Abstract
Spastin, the most commonly mutated protein in the autosomal dominant form of hereditary spastic paraplegia (AD-HSP) has been suggested to be involved in vesicular cargo trafficking; however, a comprehensive function of spastin has not yet been elucidated. To characterize the molecular function of spastin, we used the yeast two-hybrid approach to identify new interacting partners of spastin. Here, we report ZFYVE27, a novel member of the FYVE-finger family of proteins, as a specific spastin-binding protein, and we validate the interaction by both in vivo coimmunoprecipitation and colocalization experiments in mammalian cells. More importantly, we report a German family with AD-HSP in which ZFYVE27 (SPG33) is mutated; furthermore, we demonstrate that the mutated ZFYVE27 protein shows an aberrant intracellular pattern in its tubular structure and that its interaction with spastin is severely affected. We postulate that this specific mutation in ZFYVE27 affects neuronal intracellular trafficking in the corticospinal tract, which is consistent with the pathology of HSP.
Impaired axonal transport has been implicated in many neurodegenerative disorders. One such group of neurodegenerative disorders that primarily affects lower limbs is hereditary spastic paraplegia (HSP). HSPs are characterized by progressive spastic paralysis of the legs, usually caused by length-dependent distal degeneration of the corticospinal tract axons.1 Clinically, HSPs are classified as either the pure or the complex form.2 In the pure form of HSP, spastic paraplegia occurs in relative isolation and is often accompanied by urinary urgency. The complex form of HSP is associated with additional neurological or nonneurological features. HSPs are genetically complex: autosomal dominant, recessive, and X-linked modes of inheritance are reported for both pure and complex HSP.2 Within each inheritance pattern group, there is further locus heterogeneity. For autosomal dominant HSP (AD-HSP), 13 different loci are reported; also, 13 loci for the autosomal recessive form and 3 loci for the X-linked disease are known. It is likely that more loci will be identified in the near future. Eleven of the HSP genes are now identified. However, mutation in spastin (MIM 604277) is the most common cause of HSP and accounts for up to 40% of AD-HSP cases.3–6
Spastin is a member of the AAA family of proteins and is ubiquitously expressed in all tissues. The AAA proteins are proven or putative ATPases, and they are characterized by a conserved C-terminal domain containing one or two AAA cassettes.7 The N-terminal domain of the protein, which contains recognition sites for substrates or adaptors, is highly divergent and, therefore, confers specificity to its cellular function.8
A comprehensive function of spastin is not yet elucidated; previous reports suggested that spastin is principally involved in microtubule dynamics and severing.9–11 However, the expression of spastin in punctate vesicles in cultured cells also suggests a role in vesicular trafficking, which is now supported by emerging reports.12,13 Furthermore, the N-terminal portion of spastin contains a microtubule-interacting and trafficking (MIT) domain, which is predicted to play a role in the endosomal trafficking process.14 The MIT domain corresponds to a slightly extended version of the End13-SNX15-Pa1B (ESP) homology domain, which is also present in VPS4, SKD1, RPK118, and SNX15, all of which have a well-established role in endosomal trafficking.15–18
Spastin is a multifaceted protein, and, to elucidate its function, we performed a yeast two-hybrid assay. In our screen, we used spastin-lacking exon 4 (GenBank accession number NM_199436) (referred to as “spastin”) as bait against a human brain cDNA library and tested 2.2×106 clones. This spastin splice variant is the major transcript in adult spinal cord and could be pathologically relevant.19 Stringent screening revealed that 31 clones were able to grow in selective media and were also positive for α-galactosidase activity (data not shown). Sequencing of these clones led to the identification of seven ORFs of five unique genes (table 1). Clone 18, which was identified as a potential spastin-interacting protein, encodes for ZFYVE27 and contains the C-terminal fragment of ZFYVE27 (amino acids 209–318) and the 3′ UTR, which corresponds to nucleotides 822–2746 of the ZFYVE27 cDNA (ZFYVE27-c isoform [GenBank accession number AK097945]). ZFYVE27 is a novel protein that belongs to the FYVE-finger family of proteins. The FYVE-finger domain is suggested to be responsible for endosomal localization of these proteins, and the majority of the FYVE-finger proteins serve as regulators of endocytic membrane trafficking.20,21 The fact that ZFYVE27 is a putative endosomal protein, combined with the recent finding of spastin interaction with endosomal protein CHMP1B,12 prompted us to investigate the interaction of ZFYVE27 with spastin and its physiological relevance.
