Abstract
Genomic imbalance is a common cause of phenotypic abnormalities. We measured the relative expression level of genes that map within the microdeletion that causes Williams-Beuren syndrome and within its flanking regions. We found, unexpectedly, that not only hemizygous genes but also normal-copy neighboring genes show decreased relative levels of expression. Our results suggest that not only the aneuploid genes but also the flanking genes that map several megabases away from a genomic rearrangement should be considered possible contributors to the phenotypic variation in genomic disorders.
Segmental aneuploidies (i.e., gain or loss of subchromosomic DNA fragments) are important contributors to human diseases1 and, potentially, to phenotypic variation,2,3 as well as a major force of evolutionary changes.4–9 There is evidence that such genomic insertions and deletions contribute to phenotypic differences by modifying the expression levels of genes within the aneuploid segments.10–13 We hypothesize that these rearrangements also induce altered expression of the genes that lie near the breakpoints, although these do not vary in copy number; this effect could be mediated by disturbances of the copy number of long-range cis regulatory elements.14–17
To test this hypothesis, we assessed whether the human chromosome 7 (HSA7) recurrent DNA deletion causing Williams-Beuren syndrome (WBS [MIM 194050])18 influences the transcription levels of both the hemizygous genes within the deleted region and the nonhemizygous genes in the WBS flanking regions. WBS is a neurodevelopmental disorder characterized by numerous clinical aspects, including mental retardation with a unique cognitive and personality profile.19 Its incidence is estimated to be between 1:7,500 and 1:20,000, and sporadic de novo inheritance is usual.20–22
Material and Methods
Cell Culture, RNA, and cDNA Preparations
Human skin fibroblasts and lymphoblastoid cell lines were grown in HAM F-10 or RPMI 1640 media, respectively, supplemented with 10% fetal bovine serum and 1% antibiotics (Invitrogen). Total RNA was prepared from logarithmic growth–phase cells, with the use of RNeasy Mini Kit (Qiagen), in accordance with the manufacturer’s instructions. After DNAse treatment (Qiagen), the quality of all RNA samples were checked using an Agilent 2100 Bioanalyzer (Agilent Technologies). Total RNA was converted to cDNA with the use of Superscript II (Invitrogen) primed with poly d(T). For each cell line included in the study, 4.5 μg of total RNA was converted to cDNA in three individual reactions; these were then pooled and were diluted 1:14.
Sample Population
Lymphoblastoid cell lines from 10 individuals with WBS and from 40 control individuals, as well as skin fibroblasts from 7 control individuals, were acquired from the cell culture collection of the Coriell Institute for Medical Research, and skin fibroblasts from 14 individuals with WBS and from 6 control individuals were obtained from the cell culture collections of the Centre de Biotechnologie Cellulaire, Hospices Civils de Lyon, Hôpital Debrousse, in Lyon, France. One more control was received from the Galliera Genetic Bank in Genova, Italy (table 1). Appropriate informed consent was obtained for each sample by the physicians in charge. DNA was extracted from each cell line of the sample population, with the use of PureGene (Gentra Systems), in accordance with the manufacturer’s instructions. We assayed each DNA with a quantitative PCR approach, using SybrGreen dye and probes, mapping the region from the BAZ1 locus to the CYLN2 locus and the flanks of the commonly deleted region,23 to determine the size of the deletions and to ensure (1) that none of the patients with WBS presented an atypical deletion23–29 or an inversion at 7q11.23 18,30 and (2) that none of the controls were hemizygous for that same region. The results are presented in table 2.
Table 1. .
Cell Lines Employed
| Cell Line Identification Number | Sex | Phenotype | Source | Usea | Biobank |
| GM13461 | M | WBS | B-Lymphocyte | 1 | Coriell Institute for Medical Research |
| GM13472 | F | WBS | B-Lymphocyte | 1 | Coriell Institute for Medical Research |
| GM13473 | F | WBS | B-Lymphocyte | 1 | Coriell Institute for Medical Research |
| GM13475 | M | WBS | B-Lymphocyte | 1 | Coriell Institute for Medical Research |
| GM13481 | F | WBS | B-Lymphocyte | 1 | Coriell Institute for Medical Research |
| GM13482 | F | WBS | B-Lymphocyte | 1 | Coriell Institute for Medical Research |
| GM13483 | M | WBS | B-Lymphocyte | 1 | Coriell Institute for Medical Research |
| GM14162 | M | WBS | B-Lymphocyte | 1 | Coriell Institute for Medical Research |
| GM14182 | M | WBS | B-Lymphocyte | 1 | Coriell Institute for Medical Research |
| GM14268 | F | WBS | B-Lymphocyte | 1 | Coriell Institute for Medical Research |
| GM06985 | F | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM06993 | M | Unaffected | B-Lymphocyte | 1, 2 | Coriell Institute for Medical Research |
| GM06994 | M | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM07000 | F | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM07002 | F | Unaffected | B-Lymphocyte | 1, 2 | Coriell Institute for Medical Research |
| GM07017 | M | Unaffected | B-Lymphocyte | 1, 2 | Coriell Institute for Medical Research |
| GM07022 | M | Unaffected | B-Lymphocyte | 1, 2 | Coriell Institute for Medical Research |
| GM07031 | F | Unaffected | B-Lymphocyte | 1, 2 | Coriell Institute for Medical Research |
| GM07034 | M | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM07049 | M | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM07051 | M | Unaffected | B-Lymphocyte | 1, 2 | Coriell Institute for Medical Research |
| GM07055 | F | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM07056 | F | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM07341 | F | Unaffected | B-Lymphocyte | 1, 2 | Coriell Institute for Medical Research |
| GM07345 | F | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM07346 | F | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM07347 | M | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM07357 | M | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM11879 | M | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM11880 | F | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM11881 | M | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM11882 | F | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM11992 | M | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM11993 | F | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM11994 | M | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM11995 | F | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM12043 | M | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM12044 | F | Unaffected | B-Lymphocyte | 1, 2 | Coriell Institute for Medical Research |
| GM12045 | M | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM12046 | F | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM12144 | M | Unaffected | B-Lymphocyte | 1, 2 | Coriell Institute for Medical Research |
| GM12145 | F | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM12146 | M | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM12154 | M | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM12155 | M | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM12156 | F | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM12236 | F | Unaffected | B-Lymphocyte | 1, 2 | Coriell Institute for Medical Research |
| GM12239 | F | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| GM13047 | M | Unaffected | B-Lymphocyte | 1, 2 | Coriell Institute for Medical Research |
| GM13050 | F | Unaffected | B-Lymphocyte | 2 | Coriell Institute for Medical Research |
| 2219 | M | WBS | Skin fibroblast | 1 | Hospices Civils de Lyon, Hôpital Debrousses |
| 4319 | F | WBS | Skin fibroblast | 1 | Hospices Civils de Lyon, Hôpital Debrousses |
| 11808 | M | WBS | Skin fibroblast | 1 | Hospices Civils de Lyon, Hôpital Debrousses |
| D777 | M | WBS | Skin fibroblast | 1 | Hospices Civils de Lyon, Hôpital Debrousses |
| D890 | F | WBS | Skin fibroblast | 1 | Hospices Civils de Lyon, Hôpital Debrousses |
| D891 | M | WBS | Skin fibroblast | 1 | Hospices Civils de Lyon, Hôpital Debrousses |
| D974 | M | WBS | Skin fibroblast | 1 | Hospices Civils de Lyon, Hôpital Debrousses |
| F055 | F | WBS | Skin fibroblast | 1 | Hospices Civils de Lyon, Hôpital Debrousses |
| F222 | M | WBS | Skin fibroblast | 1 | Hospices Civils de Lyon, Hôpital Debrousses |
| F223 | F | WBS | Skin fibroblast | 1 | Hospices Civils de Lyon, Hôpital Debrousses |
| G371 | M | WBS | Skin fibroblast | 1 | Hospices Civils de Lyon, Hôpital Debrousses |
| G969 | M | WBS | Skin fibroblast | 1 | Hospices Civils de Lyon, Hôpital Debrousses |
| G970 | F | WBS | Skin fibroblast | 1 | Hospices Civils de Lyon, Hôpital Debrousses |
| H651 | F | WBS | Skin fibroblast | 1 | Hospices Civils de Lyon, Hôpital Debrousses |
| C411 | F | Unaffected | Skin fibroblast | 1, 2 | Hospices Civils de Lyon, Hôpital Debrousses |
| C444 | M | Unaffected | Skin fibroblast | 1, 2 | Hospices Civils de Lyon, Hôpital Debrousses |
| C412 | F | Unaffected | Skin fibroblast | 1, 2 | Hospices Civils de Lyon, Hôpital Debrousses |
| C416 | M | Unaffected | Skin fibroblast | 1, 2 | Hospices Civils de Lyon, Hôpital Debrousses |
| C406 | F | Unaffected | Skin fibroblast | 1, 2 | Hospices Civils de Lyon, Hôpital Debrousses |
| G548 | F | Unaffected | Skin fibroblast | 1 | Hospices Civils de Lyon, Hôpital Debrousses |
| GM00038 | F | Unaffected | Skin fibroblast | 1, 2 | Coriell Institute for Medical Research |
| GM00041 | F | Unaffected | Skin fibroblast | 1, 2 | Coriell Institute for Medical Research |
| GM00408 | F | Unaffected | Skin fibroblast | 1 | Coriell Institute for Medical Research |
| GM02038 | M | Unaffected | Skin fibroblast | 1, 2 | Coriell Institute for Medical Research |
| GM08447 | F | Unaffected | Skin fibroblast | 1, 2 | Coriell Institute for Medical Research |
| GM00969 | F | Unaffected | Skin fibroblast | 1, 2 | Coriell Institute for Medical Research |
| GM02036 | F | Unaffected | Skin fibroblast | 1, 2 | Coriell Institute for Medical Research |
| PM9726F | F | Unaffected | Skin fibroblast | 1, 2 | Galliera Genetic Bank |
1 = Cell line used to individually measure RELs. 2 = Cell line used in the mix necessary to normalize (see “Material and Methods” section).
Table 2. .
