP alleles in both populations, suggesting that those CNPs are ancestral mutations that were inherited by both populations. In general, though, the ability of HapMap SNPs to serve as surrogates for copy-number measurements at most of these CNPs was limited: only three CNP loci had nearby SNPs that explained >70% of the reproducible variation in copy-number measurements, and only five had nearby SNPs that explained >50% of this variation. (Fig. 8A shows an example of a marginally correlated SNP that explained only 24% of the variation in copy-number measurements.) In general, HapMap SNPs appeared to be less successful in serving as surrogates for copy-number measurements in these duplication-rich genomic regions than has been observed for deletions in unique regions of the genome.6,7
It should be noted that the density of HapMap SNPs was considerably lower within regions of segmental duplication than in unique regions of the genome. The density of successfully genotyped HapMap SNPs was inversely correlated with the degree of sequence identity of the duplications involved: a greater than fourfold difference in HapMap SNP density was observed between regions where the duplications were >99% identical than in regions of 90% sequence identity (fig. 8B). This results from the fact that the HapMap Consortium avoided SNPs for which it was not possible to design a uniquely mappable genotyping assay.9 The failure to find good SNP surrogates for CNPs in many of these regions may reflect the absence or low density of HapMap SNP assays near the breakpoints of the rearrangements.
Discussion
Overall, our analysis indicates that CNPs are typically heritable polymorphisms. We found only a modest level of LD between CNPs and the best available SNP markers, although an intense effort to map CNP breakpoints and type nearby SNPs might well identify better SNP markers. A combination of both BAC-based and customized high-density oligonucleotide arrays allowed unprecedented levels of resolution in mapping the breakpoints of a subset of these CNPs and allowed us to observe both diallelic and multiallelic (fig. 5) patterns of variation. In particular, cross-platform validation experiments showed that subtle differences in log2 relative hybridization data from BAC-based array CGH (fig. 5) and oligonucleotide microarray data may correlate even though the absolute intensity values differ. Therefore, we suggest that additional genetic information may be obtained from subtle differences in log2-relative signal intensity once a particular site has been confirmed as a CNV. Repetitive patterns in the oligonucleotide array profile were particularly powerful in distinguishing copy-number differences in regions of tandem segmental duplication. This study identifies and characterizes complex regions of human genetic variation for future sequence resolution.
CNPs are enriched within regions of segmental duplication.2,3 We designed the segmental duplication array to target unique regions that were between 50 kb and 10 Mb in size and were flanked by highly homologous (>95%) intrachromosomal segmental duplications16,17 (n=130). We reasoned that such areas, by virtue of their genomic architecture, would be prone to rearrangement through nonallelic homologous recombination.3 Of the 130 regions targeted in our array design, however, only 84 (65%) of those regions have been found to be polymorphic to date (>300 unrelated individuals tested with the segmental duplication array). Moreover, in most cases (75%), the variation was restricted to the segmental duplications and did not extend into the unique regions bracketed by the duplication. Only a fraction (11 of 103) of the novel sites showed >10% ACNF in any population. We propose that the majority of common (>10%) CNPs (>50 kb) associated with segmental duplication have now been detected.18 This raises the intriguing question of why some regions show no evidence of structural variation in the human population despite their predisposing genetic structure.
Although additional low-frequency variants will continue to be identified among normal individuals, differences in the genomic architecture, assay limitations, and/or selection likely account for these “invariant” regions. To provide further insight into this question, using data from both this study and our previous analysis,3 we divided the 130 regions into two categories: those that show and those that do not show evidence of copy-number variation within the normal population. We then assessed various sequence properties of these “variant” and “invariant” regions (table 6). In general, variant regions of the genome mapped to larger intervals (in part because of pericentromeric duplications), harbored slightly larger segmental duplications, and had fewer exons per megabase than invariant regions. Surprisingly, there was no significant difference in sequence identity between the two groups. Direct and inverted segmental duplications were observed at equal frequency among variant regions, whereas a bias against the inverted orientation was noted for invariant regions (data not shown). Although unique BACs within (24% [99 of 420]) and outside (19% [214 of 1,076]) an interval were variant, the most predictive (65% [167 of 257]) characteristic of variation was overlap with segmental duplication content. Duplications in tandem orientation were most significantly enriched, with 71% (90 of 126) of BACs containing these tandem duplications detected as variant.
Table 6. .