Table 1. .
Genes Identified as Putative Spastin-Interacting Proteins in our Yeast Two-Hybrid Assay
| Clonea | Geneb | Chromosomal Locationc |
Amino Acidsd |
| 3 | Reticulon3 | 11q13 | 57–236 |
| 5 | CRELD5 | 3p25.2 | 201–421 |
| 6 | COPS5 | 8q13.2 | 67–335 |
| 8 | Reticulon1 | 14q23.1 | 632–776 |
| 10 | Reticulon1 | 14q23.1 | 632–776 |
| 18 | ZFYVE27 | 10q24.2 | 209–319 |
| 22 | Reticulon3 | 11q13 | 85–236 |
Identity of the clone designated in our yeast two-hybrid experiment.
Name of the gene corresponding to the clone.
Chromosomal location of the gene.
Encoding amino acid sequence present in the clone.
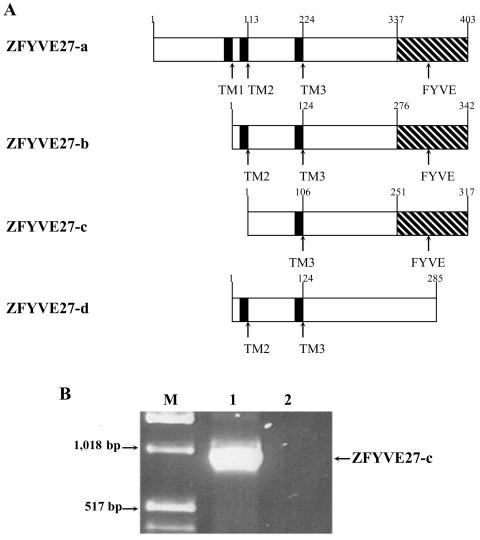
In the Ensembl database, four different splice variants of ZFYVE27 are reported, as shown in figure 1A. To amplify the complete ORF of ZFYVE27, we performed RT-PCR on human brain RNA with a primer pair flanking the complete ORF of the transcripts, and we were able to amplify a single ∼1-kb product (fig. 1B). Sequencing revealed that this product corresponds to the variant form, ZFYVE27-c (fig. 1A). We used this ZFYVE27-c product in our further investigations and referred to it as “ZFYVE27.”
Figure 1. .
Spliced isoforms of ZFYVE27. A, Schematic diagrams showing four spliced isoforms of ZFYVE27, which are reported in the Ensembl Genome browser. The amino acid positions are marked in arabic numerals. FYVE = zinc-finger FYVE domain. B, Complete ORF of the ZFYVE27-c cDNA amplified by RT-PCR from total human brain RNA. Lane M, Marker; lane 1, brain; lane 2, negative control.