Genotyping of Cell Lines[Note]
| Cell Line | 23_400kb outWBScentro | 20_Q8N8C3 | 19_WBSCR17 | 17.1_BAZ1B | 17_BAZ1B | 15_TBL2 | 14_WBSCR14 | 10_CLDN4 | 9_ELN | 6_RFC2 | 5.22 CYLN2/RCF2 | 5_CYLN2 | 4_WBSCR16 | 3_HIP1 | 2_CCL26 | .1_400kb outWBStelo |
| Lymphoblastoid: | ||||||||||||||||
| Patients with WBS: | ||||||||||||||||
| GM13461 | 1.12 | .98 | .44 | .44 | .48 | .59 | .44 | .47 | .40 | .82 | .80 | 1.10 | ||||
| GM13472 | 1.20 | .90 | .49 | .54 | .57 | .55 | .52 | .52 | .52 | 1.07 | .84 | 1.07 | ||||
| GM13473 | 1.02 | .97 | .48 | .40 | .50 | .60 | .51 | .50 | .43 | .98 | .78 | 1.23 | ||||
| GM13475 | .82 | .99 | .70 | .35 | .74 | .54 | .58 | .35 | .37 | 1.19 | .97 | 1.22 | ||||
| GM13481 | 1.05 | 1.01 | .42 | .42 | .52 | .57 | .46 | .48 | .48 | .90 | .80 | 1.15 | ||||
| GM13482 | .95 | .79 | .36 | .45 | .43 | .54 | .48 | .41 | .39 | .80 | 1.10±.11 | 1.15 | ||||
| GM13483 | 1.00 | .93 | .47 | .53 | .57 | .67 | .60 | .58 | .54 | .98 | .80 | 1.27 | ||||
| GM14162 | .95 | .89 | .61 | .35 | .59 | .95 | .65 | .54 | .63 | .41 | 1.42 | .83 | 1.67 | 1.05 | ||
| GM14182 | 1.00 | .79 | .53 | .51 | .79 | .57 | .70 | .52 | .44 | 1.11 | .89 | 1.32 | ||||
| GM14268 | .90 | .83 | .83 | .41 | .52 | .63 | .72 | .55 | .67 | .51 | 1.15 | .83 | 1.51 | 1.10 | ||
| Control: | ||||||||||||||||
| GM06993 | .77 | 1.04 | .99 | 1.24 | 1.14 | 1.30 | 1.26 | 1.12 | 1.15 | 1.10 | .93 | 1.26 | ||||
| GM07002 | 1.03 | 1.09 | 1.08 | .97 | 1.09 | 1.01 | 1.17 | 1.11 | 1.02 | 1.19 | .78 | 1.10 | ||||
| GM07017 | .94 | .88 | .96 | 1.01 | 1.10 | 1.09 | .81 | 1.08 | .90 | .99 | 1.24±.11 | .93 | ||||
| GM07022 | 1.05 | 1.00 | .84 | 1.02 | 1.10 | 1.14 | 1.08 | 1.09 | 1.00 | 1.00 | .84 | 1.11 | ||||
| GM07031 | 1.11 | .96 | .87 | 1.00 | 1.08 | 1.19 | 1.11 | 1.06 | .90 | 1.02 | .83 | 1.10 | ||||
| GM07051 | .84 | 1.11 | 1.20 | 1.07 | .98 | 1.34 | .87 | 1.13 | 1.28 | .97 | .85 | 1.20 | ||||
| GM07341 | .98 | 1.04 | .92 | 1.19 | 1.00 | .99 | 1.02 | 1.33 | 1.04 | 1.14 | 1.11 | 1.03 | 1.06 | .90 | 1.13 | 1.02 |
| GM12044 | 1.25 | 1.07 | .88 | .99 | 1.08 | 1.05 | 1.14 | .98 | .89 | .97 | .81 | .88 | ||||
| GM12144 | .92 | 1.20 | .69 | 1.05 | 1.00 | 1.03 | .98 | 1.12 | 1.05 | .95 | .91 | .98 | ||||
| GM12236 | .94 | .92 | .86 | .94 | .95 | 1.03 | .89 | .94 | .68 | .87 | 1.13 | 1.01 | ||||
| GM13047 | .92 | 1.06 | .86 | 1.08 | 1.18 | 1.18 | 1.12 | 1.12 | 1.06 | 1.02 | .88 | 1.13 | ||||
| Skin fibroblast: | ||||||||||||||||
| Patients with WBS: | ||||||||||||||||
| 2219 | .98 | .44 | .48 | .48 | .40 | 1.13 | 1.02 | 1.03 | ||||||||
| 4319 | .82 | .55 | .30 | .78 | .61 | .96 | 1.18 | |||||||||
| 11808 | 1.15 | .95 | .65 | .48 | .50 | .63 | .46 | .50 | .45 | .86 | .79 | 1.11 | ||||
| D777 | .96 | .59 | .38 | .53 | .50 | .38 | 1.05 | 1.04 | ||||||||
| D890 | 1.01 | 1.15 | .45 | .66 | .55 | .51 | .57 | .50 | 1.38 | 1.30 | ||||||
| D891 | 1.06 | .49 | .45 | .49 | .48 | .46 | .99 | .94 | ||||||||
| D974 | 1.00 | .52 | .43 | .51 | .48 | .47 | .99 | 1.00 | ||||||||
| F055 | .93 | .91 | .44 | .45 | .50 | .42 | .51 | .45 | .47 | 1.02 | 1.11±.11 | 1.05 | ||||
| F222 | 1.08 | 1.31 | .62 | .65 | .55 | .56 | .65 | .56 | 1.38 | 1.05 | ||||||
| F223 | .88 | .53 | .48 | .54 | .53 | .46 | 1.13 | 1.12 | ||||||||
| G371 | .91 | 1.09 | .47 | .60 | .57 | .52 | .60 | .51 | .50 | .90 | .82 | |||||
| G969 | .97 | .56 | .44 | .54 | .47 | .45 | 1.01 | 1.03 | ||||||||
| G970 | .90 | .54 | .41 | 2.61 | .48 | .45 | 1.04 | 1.10 | ||||||||
| H651 | 1.06 | .44 | .34 | .41 | .33 | .34 | .76 | .94 | ||||||||
| Control: | ||||||||||||||||
| C406 | .89 | 1.15 | .74 | 1.22 | .98 | .89 | .89 | .84 | .96 | .65 | 1.32 | |||||
| C411 | .91 | .97 | .90 | 1.12 | .92 | 1.14 | 1.13 | 1.09 | 1.00 | .93 | .85 | 1.27 | ||||
| C412 | 1.03 | 1.26 | 1.69 | .92 | 1.01 | .93 | .92 | .92 | .80 | 1.05 | 1.26 | |||||
| C416 | .99 | 1.10 | 1.42 | 1.08 | 1.10 | 1.14 | 1.16 | 1.14 | 1.08 | 1.17 | 1.36 | |||||
| C444 | 1.04 | 1.28 | 1.59 | 1.00 | 1.03 | .99 | .92 | 1.11 | .87 | .84 | 1.14 | |||||
| G548 | 1.02 | 1.22 | 1.03 | .99 | 1.09 | 1.01 | 1.20 | 1.15 | 1.02 | 1.09 | .92 | 1.10 | ||||
| GM00038 | .86 | 1.18 | .70 | 1.14 | 1.08 | 1.07 | 1.06 | 1.23 | 1.09 | 1.05 | .94 | 1.02 | ||||
| GM00041 | 1.08 | .92 | .86 | 1.01 | 1.14 | 1.18 | 1.14 | 1.01 | .93 | 1.00 | .84 | 1.16 | ||||
| GM00408 | 1.02 | 1.14 | .99 | 1.10 | 1.06 | 1.05 | 1.03 | 1.10 | 1.07 | 1.00 | .88 | .96 | ||||
| GM00969 | .92 | .71 | .85 | 1.01 | 1.23 | .88 | .87 | .69 | .81 | 1.42 | .93 | 1.52 | ||||
| GM02036 | .92 | .79 | 1.24 | 1.08 | 1.49 | 1.14 | 1.21 | 1.03 | .86 | 1.26 | .87 | 1.42 | ||||
| GM02038 | 1.03 | .95 | 1.00 | .96 | 1.18 | 1.25 | 1.16 | 1.04 | .97 | 1.23 | .89 | 1.13 | ||||
| GM08447 | .94 | 1.24 | .72 | 1.10 | 1.15 | 1.03 | .97 | 1.14 | 1.09 | .89 | .93 | .89 | ||||
| PM9726F | .93 | 1.12 | .84 | 1.19 | 1.00 | .86 | .85 | .94 | 1.08 | .86 | 1.07 | 1.07 |
Note.— See Howald et al.23
To make sure that differences in expression levels measured in lymphoblastoid cells were not merely due to transformation, we established six lymphoblastoid cell lines from two blood samples collected at 1-wk intervals from the same individual, after informed consent. We measured expression levels of 25 HSA21 genes (ITBG2, CBS, APP, PFKL, U2AF1, PRDM15, LSS, PDXK, SLC19A1, SLC37A1, PWP2H, MCM3AP, GART, CBR1, TMEM1, BTG3, DSCR1, ETS2, IFNAR2, ANKRD3, WRB, GABPA, SON, IFNAR1, and CCT8) that show a large variation in transcript levels in the normal population and found no significant differences in their expression levels in the assayed samples (Pearson 0.8<r<0.92 [mean 0.87]; P<.001). The observed differences correspond to the experimental variation we measure between replicates.
Real-Time Quantitative PCR and Data Analysis
We opted for Taqman real-time quantitative PCR, to measure any small differences in gene expression levels. Primers and probes were designed using the PrimerExpress program (Applied Biosystems), with default parameters in every case for all the confirmed genes mapping on HSA7, from the centromere to the beginning of band 7q21.11. They can be divided into three groups of genes: 23 HSA7 test genes that are hemizygous in patients with WBS, 2 genes mapping in the low-copy repeats (LCRs) flanking the WBS deletion, and 24 HSA7 test genes that are nonhemizygous in patients with WBS. We also designed assays in 2 HSA7p genes, in 13 control genes, and in 3 normalization genes. The complete list of tested genes, their accession numbers and mapping positions, and the primers and probes used are indicated in table 3. Amplicon sequences were checked by both BLAST and BLAT against the human genome, to ensure specificity. Whenever possible (in 94% of cases), oligos were designed to span an intron. Non–intron-spanning assays were tested in standard ± reverse transcriptase reactions of RNA samples for genomic contamination; in all cases, no amplification was observed in the absence of reverse transcriptase. High-performance liquid chromatography–purified, FAM-TAMRA–labeled, double-dye Taqman probes and qPCR mastermix (RT-QP2X-03) were obtained from Eurogentec.
Table 3. .