Sequence Properties of Variant and Invariant Regions[Note]
| Average |
||||||
| Interval | No. of Regions | Length (kb) |
Exon Density (Mb) |
Duplication Depth (copies) | Alignment Length (kb)a |
Identitya |
| Variant | 84 | 2,923 | 103.4 | 2.4 | 54 | .9819 |
| Invariant | 46 | 615 | 132.6 | 1.8 | 41 | .9819 |
Note.— Of the 130 regions targeted by the segmental duplication array, those that have been associated with a CNP in both this study and our previous work3 were considered “variant”; those regions in which no CNPs were detected were labeled “invariant.” It should be emphasized that, in most cases, copy-number variation does not extend over the entire interval but is restricted to the segmental duplication embedded within the boundaries of the interval. The regions targeted by the segmental duplication array were selected using the criteria of flanking paired intrachromosomal segmental duplications >50 kb and <10 Mb apart, >10 kb in length, and >95% sequence identity, resulting in the identification of 1,124 intervals across the hg16 genome assembly. The 1,124 intervals, however, extensively overlapped because of the clustered nature of segmental duplications in the genome; thus, the 1,124 intervals were collapsed into 130 nonredundant regions.
Average alignment length and identity were calculated on the basis of the total set of redundant intervals (n=1,124).
We found only modest evidence of LD between CNPs and HapMap SNPs, and, for a subset of CNP loci, we were unable to identify any SNPs that were significantly correlated with copy-number measurements (tables 5 and 7). This contrasts with the finding that deletion polymorphisms in unique regions of the genome generally have good SNP markers.6,7 There are several possible reasons for the failure to observe strong LD between CNPs and available SNPs in duplication-rich regions, including the low density of informative SNP assays in those regions, the frequency and nature of the CNP, and the possibility that many CNPs involve recurrent structural mutations. One must be cautious not to overinterpret these results, since an analysis of the data suggests the potential for all three.
Table 7. .
LD at Discrete CNP Sites in YRI Population[Note]
| CNP BAC ID | Chromosome and hg16 Coordinates | ACNFa | Best Available SNP | SNP R2 | LOD |
| RP11-97F19 | 2: 89779383–89954029 | .04 | rs7583495 | .15 | 2.49 |
| CTD-3065P9 | 4: 70432218–70591332 | .41 | rs11249532 | 1.00 | 23.93 |
| RP11-177L24 (CNP loss) | 10: 46859266–47011568 | .03 | rs7901145 | .11 | 1.85 |
| RP11-177L24 (CNP gain) | 10: 46859266–47011568 | .03 | rs11259816 | .33 | 1.65 |
| RP11-958F14 | 11: 55221653–55393983 | .07 | rs7950741 | .09 | 2.26 |
| RP11-261P7+RP11-1068E21b | 15: 28207785–28499440 | .17 | rs2140173 | .18 | 3.15 |
| RP11-141H9 | 17: 45061319–45230413 | .42 | rs199455 | .29 | 6.36 |
| RP11-1143M16 | 22: 24050173–24204080 | .02 | rs2092184 | 1.00 | 4.40 |
Note.— Analysis of LD across the CNP regions allowed the identification of a best available SNP in a subset of the discrete CNP sites. Significant LOD scores >3.0 (shown in bold italics) were observed for only 4 of the 19 sites and/or allelic states tested.
ACNF is within the YRI subpopulation.
Note that the two overlapping BAC clones RP11-261P7 and RP11-1068E21 were analyzed as a single variant site.
It is likely that both the nature and frequency of the CNPs themselves also affect the levels of LD observed. Multiallelic sites, such as VNTR regions (fig. 5), seem most likely to have undergone recurrent rearrangements and may be particularly difficult to “tag” using SNPs. The chance to observe LD at a CNP locus is also strongly influenced by frequency; as is seen with SNPs, we observe that common CNP variants tend to have good proxies, whereas rare variants (typically of more recent origin) are less likely to have good proxies (table 7).
Perhaps most importantly, we observed that HapMap SNP density in regions of segmental duplication was significantly lower than the genome average, in terms of both attempted SNP assays and converted SNP assays generated by the HapMap project (fig. 8B and data set 1 [online only]). The reduced density of HapMap SNP data in duplicated regions results from the difficulty of designing unique SNP assays in such regions. This effect was particularly acute in the most recently duplicated regions (>98.5% identity) (fig. 8B), which may also be the most rearrangement prone. For whole-genome SNP-association studies, this raises the concern that many of the genomic regions most prone to rearrangement have insufficient SNP density to successfully discern the state of many CNPs with use of available SNP markers. This limitation may also apply to many commercial whole-genome–genotyping resources, which appear to undersample variation in structurally complex and structurally variant regions of the genome.19 A targeted effort to resolve the structure of these regions at the base-pair level will be required to fully assess their contribution to disease and other clinical phenotypes. Our results underscore the need for complete maps of genetic variation in duplication-rich regions of the genome. Targeted BAC-based and oligonucleotide-array studies to investigate human variation are an important first step in this effort.