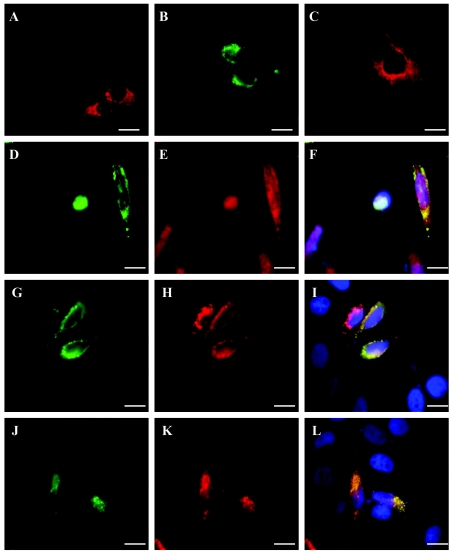
To determine the intracellular distribution of ZFYVE27, we generated epitope-tagged ZFYVE27 constructs. Analysis of cells, which were transfected with E2-ZFYVE27 or green fluorescent protein (GFP)–ZFYVE27 construct, showed that ZFYVE27 is predominantly expressed in punctate vesicles (fig. 2A and 2B). However, in a small proportion of cells, a tubular pattern of expression was also observed (fig. 2C). Overall, the intracellular distribution of ZFYVE27 in punctate vesicles was comparable to the expression pattern reported for other well-characterized members of the FYVE-finger family of proteins.22–24 Colocalization studies with subcellular markers revealed an overlapping expression with the endosomal marker, EEA122,23 (fig. 2D–2F), and the endoplasmic reticulum (ER) marker, RTN125,26 (fig. 2G–2I). ZFYVE27 expression in the vesicles originating from both ER and endosome could be dependent on the metabolic stage of the cell after the transient transfection. Presumably, the biogenesis of ZFYVE27 occurs in ER, and then it could be sorted to the endosome through its FYVE-finger domain interaction with the endosomal membrane. Examination of HeLa cells coexpressing GFP-ZFYVE27 and c-Myc-spastin by immunofluorescence microscopy revealed an obvious colocalization in vesicular-like structures in the cytoplasm (fig. 2J–2L). The colocalization of ZFYVE27 and spastin in discrete vesicles in the cytosol also highlights the physiological interaction between spastin and ZFYVE27.
Figure 2. .
Intracellular distribution of ZFYVE27 and colocalization with endosomal marker (EEA1), ER marker (RTN1), and spastin in mammalian cells. HeLa cells were transiently transfected with either E2-tagged ZFYVE27 or GFP-ZFYVE27 constructs. A, E2-tagged ZFYVE27 expressed in the punctate vesicles distributed in the cytoplasm. B, GFP-ZFYVE27 also showing vesicular expression in the cytosol. C, A small proportion of cells revealing a tubular pattern of expression, which seems to arise from the perinuclear region and is then distributed in the peripheral region of the cytoplasm. D–F, HeLa cells transiently transfected with GFP-ZFYVE27 and immunostained with EEA1. D, HeLa cell expressing GFP-ZFYVE27 in cytoplasmic vesicles. E, Staining of the same cell, with EEA1 antibody showing expression in punctate vesicles in the cytoplasm. F, Subset of cytoplasmic vesicles showing coexpression of GFP-ZFYVE27 and EEA1, as observed in the superimposed image. The nuclei of the cells were stained with 4′,6-diamidino-2-phenylindole (DAPI). G–I, Colocalization of GFP-ZFYVE27 and ER marker, E2-RTN1, in the HeLa cells. G, GFP-ZFYVE27; H, E2-RTN1; I, overlay image. An overlapping expression pattern in vesicular structure around the perinuclear region can be seen between GFP-ZFYVE27 and E2-RTN1 in the cytoplasm. J-L, Expression of GFP-ZFYVE27 and c-Myc-spastin. J, GFP-ZFYVE27; K, c-Myc-spastin; L, overlay image. A striking level of colocalization between GFP-ZFYVE27 and c-Myc-spastin can be observed in cytoplasmic vesicles. Scale bar is 20 μm.