Assayed Genes, Primers, Probes, and Efficiencies
| Primer |
Efficiencyc |
||||||||||
| Gene | Accession Number | Source | Chromosome | Positiona (kb) |
Categoryb | Forward | Reverse | Probe | BLTd | Lymphoblasts | Fibroblasts |
| GBAS | NM_001483 | RefSeq | HSA7 | 55′807 | 2 | GGGCAGCTGTACATGGTGC | GCATTCCGTATGTCTTCCCTG | CCATCTTTGGGCTTACAGGGATCTTCAGA | ND | 1.04 | 1.00 |
| PSPH | NM_004577 | RefSeq | HSA7 | 55′853 | 2 | ATCATGATTGGAGATGGTGCC | TTCCTCCAAATCCAATGAAAGC | CAGATATGGAAGCCTGTCCTCCTGCTGA | ND | 1.00 | 1.02 |
| ZFD25 | NM_016220 | RefSeq | HSA7 | 63′596 | 2 | CTCACATCTGTAATGCCAGCACT | TGGTCTCAGACTCCTGCCCT | CCACCCACCTTGGCCTCCCA | ND | .95 | 1.00 |
| VKORC1L1 | NM_173517 | RefSeq | HSA7 | 64′782 | 2 | GGCCCTGGGTGAAGTGCT | AATGGAACCCAAAAGACCAAATC | CGCCGCCCTTGCCTCCAG | ND | .96 | .99 |
| GUSB | NM_000181 | RefSeq | HSA7 | 64′870 | 2 | GTGTCCCGGCGTGGG | TGGTGATGCAGAGAAACGTTG | CTGGCGCTGCCGCAGTTCTTC | ND | .99 | 1.04 |
| ASL | NM_000048 | RefSeq | HSA7 | 64′985 | 2 | AACCAGCTGTCACTGCAGGA | TCCCACACGCAGATCACGT | CTGCAGACCATCAGCCCCCTGTTC | ND | .98 | .99 |
| RCP9 | NM_014478 | RefSeq | HSA7 | 65′024 | 2 | GCCTGTGACTGCTGTGGAGA | TTCAATCTGCTCCTCCGTGAG | GCTCTTCACTCTCTTCCACCATCAGCTGG | ND | BE/NE | BE/NE |
| TPST1 | NM_003596 | RefSeq | HSA7 | 65′163 | 2 | TGCACCATGAAGAGATGATTGG | GCTCCTACATTGACTGGCTTGA | AGTGTCTCTGTCAAAAGTGGAGAGATCTACAGACCA | .99 | BE/NE | BE/NE |
| RABGEF1 | NM_014504 | RefSeq | HSA7 | 65′315 | 2 | CCAGGGTCGGATCAAAGAAG | CTGGTTTGCCGGTTTATGGA | AATTCAGGAAGCAAAAGCTCCCAGTC | BE/NE | BE/NE | BE/NE |
| KCTD7 | NM_153033 | RefSeq | HSA7 | 65′510 | 2 | TCCGAGGGCCGGTACTTC | GCGCAGGAAATTCAGCACAT | CGACCGAGATGGCACACACTTTGG | 1.00 | .95 | .95 |
| NM_017994 | NM_017994 | RefSeq | HSA7 | 65′827 | 2 | TGGAAAAGTCTATCATGCTTTAAATCC | GGTGTGCATGAGATGCAAATTTAT | CAGTGATTGTTCCAGATGATGACCGTTCA | .95 | .96 | .95 |
| RSAFD1/NM_018264 | NM_018264 | RefSeq | HSA7 | 65′906 | 2 | CCTGGGCCTCTTCCCCTT | GTGCAATTCAATGCTAAGTGGC | CCCGGCCCTTAGATTTCATGGAGC | .96 | 1.01 | 1.00 |
| AUTS2 | NM_015570 | RefSeq | HSA7 | 69′662 | 2 | TGGGCCACCTCCTCATCA | CGGCCGATTAAAAGGCTCTA | AGCAACTTCCTCAACCCTGCTGCCC | BE/NE | 1.03 | .98 |
| WBSCR17 | NM_022479 | RefSeq | HSA7 | 70′542 | 2 | GATGGACGATTACAAGTCTCATGTG | CGATGTCAATTCCCGGATTC | ACATAGCGTGGAACCTGCCGC | .97 | BE/NE | BE/NE |
| CALN1 | NM_031468 | RefSeq | HSA7 | 70′679 | 2 | CAGCATATTCTGGCAGTTTGACA | GTGGTCTCGGAAGGCATGAT | AGAATGTGCTTCAACTCTTCCAGAGTTATCCTTTG | 1.05 | BE/NE | BE/NE |
| WBSCR20C | AF416611 | GenBank | HSA7 | 71′830 | 6 | CCTGTTGGAAGTGGGATCCA | TGTTCACACGCACAAATCGA | CCTGGTCCAGCCTCCCAGCT | 1.05 | 1.00 | .97 |
| TRIM74 (TRIM50C) | AF498999 | GenBank | HSA7 | 71′843 | 6 | CTCATCGCCAAACTGGTGAA | GGATCACCCAGCTGAAGACATC | AACCGGACCCGAATCGTCAATGAGTC | 1.00 | .99 | BE/NE |
| Proximal breakpoint | Bayese | HSA7 | 72′013 | BE/NE | BE/NE | BE/NE | |||||
| WBSCR20A | AF412028 | GenBank | HSA7 | 72′129 | 6 | CCTGTTGGAAGTGGGATCCA | TGTTCACACGCACAAATCGA | CCTGGTCCAGCCTCCCAGCT | 1.05 | 1.00 | .97 |
| TRIM50 (TRIM50A) | AY081948 | GenBank | HSA7 | 72′146 | 6 | CTCATCGCCAAACTGGTGAA | GGATCACCCAGCTGAAGACATC | AACCGGACCCGAATCGTCAATGAGTC | 1.00 | .99 | BE/NE |
| FKBP6 | NM_003602 | RefSeq | HSA7 | 72′156 | 1 | AATGTGGCACCGCATGTTC | AGCTCTCTCCCCTCGTTGGT | GGTGAAGAACCTTCAACTTTCTCCTGCTGTAGA | BE/NE | BE/NE | BE/NE |
| FZD9 | NM_003508 | RefSeq | HSA7 | 72′262 | 1 | GTTGCGTTCCTCTGGAAGCT | TCCCAGCCAGAGAGGATCAG | ACTGGAATAAACCCCCGCGTGGC | BE/NE | BE/NE | BE/NE |
| BAZ1B | NM_032408 | RefSeq | HSA7 | 72′269 | 1 | AGTGTGAAGAGATCCTCCACAAGA | TGGTCACAGGCTCCCTGAA | TGAAGTACCGCTTCAGCTGGCC | BE/NE | .96 | 1.01 |
| BCL7B | NM_138707 | RefSeq | HSA7 | 72′369 | 1 | TGATGCCTCAGCCAATTCCT | CACGGAACTCTGGTTGCTGTT | TCTCCTTCTTGAATTCCAGGACG | .99 | .95 | 1.01 |
| TBL2 | NM_032988 | RefSeq | HSA7 | 72′397 | 1 | CAACACACACGCTGCTGTATCTC | GACTTCCCAAACCTTCACATCTG | ACGAGGCTACAAATCTGCCACAG | 1.04 | .98 | .97 |
| WBSCR14 | NM_032994 | RefSeq | HSA7 | 72′420 | 1 | GTACGCTGCACAACTGGAAGTT | CCCGTTGAAGGACTCAAACAG | CCCGTTGAAGGACTCAAACAG | BE/NE | BE/NE | BE/NE |
| WBSCR24 | NM_152560 | RefSeq | HSA7 | 72′495 | 1 | GCCCTGGCCATCAAATACC | CAGCGATGCATGCTTTCCT | CAGTTCTCGGCCACCTCACGAAGCT | 1.01 | BE/NE | 1.01 |
| WBSCR18 | NM_032317 | RefSeq | HSA7 | 72′509 | 1 | ACGCAGGCCCAAATCAAG | GCGGTCCGGGTGGTAGA | CGGCTTACTACCGTCAGTGCTTT | BE/NE | BE/NE | 1.00 |
| WBSCR22 | NM_017528 | RefSeq | HSA7 | 72′520 | 1 | TGAAGTTGAACCCAGGGAGTCT | CCCGCCTCGACATCCTTA | TGTTCACCAATGAGAGGTTCCCA | BE/NE | .98 | .97 |
| STX1A | NM_004603 | RefSeq | HSA7 | 72′526 | 1 | GCGCGCCGGAAGAAA | CCAACAGTGGAGGCGATGA | CATCATCTGCTGTGTGATCCTGGGCAT | 1.05 | 1.05 | 1.01 |
| WBSCR21 | NM_148916 | RefSeq | HSA7 | 72′563 | 1 | CCTTGGTGGAAACTCCCAGTT | AGGGAAGAGCCGCATAATCTC | TGCATCCCAGCCACCACCC | .97 | .95 | .98 |
| CLDN4 | NM_001305 | RefSeq | HSA7 | 72′568 | 1 | TCTGGCCCACTCGGACAA | GAGGGTGGACTCTGTTCTTGCT | TTCCCAAGGCCGCCTCCTGC | BE/NE | BE/NE | BE/NE |
| CLDN3 | NM_001306 | RefSeq | HSA7 | 72′596 | 1 | TTCATCGGCAGCAACATCA | CTCTGCACCACGCAGTTCA | CCCTCCCAGATGTTCTGCGACGTG | .97 | BE/NE | 1.05 |
| WBSCR27 | NM_152559 | RefSeq | HSA7 | 72′661 | 1 | TGCGATACCTGAGCTACATGTCA | TTGAAGGTTGGACGAGTTGGT | GACACACCAGCCCACCTGGCTTG | .96 | BE/NE | BE/NE |
| WBSCR28 | NM_182504 | GenBank | HSA7 | 72′687 | 1 | CCAGTCAGATCCAGCCTTTTG | TGATCTCGGTTCTGAACCAACA | CTGAGAGCCTCGTAACCTGCAACAGGAT | 1.04 | BE/NE | BE/NE |
| ELN | NM_000501 | RefSeq | HSA7 | 72′879 | 1 | GCTAAGGCAGCCAAGTATGGA | CAACTACTCCCGGGCCAAA | TGGCCCACCAGGCACTAAGCCTG | .97 | BE/NE | .99 |
| LIMK1 | NM_016735 | RefSeq | HSA7 | 72′947 | 1 | GTGCGCTGTTGCGATCTG | GAGGGTCTCCAGCCAGTGTTC | ACCCCGAGAAGAGGCCATCCTTTG | BE/NE | 1.04 | 1.02 |
| WBSCR1/EIF4H | NM_031992 | RefSeq | HSA7 | 73′021 | 1 | CCCTCCCCTCGGTGGAT | GAGTCGTGGTCTCTGTGCGCTTT | CAACATGGATTTCAGAGAACCCACAGAAGAG | 1.02 | 1.05 | 1.00 |
| WBSCR5 | NM_032464 | RefSeq | HSA7 | 73′046 | 1 | GGAAGGACAAGCTGTTGCAATT | GCTGAAGTTCTGGTACCTGGAAGA | TACCCCAGCCTGGAGGATCCAGC | BE/NE | 1.01 | 1.03 |
| RFC2 | NM_181471 | RefSeq | HSA7 | 73′062 | 1 | CCAACATTGACGAAGCCTACAA | TGCCAATGATATCTTCTGGTGAGT | ATTCTTGCTCACTTGTGGCATCTGGGC | BE/NE | 1.02 | 1.01 |
| CYLN2 | NM_032719 | RefSeq | HSA7 | 73′226 | 1 | AAGCTGATGGAGGCCATGAG | TTTGCAGAACCGGAATTGC | CTGCCCTGACAAGGCCCAGACC | BE/NE | .99 | .99 |