Supplementary Material
Acknowledgments
S.A.M. contributed equally with D.P.L. and A.J.S. to this manuscript but, because of journal policy, could not be listed as a joint primary author. We thank Joshua Akey for critical comments in the preparation of this manuscript. This work was supported in part by National Institutes of Health (NIH) grant HD043569 and supplemental funds from NIH grant HG002385 (to E.E.E.) and by a fellowship from Merck Research Laboratories (to A.J.S.). Somatic tissue samples were provided by the Cooperative Human Tissue Network, which is funded by the National Cancer Institute. E.E.E. is an Investigator of the Howard Hughes Medical Institute.
Web Resources
The URLs for data presented herein are as follows:
- Cooperative Human Tissue Network, http://www-chtn.ims.nci.nih.gov/
- Eicherlab Human Structural Variation Database, http://humanparalogy.gs.washington.edu/structuralvariation/
References
- 1.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C (2004) Detection of large-scale variation in the human genome. Nat Genet 36:949–951 10.1038/ng1416 [DOI] [PubMed] [Google Scholar]
- 2.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M (2004) Large-scale copy number polymorphism in the human genome. Science 305:525–528 10.1126/science.1098918 [DOI] [PubMed] [Google Scholar]
- 3.Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Segraves R, Oseroff VV, Albertson DG, Pinkel D, Eichler EE (2005) Segmental duplications and copy-number variation in the human genome. Am J Hum Genet 77:78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, Olson MV, Eichler EE (2005) Fine-scale structural variation of the human genome. Nat Genet 37:727–732 10.1038/ng1562 [DOI] [PubMed] [Google Scholar]
- 5.Buckland PR (2003) Polymorphically duplicated genes: their relevance to phenotypic variation in humans. Ann Med 35:308–315 10.1080/07853890310001276 [DOI] [PubMed] [Google Scholar]
- 6.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, Dallaire S, Gabriel SB, Lee C, Daly MJ, Altshuler DM, The International HapMap Consortium (2006) Common deletion polymorphisms in the human genome. Nat Genet 38:86–92 [DOI] [PubMed] [Google Scholar]
- 7.Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA (2006) Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat Genet 38:82–85 [DOI] [PubMed] [Google Scholar]
- 8.International HapMap Consortium (2003) The International HapMap Project. Nature 426:789–796 10.1038/nature02168 [DOI] [PubMed] [Google Scholar]
- 9.International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437:1299–1320 10.1038/nature04226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B, Kimura K, Law S, Myambo K, Palmer J, Ylstra B, Yue JP, Gray JW, Jain AN, Pinkel D, Albertson DG (2001) Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet 29:263–264 10.1038/ng754 [DOI] [PubMed] [Google Scholar]
- 11.Fisher R (1930) The genetical theory of natural selection. Clarendon Press, Oxford, United Kingdom [Google Scholar]
- 12.Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 13.de Vries BBA, Pfundt R, Leisink M, Koolen DA, Vissers LELM, Janssen IM, van Reijmersdal S, Nillesen WM, Huys EHLPG, de Leeuw N, Smeets D, Sistermans EA, Feuth T, van Ravenswaaij-Arts CMA, van Kessel AG, Schoenmakers EFPM, Brunner HG, Veltman JA (2005) Diagnostic genome profiling in mental retardation. Am J Hum Genet 77:606–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK (2006) A high-resolution survey of deletion polymorphism in the human genome. Nat Genet 38:75–81 [DOI] [PubMed] [Google Scholar]
- 15.Bhangale TR, Rieder MJ, Livingston RJ, Nickerson DA (2005) Comprehensive identification and characterization of diallelic insertion-deletion polymorphisms in 330 human candidate genes. Hum Mol Genet 14:59–69 10.1093/hmg/ddi006 [DOI] [PubMed] [Google Scholar]
- 16.Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE (2002) Recent segmental duplications in the human genome. Science 297:1003–1007 10.1126/science.1072047 [DOI] [PubMed] [Google Scholar]
- 17.She X, Jiang Z, Clark RA, Liu G, Cheng Z, Tuzun E, Church DM, Sutton G, Halpern AL, Eichler EE (2004) Shotgun sequence assembly and recent segmental duplications within the human genome. Nature 431:927–930 10.1038/nature03062 [DOI] [PubMed] [Google Scholar]
- 18.Kruglyak L, Nickerson DA (2001) Variation is the spice of life. Nat Genet 27:234–236 10.1038/85776 [DOI] [PubMed] [Google Scholar]
- 19.Wirtenberger M, Hemminki K, Burwinkel B (2006) Identification of frequent chromosome copy-number polymorphisms by use of high-resolution single-nucleotide–polymorphism arrays. Am J Hum Genet 78:520–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fredman D, White SJ, Potter S, Eichler EE, Den Dunnen JT, Brookes AJ (2004) Complex SNP-related sequence variation in segmental genome duplications. Nat Genet 36:861–866 10.1038/ng1401 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.