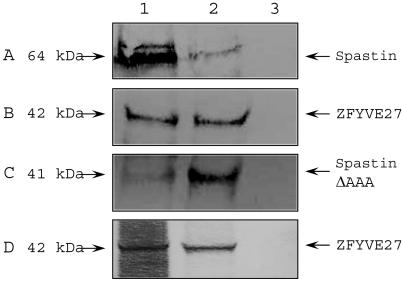
To investigate the interaction of ZFYVE27 with spastin, we performed coimmunoprecipitation studies by cotransfecting NIH 3T3 cells with E2-ZFYVE27 and c-Myc-spastin constructs. When E2-ZFYVE27 was immunoprecipitated from total protein lysate of the transfected cells, c-Myc-spastin (64 kDa) could also be detected in the precipitated fraction (fig. 3A). Similarly, E2-ZFYVE27 (42 kDa) could be coimmunoprecipitated with c-Myc-spastin (fig. 3B). To determine the domain of spastin, which is responsible for the interaction, we generated N-terminal (c-Myc-spastinΔNT) and C-terminal (c-Myc-spastinΔAAA) deletion constructs of spastin for immunoprecipitation experiments. The c-Myc-spastinΔNT lacks the first 300 amino acids, including the MIT domain, whereas, in the c-Myc-spastinΔAAA construct, amino acids 398–583 were deleted.
Figure 3. .
In vivo interaction between spastin and ZFYVE27 in mammalian cells. NIH 3T3 cells were cotransfected with c-Myc-spastin and E2-ZFYVE27 constructs. Soluble protein lysate was prepared after 24–48 h of transfection. A, Immunoprecipitation performed with E2-tag antibody, and, subsequently, Western blot performed with c-Myc antibody. Lane 1, standard protein lysate, which serves as a control for transfection efficiency; lane 2, immunoprecipitated fraction; lane 3, mock precipitation done without E2-tag antibody. A 64-kDa band corresponding to c-Myc-spastin can be seen in lane 1 and lane 2 but not in lane 3. B, Precipitation done with c-Myc antibody and the immunoblot done with E2 antibody. A 42-kDa band corresponding to E2-ZFYVE27 can be detected in protein standard (lane 1) and precipitated fraction (lane 2) but not in mock experiment (lane 3). C, Precipitation performed with cells cotransfected with c-Myc-spastinΔAAA, E2-ZFYVE27 with E2 antibody, and the Western blot with c-Myc antibody. A c-Myc-spastinΔAAA protein can be detected as a 41-kDa band in the precipitated fraction (lane 1) as well as in the standard protein fraction (lane 2) but not in the mock precipitate (lane 3). D, Immunoprecipitation assay between endogenous spastin and E2-ZFYVE27, with the use of antispastin antibody. Lane 1, immunoprecipitated fraction; lane 2, protein standard; lane 3, mock precipitation. The endogenous spastin was able to coprecipitate E2-ZFYVE27, since, in the immunoblot, a 42-kDa band corresponding to E2-ZFYVE27 could be detected in both lanes 1 and 2 with the use of E2 antibody.
Our results revealed that E2-ZFYVE27 was able to interact with c-Myc-spastinΔAAA (fig. 3C) but not with c-Myc-spastinΔNT (data not shown), suggesting that spastin mediates interaction with ZFYVE27 through its N-terminal domain, which consists of a MIT motif. Interestingly, the N-terminal spastin domain is also responsible for the interaction with other proteins—for example, the endosomal protein CHMP1B,12 microtubules,9 and the centrosomal protein NA14.27 To validate the interaction between spastin and ZFYVE27 at the endogenous level, we performed an immunoprecipitation assay, using antispastin antibodies with protein extract derived from HeLa cells transfected with E2-ZFYVE27. Our analysis confirmed that endogenous spastin interacts with E2-ZFYVE27 (fig. 3D).