| GTF2IRD1 | NM_016328 | RefSeq | HSA7 | 73′428 | 1 | GCATCGGCCAACCAGATCT | CGTTCAGGCCGGCATAGT | ACTCGTGCAATGGCCAATGTACA | .95 | 1.05 | .98 |
| GTF2I | NM_033003 | RefSeq | HSA7 | 73′585 | 1 | CAGCTGAACCAAGCCAGTTG | TTGCTTGATCTGAGAGCTTCCA | AAGTTCCAGCCACAGAAGAAATAAAAGAGACT | BE/NE | .99 | 1.05 |
| Distal breakpoint | Bayese | HSA7 | 74′032 | BE/NE | BE/NE | BE/NE | |||||
| WBSCR16 | NM_030798 | RefSeq | HSA7 | 74′388 | 2 | CCGCATCCGATGTGGACT | TGCCCCATACAAACAGCTCTC | AGCCACTTTGCTGCACTGACCAACAA | .95 | BE/NE | BE/NE |
| TRIM73 (TRIM50B) | AF498998 | GenBank | HSA7 | 74′645 | 6 | CTCATCGCCAAACTGGTGAA | GGATCACCCAGCTGAAGACATC | AACCGGACCCGAATCGTCAATGAGTC | 1.00 | .99 | BE/NE |
| WBSCR20B | AF416610 | GenBank | HSA7 | 74′657 | 6 | CCTGTTGGAAGTGGGATCCA | TGTTCACACGCACAAATCGA | CCTGGTCCAGCCTCCCAGCT | 1.05 | 1.00 | .97 |
| HIP1 | NM_005338 | RefSeq | HSA7 | 74′780 | 2 | CACTACGAGCTTGCTGGTGTTG | TTCTTGCAGTGTAGGTGGAGATG | TCTGTTCCTTCTTCCCAGCCCTCA | BE/NE | 1.05 | .98 |
| RHBDL7/NPD007 | NM_020684 | RefSeq | HSA7 | 75′120 | 2 | CGGGCCTCACGCTGAA | ACAAAGATGTAGGTTACCAGCCTGTA | TCCGAGGCCCTTCGCAACTGG | BE/NE | 1.01 | 1.00 |
| POR | NM_000941 | RefSeq | HSA7 | 75′227 | 2 | CGAGAGCACCTGTGGAAGTTG | CCGTGCATCCCCACAGA | CGAAGGCGGTGCCCACATCTAC | BE/NE | 1.04 | 1.04 |
| MK-STYX | NM_016086 | RefSeq | HSA7 | 75′270 | 2 | CCGCCATCATAGCCTACCTC | CACTTCTTGACATAGGCCCAGG | CCTCTGCAAGGTCTGCTCGTTACTATGCA | ND | BE/NE | BE/NE |
| MDH2 | NM_005918 | RefSeq | HSA7 | 75′322 | 2 | CTCCACACCGCTGCTGC | AGGAGACTTTGCCGATGCC | AGGTTCTTCTCGATGCCCTTTTTCCCA | ND | .99 | 1.00 |
| DTX2 | NM_020892 | RefSeq | HSA7 | 75′736 | 2 | CAGTGCTACCTTCCAGACAACG | TTCCAGGCCACCTTCAGG | CCAGGGCCGCAAGGTCCTAGAGC | ND | .98 | 1.05 |
| FGL2 | NM_006682 | RefSeq | HSA7 | 76′470 | 2 | AATTAAAGATGAAAGAGCAAAGGATGT | CCCCTGCCTCTTCGCAT | TCCCTCTGCTTTCTAGTCTCACTGGGCA | ND | BE/NE | BE/NE |
| AIP1 | NM_012301 | RefSeq | HSA7 | 77′291 | 2 | GGCGATCAGGTCCATCTCA | GCAATTACAAGGAGAGGGAATGA | AGGTCACACGTAATAGCCTCCCTGGGC | ND | BE/NE | BE/NE |
| CACNA2D1 | NM_000722 | RefSeq | HSA7 | 81′224 | 2 | CTTGAGTTTGACCTTTCCACGAC | ACAGGGAGGCCGTGAAGTC | TCATCCTCCATCTCAACTGCCTCAAGG | ND | BE/NE | 1.00 |
| GRM3 | NM_000840 | RefSeq | HSA7 | 85′918 | 2 | CGCTCAGAGGCCAAAATTCA | TGCACCAGGATCAGACCCA | CAGCCCCAGTTCTCAGGTTTTCATCTGC | ND | BE/NE | BE/NE |
| PSMA5 | NM_002790 | RefSeq | HSA1 | N | AGGAGAAGCTGAATGCAACAAAC | TTCCTTTGTGAACATGTGGAAATT | CCAGGCTGCACTGTGGCTAGCTCAA | 1.01 | .98 | .99 | |
| AGPAT1 | NM_032741 | RefSeq | HSA6 | N | CTCCTACCAAGACTTCTACTGCAAGA | AGCACCCGCACCTGACA | AGCGTCGCTTCACCTCGGGACA | 1.00 | 1.00 | 1.00 | |
| EEF1A1 | NM_001402 | RefSeq | HSA6 | N | AGCAAAAATGACCCACCAATG | GGCCTGGATGGTTCAGGATA | CACCTGAGCAGTGAAGCCAGCTGCTT | .98 | .98 | .98 | |
| B2M | NM_004048 | RefSeq | HSA15 | C | ACTTTGTCACAGCCCAAGATAGTTAA | CGGCATCTTCAAACCCTCC | TGGGATCGAGACATGTAAGCAGCATCA | .98 | .98 | .98 | |
| BTG3 | NM_006806 | RefSeq | HSA21 | C | GCTCACTCTCTGGGTGGACC | GCTGGCAACAATGAATGCAT | ATGTGAGGTGTGCTGTCGGTATGGAGAGA | .98 | .98 | .98 | |
| SON | NM_138927 | RefSeq | HSA21 | C | CAGCAATTTGCCCTCAGAGG | CGGGTTGTTTCATCTGTCGTC | CCGGGTTAAACGGCAGGGCC | .98 | .98 | .98 | |
| ATP50 | NM_001697 | RefSeq | HSA21 | C | TCCATCGCGGAGAGGTACC | TGAGGACAGTTTTTAATTCAGAGAGTG | CACAGTGACCTCTGCATCTCCTTTAGAAGAAGC | .97 | .97 | BE/NE | |
| SIM2 | NM_009586 | RefSeq | HSA21 | C | AAGCTGAGAACAAACCCTTACCC | GCTGGCCGCATTCCAGT | CCACAGCAATACAGCTCGTTCCAAATGG | BE/NE | .96 | BE/NE | |
| DSCR2 | NM_203433 | RefSeq | HSA21 | C | TCCATTGCTAGAACAACCGAATATAG | GCTGGGATTTTCCATACTTGACA | ACACGACCTTCCTGCAGCAGTTCTAAGC | .97 | .97 | .97 | |
| GABPA | NM_002040 | RefSeq | HSA21 | C | CAAAGAGCGCCGAGGATTT | TGGATTTGGCCATTGTTTCC | AGGAGAAGATAGAAGCTCACCTGGGAACAGAA | .98 | BE/NE | .98 | |
| IL10RB | NM_000628 | RefSeq | HSA21 | C | GACAAAGTACGCCTTCTCCCC | GTTATGATGAGGATGGCCCAA | AGGAATTCTCTTCCACAGCACCTGAAAGAGTT | .99 | .99 | .99 | |
| IFNAR1 | NM_000629 | RefSeq | HSA21 | C | TGAAAAGCTGAATAAAAGCAGTGTTT | TCCAACTATAAGCCAAATTTTAGAGGT | TAGTGACGCTGTATGTGAGAAAACAAAACCAGG | 1.03 | 1.03 | 1.03 | |
| IFNGR2 | NM_005534 | RefSeq | HSA21 | C | GGCCTGATTAAATACTGGTTTCACA | TGGGCTGAGTTGGGTCTTTT | TCCACCAAGCATCCCATTACAGATAGAAGAGTATT | .97 | BE/NE | .97 | |
| USP18 | NM_017414 | RefSeq | HSA22 | C | CGAGAGTCTTGTGATGCTGAGG | TTCCCACGTGCGCAATC | CAGTCTGGAGGGCAGTATGAGCTTTTTGC | ND | 1.02 | .98 | |
| DGCR8 | NM_022720 | RefSeq | HSA22 | C | GAGATGAAGAGGCTAGCTGAGGA | GCGGACGCCACAATGG | AGGGAGGAGACTCGAAAGAAGCCCAAGAT | ND | BE/NE | .96 | |
| UFD1L | NM_005659 | RefSeq | HSA22 | C | TCATCAGAAATTCACGTCCCC | GAGAAAGCGACGAATCTGCC | TGTCAAAAAGGTTGAAGAGGATGAAGCTGG | ND | .98 | BE/NE | |
On NCBI 34/Ensembl.
1 = HSA7 hemizygous in WBS; 2 = HSA7 nonhemizygous in WBS; 6 = genes mapping in the LCR flanking the WBS commonly deleted region (i.e., present in six copies/genome ([LCRs]); C = control genes; N = normalization genes.
ND = Not done; BE/NE = bad efficiency (efficiency⩽0.95 or ⩾1.05) or not expressed.
BLT = mix of cDNA samples from brain, liver, and testis.
Most common breakpoint, as described in Bayes et al.18
The efficiency of each Taqman assay was measured using a dilution series of fibroblast cDNA and lymphoblastoid cells or a pool of cDNA samples of brain, liver, and testis, as described elsewhere31 (see table 3 for results). A working Taqman assay was obtained for 57 (85%) of the 67 assayed genes. We were unsuccessful for RCP9, RABGEF1, FKBP6, FZD9, WBSCR14, CLDN4, MK-STYX, FGL2, AIP, and GRM3. Six more genes (TPST1, WBSCR17/GALNT9, CALN1, WBSCR27, WBSCR28, and WBSCR16) were excluded because of a lack of expression in both fibroblasts and lymphoblastoid cell lines (see efficiencies in table 3). Note that TRIM50/73/74, ATP50, SIM2, and UFD1L are not expressed in skin fibroblasts, whereas WBSCR24, WBSCR18, CLDN3, ELN, CACNA2D1, GABPA, IFNGR2, and DGCR8 are not expressed in lymphoblastoids. Thus, in at least one of the two studied cell lines, we were able to assess the relative expression level (REL) of 76% (51/76) of the selected genes, a proportion significantly above the one expected with genomewide technologies. Typically, microarrays hybridized with fibroblast or lymphoblastoid cell cDNA measure the expression of 30%–40% of human genes.
All RT-PCRs were performed in a 10-μl final volume and in five replicates per sample and were set up in a 384-well plate format, with the use of a Biomek 2000 robot (Beckman). They were run in an ABI 7900 Sequence Detection System (Applied Biosystems) with the following amplification conditions: 50°C for 2 min, 95°C for 10 min, and 50 cycles at 95°C for 15 s and 60°C for 1 min. Each plate contained the appropriate normalization genes to control for any variability between the different plate runs.
Raw threshold-cycle (CT) values were obtained using SDS2.2 (Applied Biosystems). To calculate the normalized relative expression ratio between individuals with WBS and controls, we followed methods described elsewhere.31 We exploited the geNorm method32 to select the three normalization genes: AGPAT1, EEF1A1, and PSMA5. They were used to normalize input cDNA for each sample, whereas mixes of 40 lymphoblastoid and 12 fibroblast control-cell-line cDNA samples (table 1) were used to define normal RELs.
Results
We used the high sensitivity of real-time quantitative PCR to accurately measure the expression of all the HSA7q genes mapping in the region 11.9 Mb downstream (band 7q21.11) to 8.4 Mb upstream (centromere position) of the WBS deleted region, in which we could design a working Taqman assay (i.e., efficiency ⩾0.95 and ⩽1.05 for expression in fibroblasts and/or lymphoblastoid cell lines) (see “Material and Methods” and table 3 for details). These genes can be divided into 17 HSA7q hemizygous genes that map within the WBS deletion, 14 HSA7q nonhemizygous genes, and 2 genes that map within the LCR flanking the deletion. This panel of genes was completed with 2 HSA7 nonhemizygous genes that map on the short arm of the chromosome (band 7p11.2) and 19 control, non-HSA7 genes. We compared the mRNA expression levels of these genes in nontransformed skin fibroblast cells obtained from 14 subjects with WBS and from 14 controls and in transformed lymphoblastoid cells obtained from 10 subjects with WBS and from 11 controls (see table 1 for cell lines; see table 2 for genotyping23; and see table 3 for a complete list of assayed genes, primers, and probes).
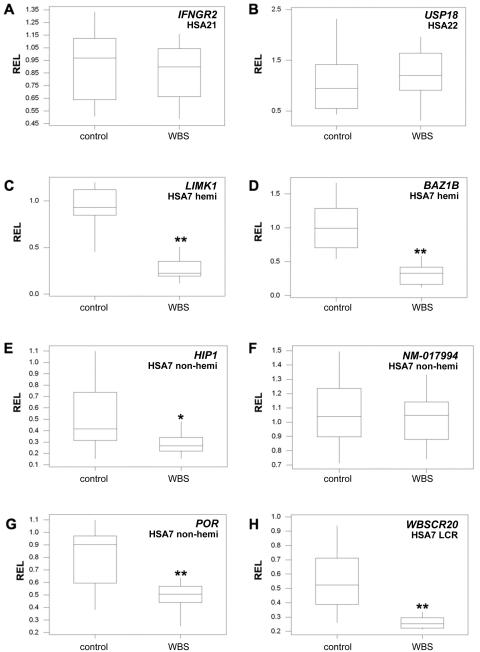
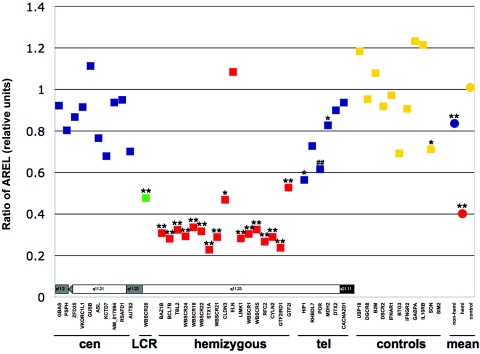
The results of these analyses are summarized in table 4. We found extensive variability in gene RELs in humans (table 4, fig. 1, and data not shown), which is consistent with previous reports.16,17,33 In the population with WBS, all but two of the genes that map to the common deletion interval and that are hemizygous in patients with WBS show average relative expression levels (ARELs) that are approximately half of the normal ARELs (see table 4 and figs. 1C, 1D, and 2), which is consistent with partial results published elsewhere.34 In contrast, the control genes show no significant variation in RELs between the patients and the controls (see table 4 and figs. 1A, 1B, and 2). Interestingly, one hemizygous gene per cell type deviates from this general pattern; the GTF2IRD1 gene in lymphoblastoid cells and the ELN gene in fibroblasts show no significant changes in their ARELs between control and patient samples.
Table 4. .
ARELs[Note]
| Controls |
WBS |
WBS/Controls |
|||||||
| Gene | Categorya | AREL | SD | AREL | SD | t Test P | Mann- Whitney P |
AREL Ratiob | Pairwise t Test P |
| Lymphoblastoid cell lines: | |||||||||
| GBAS | 2 | 1.38 | .37 | 1.01 | .20 | .02 | .04 | .74 | … |
| PSPH | 2 | 1.19 | .31 | .99 | .37 | .23 | .16 | .83 | … |
| ZFD25 | 2 | 1.15 | .28 | .98 | .33 | .24 | .15 | .85 | … |
| VKORC1L1 | 2 | 1.31 | .35 | 1.05 | .42 | .15 | .15 | .80 | … |
| GUSB | 2 | .95 | .18 | .92 | .36 | .82 | .35 | .97 | … |
| ASL | 2 | .81 | .15 | 1.29 | .39 | .004 | .004 | 1.59 | … |
| KCTD7 | 2 | .91 | .42 | .36 | .08 | .004 | .005 | .39 | … |
| NM_017994 | 2 | .99 | .19 | .89 | .14 | .18 | .13 | .89 | … |
| RSAFD1/NM_018264 | 2 | 1.13 | .23 | 1.20 | .53 | .71 | .54 | 1.06 | … |
| AUTS2 | 2 | .57 | .50 | .20 | .21 | .06 | .06 | .35 | … |
| WBSCR20 | 6 | .63 | .09 | .30 | .11 | 1.0×10−6 | .0003 | .48 | … |
| TRIM50/73/74 | 6 | 1.07 | .71 | .71 | .29 | .22 | .36 | .66 | … |
| BAZ1B | 1 | 2.29 | .92 | .64 | .22 | .0006 | .0004 | .28 | … |
| BCL7B | 1 | .74 | .31 | .26 | .09 | .002 | .0004 | .35 | … |
| TBL2 | 1 | .64 | .14 | .24 | .08 | 5.0×10−6 | .0003 | .38 | … |
| WBSCR24 | 1 | NE | NE | NE | NE | NE | NE | NE | … |
| WBSCR18 | 1 | NE | NE | NE | NE | NE | NE | NE | … |
| WBSCR22 | 1 | .65 | .20 | .28 | .09 | .0003 | .0003 | .43 | … |
| STX1A | 1 | .92 | .33 | .27 | .10 | .0003 | .0003 | .29 | … |
| WBSCR21 | 1 | 1.18 | .48 | .44 | .15 | .001 | .0009 | .37 | … |
| CLDN3 | 1 | NE | NE | NE | NE | NE | NE | NE | … |
| ELN | 1 | NE | NE | NE | NE | NE | NE | NE | … |
| LIMK1 | 1 | 2.24 | .80 | .42 | .14 | .0001 | .0004 | .19 | … |
| WBSCR1/EIF4H | 1 | .73 | .15 | .25 | .09 | 1.1×10−6 | .0003 | .34 | … |
| WBSCR5 | 1 | .68 | .21 | .24 | .07 | .0001 | .0003 | .36 | … |
| RFC2 | 1 | .86 | .28 | .40 | .12 | .0009 | .0003 | .46 | … |
| CYLN2 | 1 | .81 | .37 | .23 | .08 | .001 | .0003 | .29 | … |
| GTF2IRD1 | 1 | .28 | .21 | .32 | .19 | .68 | .65 | 1.14 | … |
| GTF2I | 1 | .95 | .19 | .35 | .11 | 2.0×10−6 | .0003 | .37 | … |
| HIP1 | 2 | .90 | .46 | .42 | .17 | .015 | .02 | .47 | … |
| RHBDL7/NPD007 | 2 | .62 | .23 | .50 | .20 | .23 | .31 | .80 | … |
| POR | 2 | .56 | .14 | .50 | .15 | .37 | .71 | .89 | … |
| MDH2 | 2 | .86 | .08 | 1.05 | .14 | .002 | .005 | 1.23 | … |
| DTX2 | 2 | .78 | .29 | .83 | .17 | .62 | .18 | 1.07 | … |
| CACNA2D1 | 2 | NE | NE | NE | NE | NE | NE | NE | … |
| USP18 | C | 1.19 | .71 | 1.32 | .62 | .26 | .66 | 1.11 | … |
| DGCR8 | C | NE | NE | NE | NE | NE | NE | NE | … |
| ATP50 | C | .45 | .11 | .47 | .16 | .71 | .21 | 1.05 | … |
| B2M | C | .63 | .12 | .60 | .28 | .76 | .54 | .95 | … |
| DSCR2 | C | 1.58 | .59 | 1.99 | .95 | .29 | .45 | 1.25 | … |
| IFNAR1 | C | 1.08 | .33 | 1.34 | .66 | .28 | .54 | 1.25 | … |
| BTG3 | C | .79 | .25 | .56 | .19 | .04 | .02 | .71 | … |
| IFNGR2 | C | NE | NE | NE | NE | NE | NE | NE | … |
| GABPA | C | NE | NE | NE | NE | NE | NE | NE | … |
| IL10RB | C | .94 | .11 | 1.19 | .65 | .24 | .65 | 1.28 | … |
| SON | C | 1.00 | .26 | .86 | .27 | .26 | .49 | .86 | … |
| SIM2 | C | 1.04 | .47 | .81 | .55 | .35 | .31 | .78 | … |
| UFD1L | C | 1.51 | .49 | 1.84 | .40 | .04 | .08 | 1.22 | … |
| Nonhemizygous: | |||||||||
| Allc | 2 | .89 | .23 | .78 | .35 | … | … | .88 | .20 |
| Centromerec | 2 | .98 | .23 | .86 | .39 | … | … | .88 | .32 |
| Telomere | 2 | .74 | .14 | .66 | .27 | … | … | .89 | .51 |
| Closed | 2 | .81 | .23 | .58 | .34 | … | … | .71 | .04 |
| LCRs | 6 | .85 | .30 | .51 | .29 | … | … | .60 | .03 |
| Hemizygous | 1 | 1.00 | .60 | .33 | .12 | … | … | .33 | .0005 |
| Controls | C | 1.00 | .34 | 1.12 | .51 | … | … | 1.12 | .16 |
| Skin fibroblasts: | |||||||||
| GBAS | 2 | .99 | .35 | .91 | .23 | .53 | .89 | .92 | … |
| PSPH | 2 | .94 | .34 | .76 | .32 | .18 | .31 | .80 | … |
| ZFD25 | 2 | .90 | .37 | .78 | .38 | .45 | .34 | .87 | … |
| (continued) | |||||||||
| VKORC1L1 | 2 | 1.10 | .33 | 1.00 | .41 | .55 | .67 | .92 | … |
| GUSB | 2 | .95 | .31 | 1.06 | .16 | .29 | .31 | 1.11 | … |
| ASL | 2 | 1.11 | .32 | .85 | .23 | .03 | .08 | .77 | … |
| KCTD7 | 2 | .87 | .43 | .59 | .13 | .06 | .13 | .68 | … |
| NM_017994 | 2 | 1.09 | .24 | 1.02 | .17 | .44 | .69 | .94 | … |
| RSAFD1/NM_018264 | 2 | .98 | .19 | .93 | .16 | .50 | .47 | .95 | … |
| AUTS2 | 2 | 1.02 | .46 | .72 | .43 | .11 | .13 | .70 | … |
| WBSCR20 | 6 | .54 | .21 | .26 | .04 | .002 | .0005 | .48 | … |
| TRIM50/73/74 | 6 | NE | NE | NE | NE | NE | NE | NE | … |
| BAZ1B | 1 | 1.02 | .34 | .31 | .15 | 8.5×10−6 | .00001 | .31 | … |