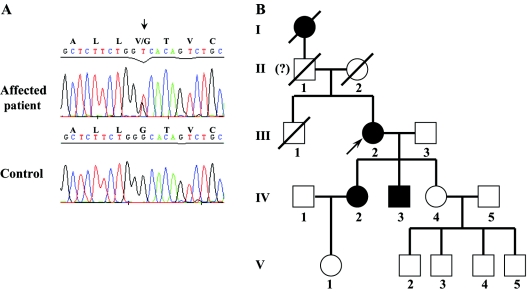
The fact that ZFYVE27 is a spastin-interacting protein and its expression in brain led us to consider ZFYVE27 as a candidate gene for HSP. The ZFYVE27 gene is located on chromosome 10q24.2 and consists of 13 exons. We investigated genomic DNA of 43 patients with AD-HSP for mutation in ZFYVE27 (for primer sequences, see table 2). Our mutational analysis detected a single sequence variant in the exon 6 of ZFYVE27 in the genomic DNA of one patient with AD-HSP (III.2 in fig. 4B). We identified a transversion, G→T, at position 572 of ZFYVE27 cDNA (ZFYVE27-a isoform [GenBank accession number NM_001002261]), which leads to a missense mutation, G191V, in the protein (fig. 4A). Sequencing of the ZFYVE27 gene in the genomic DNA of other family members (IV.2, IV.3, and IV.4) revealed that the mutation c.572G→T (p.G191V) segregated with the clinical phenotype in the affected individuals (IV.2 and IV.3) and was not detected in the clinically unaffected individual (IV.4) (fig. 4B). An extensive GenBank search of ZFYVE27 EST clones for the presence of this base exchange failed to detect any EST containing the identified sequence variant. Furthermore, this mutation was not found in 210 control chromosomes (data not shown). Thus, the detected base exchange is, most probably, causative for the HSP in this family and is not a rare polymorphism.
Table 2. .
Primer Sequences for Amplifying and Sequencing of ZFYVE27
| Primer Name | Sequence 5′–3′ |
| FL-1-F | GCAGGCGCACTGCTGGCAAGGGTGCTTG |
| FL-1-R | GGAGCCCGTGAGGCTGGACAGGGAAGGCCAT |
| FL-2-F | GCTCACAGGTTTACTGTTGGCACCACCTG |
| FL-2-R | GGACAGCCCAGCCACTGCCTAATGGTC |
| FL-3-F | GACCATCCCTAGCCAACTGTACCTGCAAG |
| FL-3-R | GCTCTTCCCCTGAGGTAAGAGGGGCAGAG |
| FL-4-F | GAGCGGCTCTTGTCAGTTTATTAATGTGC |
| FL-4-R | CAGCAAACTGATGATTAGCTGTAGAGAC |
| FL-5-F | GCTCAGAGTTAGGAGGGTGACTCTTTC |
| FL-5-R | GGCCTGGGAAATAGCAGGAACTACAACAGA |
| FL-6-F | GCCATTTCGTGGTGTTTTGGGTAGCCTGG |
| FL-6-R | GGCACTCTTTCAACCAGTGAGTTCTAGC |
| FL-7-F | GGCTGAACCGGCTGCTGCCACACCTCGT |
| FL-7-R | CCCACAGTTCTGAGACTCTGCCCCTGTGG |
| FL-8/9-F | CTGGCCTTGCTCCCTGGCTCACTGTGTCC |
| FL-8/9-R | GGAGGAAACGTTCCCGTAGGCCTGACTAC |
| FL-10-F | CGGGCAGGCTAGCAAGCTGGGCAGGGAG |
| FL-10-R | CAGCTCCAGAATCCAGCTGGAGAGAGT |
| FL-A-F | CCGTCTGTGCTGCCTGGTCTCCCCTCCCCA |
| FL-A-R | TAGGGTGGGGCTGCATCCAGGTGTTCTTCC |
| FL-B-R | GAGGAACAAGCAGTGGGCGTCTGGGTGGGC |
| FL-B-R | GCCTAATATGATGTCGAGCACACAAAGCTAG |
| FL-C-F | CTTGGTCTTCCAAAGTGTTAGGATTACAAG |
| FL-C-R | TCAGACAACCTGAATTTGAGTCACAGCTCC |
Figure 4. .
Identification of a mutation in the ZFYVE27 gene in a family with AD-HSP. A, Forward sequence of a PCR product of exon 6 of the ZFYVE27 gene in genomic DNA from an affected patient and a control. The mutated nucleotide is indicated by a black arrow; the identified mutation G→T leads to the amino acid substitution G191V in ZFYVE27. B, Pedigree of a five-generation German family with AD-HSP, showing segregation of G191V in affected family members. We could not assert whether individual II.1 was symptomatic, because of limited family history.
The clinical features of the affected members suggest that theirs is a pure form of HSP. The index patient, III.2, had had pes equinus for many years when she noticed a gait disorder at age 50 years. She then manifested hyperreflexia and spasticity of the lower limbs, a positive Babinski sign for an ankle clonus. Sensitivity was normal. Six years ago, she needed walking sticks, some years later a wheelchair was necessary, and now she is bedridden. Her daughter (IV.2), aged 47 years, also had pes equinus for many years. Five years ago, she developed a gait disorder. She also has hyperreflexia and spasticity of the legs, as well as an ankle clonus. The index patient’s son (IV.3) is reported to have pes equinus, spasticity of the legs, and a gait disorder. However, the definite age of onset of the symptoms could not be determined from the family history. Cognitive deficits or additional neurological symptoms are not present in this family. Thus, this SPG form seems to be a pure form of HSP.
ZFYVE27 is located at 10q24.2. In the vicinity of this locus, two SPG loci were previously mapped—namely, SPG9 (MIM 601162), which is located between 10q23.3 and 10q24.2,28 and SPG27 (MIM 609041), which is mapped within 10q22.1-q24.1.29 ZFYVE27 could be excluded as a candidate gene for SPG27 subtype, since ZFYVE27 is located distal to the critical region of the locus. Recent updated mapping of the SPG9 locus (M. Seri, personal communication) also excluded ZFYVE27 from the refined region. Therefore, the ZFYVE27 locus should be considered a new subtype of HSP, which we termed “SPG33.”
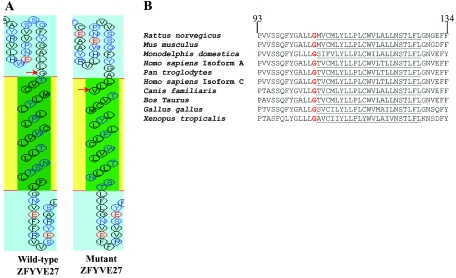
The identified mutation G191V of ZFYVE27 isoform-a (position G105V of isoform-c) is located near the vicinity of the third transmembrane (TM) motif. In silico topology prediction, with the use of SOSUI software, revealed that the G105V mutation shifts the TM motif forward by three amino acids (fig. 5A), which could have a conformational effect in the three-dimensional structure of ZFYVE27, particularly on the FYVE-finger domain. Phylogenetic analysis revealed that the G105 residue of ZFYVE27-c is conserved in most vertebrates (fig. 5B). To study the functional implication of the G105V exchange mutation on the ZFYVE27 protein, we introduced this mutation in the GFP-ZFYVE27 construct and performed in vivo immunoprecipitation assay. We used c-Myc antibody to precipitate spastin from the protein lysate derived from cells cotransfected with c-Myc-spastin and GFP-ZFYVE27(G105V). Immunoblot analysis with GFP antibody detected either no band (fig. 6A, panel i) or, by sample overloading, a very weak band (fig. 6A, panel ii) equivalent to protein size of GFP-ZFYVE27(G105V). The ZFYVE27 interacts with spastin through its C-terminal domain, and the G105V mutation could lead to misfolding of the C-termini of the FYVE-finger domain, which could be consequential to this interaction.
Figure 5. .