| BCL7B | 1 | .95 | .37 | .27 | .06 | 4.6×10−5 | .00001 | .28 | … |
| TBL2 | 1 | .91 | .30 | .30 | .08 | 1.7×10−5 | .0001 | .32 | … |
| WBSCR24 | 1 | .62 | .45 | .18 | .04 | .006 | .0001 | .29 | … |
| WBSCR18 | 1 | 1.02 | .33 | .34 | .13 | 9.9×10−6 | .00001 | .34 | … |
| WBSCR22 | 1 | .92 | .22 | .29 | .07 | 3.9×10−7 | .00001 | .32 | … |
| STX1A | 1 | .98 | .59 | .22 | .06 | .001 | .0001 | .23 | … |
| WBSCR21 | 1 | .92 | .33 | .27 | .08 | 1.9×10−5 | .0001 | .29 | … |
| CLDN3 | 1 | .81 | .56 | .38 | .17 | .03 | .04 | .47 | … |
| ELN | 1 | 1.60 | 1.63 | 1.74 | 1.74 | .85 | .62 | 1.08 | … |
| LIMK1 | 1 | .94 | .21 | .26 | .11 | 2.5×10−8 | .00001 | .28 | … |
| WBSCR1/EIF4H | 1 | .91 | .31 | .28 | .08 | 1.5×10−5 | .00001 | .30 | … |
| WBSCR5 | 1 | 1.15 | .48 | .38 | .12 | .0001 | .0001 | .33 | … |
| RFC2 | 1 | .65 | .24 | .17 | .06 | 2.4×10-5 | .00001 | .27 | … |
| CYLN2 | 1 | .61 | .19 | .18 | .05 | 6.7×10–6 | .00001 | .29 | … |
| GTF2IRD1 | 1 | .69 | .27 | .16 | .04 | 3.2×10−5 | .00001 | .24 | … |
| GTF2I | 1 | 1.36 | .38 | .72 | .18 | 8.3×10−5 | .0001 | .53 | … |
| HIP1 | 2 | .51 | .29 | .29 | .09 | .03 | .04 | .56 | … |
| RHBDL7/NPD007 | 2 | .88 | .38 | .64 | .17 | .06 | .10 | .73 | … |
| POR | 2 | .81 | .23 | .50 | .10 | .0006 | .005 | .62 | … |
| MDH2 | 2 | 1.09 | .18 | .90 | .16 | .01 | .01 | .83 | … |
| DTX2 | 2 | 1.06 | .27 | .95 | .20 | .29 | .44 | .90 | … |
| CACNA2D1 | 2 | 1.01 | .61 | .94 | .62 | .80 | .98 | .94 | … |
| USP18 | C | 1.05 | .55 | 1.24 | .47 | .36 | .21 | 1.18 | … |
| DGCR8 | C | .98 | .63 | .94 | .32 | .83 | .84 | .95 | … |
| ATP50 | C | NE | NE | NE | NE | NE | NE | NE | … |
| B2M | C | 1.07 | .57 | 1.15 | .35 | .67 | .29 | 1.08 | … |
| DSCR2 | C | .87 | .26 | .80 | .28 | .53 | .56 | .92 | … |
| IFNAR1 | C | .85 | .45 | .82 | .20 | .42 | .49 | .97 | … |
| BTG3 | C | .86 | .44 | .60 | .21 | .08 | .09 | .69 | … |
| IFNGR2 | C | .93 | .27 | .85 | .22 | .41 | .56 | .91 | … |
| GABPA | C | 1.11 | .57 | 1.37 | .61 | .30 | .31 | 1.23 | … |
| IL10RB | C | 1.03 | .40 | 1.25 | .29 | .15 | .10 | 1.21 | … |
| SON | C | .61 | .24 | .43 | .10 | .03 | .03 | .71 | … |
| SIM2 | C | NE | NE | NE | NE | NE | NE | NE | … |
| UFD1L | C | NE | NE | NE | NE | NE | NE | NE | … |
| Nonhemizygous: | |||||||||
| Allc | 2 | .96 | .16 | .80 | .23 | … | … | .84 | .0003 |
| Centromerec | 2 | 1.00 | .09 | .87 | .17 | … | … | .87 | .03 |
| Telomere | 2 | .89 | .22 | .71 | .28 | … | … | .79 | .004 |
| Closed | 2 | .94 | .19 | .74 | .24 | … | … | .78 | .0001 |
| LCRs | 6 | .54 | .21 | .26 | .04 | … | … | .48 | .002 |
| Hemizygous | 1 | .94 | .26 | .38 | .37 | … | … | .40 | 7.3×10−9 |
| Controls | C | .94 | .15 | .95 | .30 | … | … | 1.01 | .87 |
Note.— NE indicates that the gene is not expressed in this cell line.
1 = HSA7 hemizygous in WBS; 2 = HSA7 nonhemizygous in WBS; 6 = genes mapping in the LCR flanking the WBS commonly deleted region (i.e., present in six copies/genome); C = control genes.
Ratio of ARELs of patients with WBS compared with controls.
Does not take into account GBAS and PSPH, the two genes mapping to the HSA7 short arm.
Close nonhemizygous genes on both the centromeric and the telomeric side.
Figure 1. .
REL distributions measured in 14 patients with WBS and in 14 control skin fibroblasts. REL boxplots are shown for control genes (IFNGR2 [A] and USP18 [B]), for hemizygous genes that map to the commonly deleted WBS interval (LIMK1 [C] and BAZ1B [D]), for nonhemizygous genes that map to the flank of the commonly deleted WBS interval (HIP1 [E], NM_017994 [F], and POR [G]), and for LCR genes that map to the repeats flanking the WBS deletion (WBSCR20 [H]). Asterisks indicate P<.04 and double asterisks indicate P<.005, at both t and Mann-Whitney tests.
Figure 2. .
Differences of expression levels in patients with WBS and in controls. Ratio of ARELs from 14 patients with WBS and from 14 controls, measured in skin fibroblasts. Left to right, Two HSA7 short-arm genes, HSA7 long-arm genes from the centromere to the telomere, followed by the control genes (squares) and the mean (disks) of each tested gene category. Nonhemizygous WBS HSA7 genes (blue) map centromerically (cen) or telomerically (tel) of the deletion. LCR = Gene mapping to the repeats flanking the WBS deletion (green). hemizygous = Hemizygous WBS HSA7 genes (red). controls = Control genes mapping outside HSA7 (yellow). A schematic representation (not to scale) of the HSA7 cytogenetic bands in which the assessed genes map is presented in the lower part of the graph. Asterisks indicate P<.05 and double asterisks indicate P<.001, at both t and Mann-Whitney tests for individual genes and also at pairwise t test for categories. A double number sign (##) indicates that the t and Mann-Whitney tests are significant at P<.001 and P<.005, respectively (see table 4 for details).
ELN haploinsufficiency has been linked to supravalvular aortic stenosis (SVAS) and to other stenoses.35–39 Here, we find that the REL of the ELN gene in skin fibroblasts is not significantly different between the control population and the patients with WBS (ARELfibro=1.08 ± 0.31; P=.85) (table 4). This result is in agreement with those obtained using microarray technology (A. Quattrone and G. Merla, unpublished data) but differs substantially from the one described elsewhere.40 This discrepancy might be due to the very limited number (only one) of samples studied and/or to the less sensitive method used in the latter study.40 As demonstrated by the large SD (table 4), we observe a large variation in relative expression of ELN in the patients with WBS. It is, therefore, possible that the incomplete penetrance of the SVAS phenotype is correlated with the REL of ELN41,42—that is, that patients who are under a compensatory mechanism of expression are less likely to present the phenotype. Consistently, the AREL of the ELN gene in patients with WBS with SVAS (AREL=1.15 ± 1.08) is lower than it is in patients with WBS without this phenotype (AREL=2.56 ± 1.97); however, this difference is not significant. To confirm this hypothesis, we will need to measure the relative expression of this gene in a large number of patients.
A mouse model and recent functional data suggest that hemizygosity of CYLN2 and WBSCR14 might contribute to the cognitive profile and to impaired glucose tolerance or silent diabetes, respectively, in patients with WBS.43–46 Whereas the study of patients with WBS with atypical deletion suggests that hemizygosity of GTF2IRD1 and GTF2I is linked to their visual spatial processing deficits,23–29,47,48 Gtf2ird1-null mice display craniofacial abnormalities, thus suggesting a possible link between hemizygosity of GTF2IRD1 and craniofacial abnormalities displayed in patients with WBS.49 In this article, we find that the relative expression of GTF2IRD1 is significantly decreased in the fibroblasts of patients with WBS (ARELfibro=0.24 ± 0.02; P=3.2×10-5) but is not affected in lymphoblastoid cell lines (ARELlympho=1.14 ± 0.21; P=.68) (table 4). Thus, we cannot assess the contribution of the GTF2IRD1 gene to the WBS cognitive phenotype, because its expression might be under special control in the CNS of patients with WBS.
Remarkably, a significant decrease in relative expression was observed for the nonhemizygous genes ASL, KCTD7, HIP1, POR, and MDH2, which map outside the common deletion region (see table 4 and figs. 1E–1G and 2), although the decrease was not as large as that observed for hemizygous genes. This decrease is significant even if we consider all the tested nonhemizygous genes in fibroblasts mapping to HSA7q or the subset of closest-tested hemizygous genes in lymphoblastoid cells (table 4 and fig. 2).