In silico analysis of the ZFYVE27 mutation. A, Topological prediction of the mutant ZFYVE27 protein, with use of the SOSUI program, suggesting that the TM motif of mutant ZFYVE27 is shifted forward by three amino acids and is embedded in the membrane, when compared with the wild-type ZFYVE27 TM motif, as shown by the red arrow. B, Phylogenetic analysis using the ClustalW program shows that the G105 (bold red) residue of the ZFYVE27 isoform-c is conserved between most vertebrates. The putative TM motif is underlined.
Figure 6. .
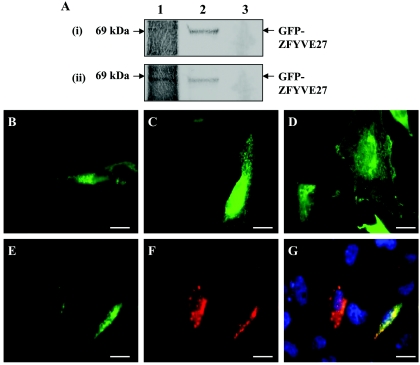
Functional analysis of the ZFYVE27 mutation. A, Immunoprecipitation assay between c-Myc-spastin and GFP-ZFYVE27(G105V), with the use of anti-Myc antibody and detection with anti-GFP antibody. Lane 1, Immunoprecipitated fraction; lane 2, protein standard; lane 3, mock precipitation. No band (i) or a very weak band (ii) corresponding to GFP-ZFYVE27 was detected in the precipitated fraction. B–G, Subcellular localization of GFP-ZFYVE27(G105V) and coexpression with c-Myc-spastin in HeLa cells. B, Transiently transfected HeLa cells showing a diffuse vesicular expression. However, a significant proportion of cells showed expression in tubular structures arising from the perinuclear region. C, GFP-ZFYVE27(G105V) localized to tubular structure in the cytoplasm. D, GFP-ZFYVE27(G105V) showing expression also in the plasma membrane. E–G, Coexpression of c-Myc-spastin and GFP-ZFYVE27(G105V) in HeLa cells showing a diffuse overlapping pattern of expression. E, GFP-ZFYVE27(G105V); F, c-Myc-spastin; G, superimposition of images E and F. Scale bar is 20 μm (B–G).
Next, we investigated the effect of this mutation on the intracellular distribution of ZFYVE27 protein. Analysis of the transfected cells by fluorescence microscopy indicated that, in addition to diffuse vesicular expression (fig. 6B), a significant proportion of cells showed expression in tubular structures or in plasma membrane (fig. 6C–6D). Examination of cells coexpressing GFP-ZFYVE27(G105V) and c-Myc-spastin by immunofluorescence microscopy revealed weak and diffuse colocalization in a subset of vesicles in the cytoplasm (fig. 6E–6G). The observation that a significant proportion of cells showed a tubular expression, presumably in the ER network, suggests that the G105V mutation might hinder or delay the biogenesis process of ZFYVE27.
Our genetic and biochemical studies strongly suggest that the G105V mutation in ZFYVE27 is responsible for the pathological phenotype observed in this family with AD-HSP. It is unclear whether this mutation results in a loss-of-function or a dominant-negative form of ZFYVE27 that is due to misfolding of the C-terminal FYVE-finger domain. Defects in the spastin–dependent/independent endosomal transport may influence the retrograde transmission and, therefore, contribute to a slow accumulation of pathological processes/substrates in HSP. Identification of ZFYVE27 as a mutated gene in HSP, therefore, will aid in the understanding of the axonal neurodegeneration in HSP and suggests that other molecules involved in this process may play a role in the pathogenesis of this group of diseases. Moreover, by elucidating the molecular mechanism and by studying the interacting partner(s) for known HSP proteins, novel genes involved in HSP pathogenesis might be identified. More insight into the pathological mechanisms will help to create new therapeutical options for patients with HSP in the future.