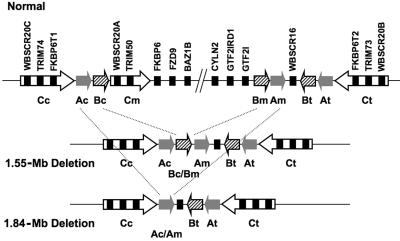
Two of the tested genes, WBSCR20 and TRIM50, map in the LCRs. Three highly similar copies (WBSCR20A, WBSCR20B, and WBSCR20C and TRIM50, TRIM73, and TRIM74; >98% identity each) of each of these genes are present within the studied region (see table 3 and fig. 3).50 TRIM50 (also known as “TRIM50A”) and WBSCR20A map centromerically to FKBP6 within repeat BLOCK-C-mid; TRIM73 (also known as “TRIM50B”) and WBSCR20B map telomerically to FKBP6T2 in BLOCK-C-tel; whereas TRIM74 (also known as “TRIM50C”) and WBSCR20C map to the BLOCK-C-cen interval centromeric to FKBP6T1 (see schematic representation in fig. 3).50–52 WBSCR20A and TRIM50, only one copy of each gene, are hemizygous in patients with WBS (fig. 3).18 We designed a Taqman assay able to simultaneously recognize all three copies and collectively measure the RELs of all copies. Both WBSCR20A/B/C and TRIM50/73/74 exhibit RELs decreased by about one-half and one-third, respectively, in patients with WBS (see table 4 and figs. 1H and 2), a result that deviates noticeably from the theoretically predicted decrease of 17%. It is possible that different levels of expression of the three copies account for this discrepancy. Consistently, the study of UniGene clusters suggests that TRIM50 is expressed more than TRIM73 and TRIM74 together. Conversely, WBSCR20A is not expressed at a higher level than the B and C copies (table 5). These observations suggest that the decrease in relative expression cannot be explained solely by copy-specific expression-level differences. A possible explanation would be that the number of BLOCK-C repeats is polymorphic in the population; however, published results suggest only that the number of BLOCK-A and BLOCK-B copies are polymorphic (see fig. 3).52,53 Thus, our results are consistent with the hypothesis that the nondeleted copies in cis with the deletion are possibly affected in their expression.
Figure 3. .
Schematic partial transcript map of the 7q11.23 region in normal chromosome (top) and in chromosomes bearing the classical WBS 1.55-Mb (center) and 1.84-Mb (bottom) deletions. The different centromeric (c), middle (m), and telomeric (t) duplicons within the LCRs are represented by specific arrows that specify their relative orientation and type. Gray arrow indicates BLOCK-A (A); striped arrow indicates BLOCK-B (B); and white arrow indicates BLOCK-C (C). Genes are depicted by black rectangles, with their names given above.
Table 5. .
Numbers of UniGene ESTs for TRIM50 and WBSCR20
| Gene | GenBank | UniGene | No. of ESTs | % |
| TRIM50 (TRIM50A) | AY081948 | Hs.404810 | 37 | 64.9 |
| TRIM73 (TRIM50B) | AF498998 | Hs.511015 | 5 | 8.8 |
| TRIM74 (TRIM50C) | AF498999 | Hs.534009 | 15 | 26.3 |
| TRIM50/73/74 | 57 | 100 | ||
| WBSCR20A | AF412028 | Hs.272820 | 231 | 29.9 |
| WBSCR20B | AF416610 | Hs .436034 | 214 | 27.7 |
| WBSCR20C | AF416611 | Hs.436034 | 328 | 42.3 |
| WBSCR20 | 773 | 100 |
Discussion
Our results suggest that, in genomic disorders, not only the aneuploid genes but also the normal-copy genes that map close to a deletion should be considered as candidate genes for features of these abnormal phenotypes, although we cannot exclude the possibility that what we observe here is only a 7q11.23 region–specific phenomenon. For example, the HIP1, POR, and KCTD7 genes, located at distances of 0.7, 1.2, and 6.5 Mb from the WBS region, respectively (table 1), show significantly disregulated patterns (t test P=.015 and P=.025, P=6.3×10-4, and P=3.8×10-3, respectively; Mann-Whitney test P=.025 and P=.04, P=5.1×10-3, and P=4.7×10-3, respectively) (table 4) and are thus good candidates for involvement in certain WBS phenotypic features. Remarkably, it appears that this deregulation is more pronounced for genes mapping closer to the breakpoint (fig. 2). This finding also suggests the presence of very distant long-range cis-regulatory elements—to an extent, undescribed elsewhere—and substantiates the notion that functional gene domains extend way beyond their transcription units.54 Although this phenomenon was observed in both a transformed and an untransformed cell line, we cannot be certain that the relative expression pattern is the same in the tissues affected with the different phenotypes. However, data obtained elsewhere, from partial Down syndrome mouse models, have shown that relative expression from aneuploid genes is significantly similar across different tissues and developmental stages.10,11
Even though deletions or duplications of large genomic regions result in significant gene expression changes, our results show that the changes are not always directly correlated to copy number, which suggests an underlying complexity that might involve the size of the deletion, the altered structure of chromatin, a dosage-compensation mechanism, or a combination of these factors. In particular, we identified two transcripts within the commonly deleted WBS region for which there were no significant expression differences. Our observations also suggest that changes in the expression levels of genes neighboring large-scale copy-number polymorphisms2,3,55–57 might play an important role in phenotypic variation in normal populations and, possibly, in evolution.
Acknowledgments
We thank B. Conrad, E. T. Dermitzakis, S. Deutsch, P. Descombes, M. Docquier, M. Gagnebin, C. Gehrig, H. Kaessmann, C. Ucla, and the members of the GRIEG (Group Interdisciplinaire pour l'étude de l'élastine et son gene) group, for assistance and/or critical reading of the manuscript, and A. Quattrone, for sharing unpublished data. This work was supported by grants from Telethon Action Suisse, the Jérôme Lejeune Foundation, the Désirée and Niels Yde Foundation, the Novartis Foundation, the Swiss National Science Foundation (to A.R.), the European Commission (grant number 037627) (to S.E.A. and A.R.), and the Italian Ministry of Health (to G.M.).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Coriell Institute for Medical Research, http://www.coriell.org/index.php/content/view/31/78/ (for cell lines)
- Galliera Genetic Bank, http://ggb.galliera.it (for cell lines)
- geNorm, http://medgen.ugent.be/~jvdesomp/genorm/ (for selection of normalization genes)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for WBS) [PubMed]
- UniGene, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene (for sets of transcript sequences that appear to come from the same transcription locus)
References
- 1.Shaw CJ, Lupski JR (2004) Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet 13:R57–R64 10.1093/hmg/ddh073 [DOI] [PubMed] [Google Scholar]
- 2.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M (2004) Large-scale copy number polymorphism in the human genome. Science 305:525–528 10.1126/science.1098918 [DOI] [PubMed] [Google Scholar]
- 3.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C (2004) Detection of large-scale variation in the human genome. Nat Genet 36:949–951 10.1038/ng1416 [DOI] [PubMed] [Google Scholar]
- 4.Frazer KA, Chen X, Hinds DA, Pant PV, Patil N, Cox DR (2003) Genomic DNA insertions and deletions occur frequently between humans and nonhuman primates. Genome Res 13:341–346 10.1101/gr.554603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu G, Zhao S, Bailey JA, Sahinalp SC, Alkan C, Tuzun E, Green ED, Eichler EE (2003) Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res 13:358–368 10.1101/gr.923303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Locke DP, Segraves R, Carbone L, Archidiacono N, Albertson DG, Pinkel D, Eichler EE (2003) Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization. Genome Res 13:347–357 10.1101/gr.1003303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koszul R, Caburet S, Dujon B, Fischer G (2004) Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J 23:234–243 10.1038/sj.emboj.7600024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do HH, Weiss G, Enard W, Heissig F, Arendt T, Nieselt-Struwe K, Eichler EE, Paabo S (2004) Regional patterns of gene expression in human and chimpanzee brains. Genome Res 14:1462–1473 10.1101/gr.2538704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller S, Finelli P, Neusser M, Wienberg J (2004) The evolutionary history of human chromosome 7. Genomics 84:458–467 10.1016/j.ygeno.2004.05.005 [DOI] [PubMed] [Google Scholar]
- 10.Lyle R, Gehrig C, Neergaard-Henrichsen C, Deutsch S, Antonarakis SE (2004) Gene expression from the aneuploid chromosome in a trisomy mouse model of down syndrome. Genome Res 14:1268–1274 10.1101/gr.2090904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahlem P, Sultan M, Herwig R, Steinfath M, Balzereit D, Eppens B, Saran NG, Pletcher MT, South ST, Stetten G, Lehrach H, Reeves RH, Yaspo ML (2004) Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of down syndrome. Genome Res 14:1258–1267 10.1101/gr.1951304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amano K, Sago H, Uchikawa C, Suzuki T, Kotliarova SE, Nukina N, Epstein CJ, Yamakawa K (2004) Dosage-dependent over-expression of genes in the trisomic region of Ts1Cje mouse model for Down syndrome. Hum Mol Genet 13:1333–1340 10.1093/hmg/ddh154 [DOI] [PubMed] [Google Scholar]
- 13.Hollox EJ, Armour JA, Barber JC (2003) Extensive normal copy number variation of a β-defensin antimicrobial-gene cluster. Am J Hum Genet 73:591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E (2003) A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12:1725–1735 10.1093/hmg/ddg180 [DOI] [PubMed] [Google Scholar]
- 15.Spitz F, Gonzalez F, Duboule D (2003) A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113:405–417 10.1016/S0092-8674(03)00310-6 [DOI] [PubMed] [Google Scholar]
- 16.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG (2004) Genetic analysis of genome-wide variation in human gene expression. Nature 430:743–747 10.1038/nature02797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT (2005) Mapping determinants of human gene expression by regional and genome-wide association. Nature 437:1365–1369 10.1038/nature04244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayes M, Magano LF, Rivera N, Flores R, Perez Jurado LA (2003) Mutational mechanisms of Williams-Beuren syndrome deletions. Am J Hum Genet 73:131–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris CA, Demsey SA, Leonard CO, Dilts C, Blackburn BL (1988) Natural history of Williams syndrome: physical characteristics. J Pediatr 113:318–326 10.1016/S0022-3476(88)80272-5 [DOI] [PubMed] [Google Scholar]
- 20.Francke U (1999) Williams-Beuren syndrome: genes and mechanisms. Hum Mol Genet 8:1947–1954 10.1093/hmg/8.10.1947 [DOI] [PubMed] [Google Scholar]
- 21.Osborne LR (1999) Williams-Beuren syndrome: unraveling the mysteries of a microdeletion disorder. Mol Genet Metab 67:1–10 10.1006/mgme.1999.2844 [DOI] [PubMed] [Google Scholar]
- 22.Stromme P, Bjornstad PG, Ramstad K (2002) Prevalence estimation of Williams syndrome. J Child Neurol 17:269–271 [DOI] [PubMed] [Google Scholar]
- 23.Howald C, Merla G, Digilio MC, Amenta S, Lyle R, Deutsch S, Choudhury U, Bottani A, Antonarakis SE, Fryssira H, Dallapiccola B, Reymond A (2006) Two high throughput technologies to detect segmental aneuploidies identify new Williams-Beuren syndrome patients with atypical deletions. J Med Genet 43:266–273 10.1136/jmg.2005.034009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korenberg JR, Chen XN, Hirota H, Lai Z, Bellugi U, Burian D, Roe B, Matsuoka R (2000) VI. Genome structure and cognitive map of Williams syndrome. J Cogn Neurosci 12:89–107 10.1162/089892900562002 [DOI] [PubMed] [Google Scholar]
- 25.Hirota H, Matsuoka R, Chen XN, Salandanan LS, Lincoln A, Rose FE, Sunahara M, Osawa M, Bellugi U, Korenberg JR (2003) Williams syndrome deficits in visual spatial processing linked to GTF2IRD1 and GTF2I on chromosome 7q11.23. Genet Med 5:311–321 [DOI] [PubMed] [Google Scholar]
- 26.Gagliardi C, Bonaglia MC, Selicorni A, Borgatti R, Giorda R (2003) Unusual cognitive and behavioural profile in a Williams syndrome patient with atypical 7q11.23 deletion. J Med Genet 40:526–530 10.1136/jmg.40.7.526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Botta A, Novelli G, Mari A, Novelli A, Sabani M, Korenberg J, Osborne LR, Digilio MC, Giannotti A, Dallapiccola B (1999) Detection of an atypical 7q11.23 deletion in Williams syndrome patients which does not include the STX1A and FZD3 genes. J Med Genet 36:478–480 [PMC free article] [PubMed] [Google Scholar]
- 28.Heller R, Rauch A, Luttgen S, Schroder B, Winterpacht A (2003) Partial deletion of the critical 1.5 Mb interval in Williams-Beuren syndrome. J Med Genet 40:e99 10.1136/jmg.40.8.e99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris CA, Mervis CB, Hobart HH, Gregg RG, Bertrand J, Ensing GJ, Sommer A, Moore CA, Hopkin RJ, Spallone PA, Keating MT, Osborne L, Kimberley KW, Stock AD (2003) GTF2I hemizygosity implicated in mental retardation in Williams syndrome: genotype-phenotype analysis of five families with deletions in the Williams syndrome region. Am J Med Genet A 123:45–59 10.1002/ajmg.a.20496 [DOI] [PubMed] [Google Scholar]
- 30.Osborne LR, Li M, Pober B, Chitayat D, Bodurtha J, Mandel A, Costa T, Grebe T, Cox S, Tsui LC, Scherer SW (2001) A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet 29:321–325 10.1038/ng753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 32.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deutsch S, Lyle R, Dermitzakis ET, Attar H, Subrahmanyan L, Gehrig C, Parand L, Gagnebin M, Rougemont J, Jongeneel CV, Antonarakis SE (2005) Gene expression variation and expression quantitative trait mapping of human chromosome 21 genes. Hum Mol Genet 14:3741–3749 10.1093/hmg/ddi404 [DOI] [PubMed] [Google Scholar]
- 34.Somerville MJ, Mervis CB, Young EJ, Seo EJ, del Campo M, Bamforth S, Peregrine E, Loo W, Lilley M, Perez-Jurado LA, Morris CA, Scherer SW, Osborne LR (2005) Severe expressive-language delay related to duplication of the Williams-Beuren locus. N Engl J Med 353:1694–1701 10.1056/NEJMoa051962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ewart AK, Morris CA, Atkinson D, Jin W, Sternes K, Spallone P, Stock AD, Leppert M, Keating MT (1993) Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet 5:11–16 10.1038/ng0993-11 [DOI] [PubMed] [Google Scholar]
- 36.Curran ME, Atkinson DL, Ewart AK, Morris CA, Leppert MF, Keating MT (1993) The elastin gene is disrupted by a translocation associated with supravalvular aortic stenosis. Cell 73:159–168 10.1016/0092-8674(93)90168-P [DOI] [PubMed] [Google Scholar]
- 37.Tassabehji M, Metcalfe K, Donnai D, Hurst J, Reardon W, Burch M, Read AP (1997) Elastin: genomic structure and point mutations in patients with supravalvular aortic stenosis. Hum Mol Genet 6:1029–1036 10.1093/hmg/6.7.1029 [DOI] [PubMed] [Google Scholar]
- 38.Li DY, Brooke B, Davis EC, Mecham RP, Sorensen LK, Boak BB, Eichwald E, Keating MT (1998) Elastin is an essential determinant of arterial morphogenesis. Nature 393:276–280 10.1038/30522 [DOI] [PubMed] [Google Scholar]
- 39.Li DY, Faury G, Taylor DG, Davis EC, Boyle WA, Mecham RP, Stenzel P, Boak B, Keating MT (1998) Novel arterial pathology in mice and humans hemizygous for elastin. J Clin Invest 102:1783–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urban Z, Michels VV, Thibodeau SN, Davis EC, Bonnefont JP, Munnich A, Eyskens B, Gewillig M, Devriendt K, Boyd CD (2000) Isolated supravalvular aortic stenosis: functional haploinsufficiency of the elastin gene as a result of nonsense-mediated decay. Hum Genet 106:577–588 10.1007/s004390050028 [DOI] [PubMed] [Google Scholar]
- 41.Pankau R, Siebert R, Kautza M, Schneppenheim R, Gosch A, Wessel A, Partsch CJ (2001) Familial Williams-Beuren syndrome showing varying clinical expression. Am J Med Genet 98:324–329 [DOI] [PubMed] [Google Scholar]
- 42.Wang MS, Schinzel A, Kotzot D, Balmer D, Casey R, Chodirker BN, Gyftodimou J, Petersen MB, Lopez-Rangel E, Robinson WP (1999) Molecular and clinical correlation study of Williams-Beuren syndrome: no evidence of molecular factors in the deletion region or imprinting affecting clinical outcome. Am J Med Genet 86:34–43 [DOI] [PubMed] [Google Scholar]
- 43.Cairo S, Merla G, Urbinati F, Ballabio A, Reymond A (2001) WBSCR14, a gene mapping to the Williams-Beuren syndrome deleted region, is a new member of the Mlx transcription factor network. Hum Mol Genet 10:617–627 10.1093/hmg/10.6.617 [DOI] [PubMed] [Google Scholar]
- 44.Hoogenraad CC, Koekkoek B, Akhmanova A, Krugers H, Dortland B, Miedema M, van Alphen A, Kistler WM, Jaegle M, Koutsourakis M, Van Camp N, Verhoye M, van der Linden A, Kaverina I, Grosveld F, De Zeeuw CI, Galjart N (2002) Targeted mutation of Cyln2 in the Williams syndrome critical region links CLIP-115 haploinsufficiency to neurodevelopmental abnormalities in mice. Nat Genet 32:116–127 10.1038/ng954 [DOI] [PubMed] [Google Scholar]
- 45.Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K (2004) Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci USA 101:7281–7286 10.1073/pnas.0401516101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merla G, Howald C, Antonarakis SE, Reymond A (2004) The subcellular localization of the ChoRE-binding protein, encoded by the Williams-Beuren syndrome critical region gene 14, is regulated by 14-3-3. Hum Mol Genet 13:1505–1514 10.1093/hmg/ddh163 [DOI] [PubMed] [Google Scholar]
- 47.Karmiloff-Smith A, Grant J, Ewing S, Carette MJ, Metcalfe K, Donnai D, Read AP, Tassabehji M (2003) Using case study comparisons to explore genotype-phenotype correlations in Williams-Beuren syndrome. J Med Genet 40:136–140 10.1136/jmg.40.2.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tassabehji M, Metcalfe K, Karmiloff-Smith A, Carette MJ, Grant J, Dennis N, Reardon W, Splitt M, Read AP, Donnai D (1999) Williams syndrome: use of chromosomal microdeletions as a tool to dissect cognitive and physical phenotypes. Am J Hum Genet 64:118–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tassabehji M, Hammond P, Karmiloff-Smith A, Thompson P, Thorgeirsson SS, Durkin ME, Popescu NC, Hutton T, Metcalfe K, Rucka A, Stewart H, Read AP, Maconochie M, Donnai D (2005) GTF2IRD1 in craniofacial development of humans and mice. Science 310:1184–1187 10.1126/science.1116142 [DOI] [PubMed] [Google Scholar]
- 50.Merla G, Ucla C, Guipponi M, Reymond A (2002) Identification of additional transcripts in the Williams-Beuren syndrome critical region. Hum Genet 110:429–438 10.1007/s00439-002-0710-x [DOI] [PubMed] [Google Scholar]
- 51.Doll A, Grzeschik KH (2001) Characterization of two novel genes, WBSCR20 and WBSCR22, deleted in Williams-Beuren syndrome. Cytogenet Cell Genet 95:20–27 10.1159/000057012 [DOI] [PubMed] [Google Scholar]
- 52.Valero MC, de Luis O, Cruces J, Perez Jurado LA (2000) Fine-scale comparative mapping of the human 7q11.23 region and the orthologous region on mouse chromosome 5G: the low-copy repeats that flank the Williams-Beuren syndrome deletion arose at breakpoint sites of an evolutionary inversion(s). Genomics 69:1–13 10.1006/geno.2000.6312 [DOI] [PubMed] [Google Scholar]
- 53.Perez Jurado LA, Wang YK, Peoples R, Coloma A, Cruces J, Francke U (1998) A duplicated gene in the breakpoint regions of the 7q11.23 Williams-Beuren syndrome deletion encodes the initiator binding protein TFII-I and BAP-135, a phosphorylation target of BTK. Hum Mol Genet 7:325–334 10.1093/hmg/7.3.325 [DOI] [PubMed] [Google Scholar]
- 54.Kleinjan DA, van Heyningen V (2005) Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet 76:8–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bignell GR, Huang J, Greshock J, Watt S, Butler A, West S, Grigorova M, Jones KW, Wei W, Stratton MR, Futreal PA, Weber B, Shapero MH, Wooster R (2004) High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res 14:287–295 10.1101/gr.2012304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Segraves R, Oseroff VV, Albertson DG, Pinkel D, Eichler EE (2005) Segmental duplications and copy-number variation in the human genome. Am J Hum Genet 77:78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, Olson MV, Eichler EE (2005) Fine-scale structural variation of the human genome. Nat Genet 37:727–732 10.1038/ng1562 [DOI] [PubMed] [Google Scholar]