Acknowledgments
The authors thank N. Doerwald, for excellent technical assistance, and all members of the family with HSP, for their willingness to participate in this study. This work was funded by a grant from the medical faculty of the University of Goettingen (to A.U.M.) and by the Deutsche Forschungsgemeinschaft, through the DFG Research Center for Molecular Physiology of the Brain (to W.E.). S.M.S. was supported by a grant from the medical faculty of the University of Goettingen.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Ensembl, http://www.ensembl.org
- ClustalW, http://www.ebi.ac.uk/clustalw
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for spastin-lacking exon 4 [accession number NM_199436], ZFYVE27-c isoform [accession number AK097945], and ZFYVE27-a isoform [accession number NM_001002261])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for spastin, SPG9, and SPG27)
- SOSUI, http://www.expasy.ch
References
- 1.Harding AE (1983) Classification of the hereditary ataxias and paraplegias. Lancet 1:1151–1154 10.1016/S0140-6736(83)92879-9 [DOI] [PubMed] [Google Scholar]
- 2.Reid E (2003) Science in motion: common molecular pathological themes emerge in the hereditary spastic paraplegias. J Med Genet 40:81–86 10.1136/jmg.40.2.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hazan J, Fonknechten N, Mavel D, Paternotte C, Samson D, Artiguenave F, Davoine CS, Cruaud C, Dürr A, Wincker P, Brottier P, Cattolico L, Barbe V, Burgunder JM, Prud’homme JF, Brice A, Fontaine B, Heilig B, Weissenbach J (1999) Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Genet 23:296–303 10.1038/15472 [DOI] [PubMed] [Google Scholar]
- 4.Lindsey JC, Lusher ME, McDermott CJ, White KD, Reid E, Rubinsztein DC, Bashir R, Hazan J, Shaw PJ, Bushby KMD (2000) Mutation analysis of the spastin gene (SPG4) in patients with hereditary spastic paraparesis. J Med Genet 37:759–765 10.1136/jmg.37.10.759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fonknechten N, Mavel D, Byrne P, Davoine CS, Cruaud C, Boentsch D, Samson D, Coutinho P, Hutchinson M, McMonagle P, Burgunder JM, Tartaglione A, Heinzlef O, Feki I, Deufel T, Parfrey N, Brice A, Fontaine B, Prud’homme JF, Weissenbach J, Durr A, Hazan J (2000) Spectrum of SPG4 mutations in autosomal dominant spastic paraplegia. Hum Mol Genet 9:637–644 10.1093/hmg/9.4.637 [DOI] [PubMed] [Google Scholar]
- 6.Sauter S, Miterski B, Klimpe S, Bönsch D, Schöls L, Visbeck A, Papke T, Hopf HC, Engel W, Deufel T, Epplen JT, Neesen J (2002) Mutation analysis of the spastin gene (SPG4) in patients in Germany with autosomal dominant hereditary spastic paraplegia. Hum Mutat 20:127–132 10.1002/humu.10105 [DOI] [PubMed] [Google Scholar]
- 7.Confalonieri F, Duguet M (1995) A 200-amino acid ATPase module in search of a basic function. Bioessays 17:639–650 10.1002/bies.950170710 [DOI] [PubMed] [Google Scholar]
- 8.Frickey T, Lupas AN (2004) Phylogenetic analysis of AAA proteins. J Struct Biol 146:2–10 10.1016/j.jsb.2003.11.020 [DOI] [PubMed] [Google Scholar]
- 9.Errico A, Ballabio A, Rugarli EI (2002) Spastin, the protein mutated in autosomal dominant hereditary spastic paraplegia, is involved in microtubule dynamics. Hum Mol Genet 11:153–163 10.1093/hmg/11.2.153 [DOI] [PubMed] [Google Scholar]
- 10.Trotta N, Orso G, Rossetto MG, Daga A, Broadie K (2004) The hereditary spastic paraplegia gene, spastin, regulates microtubule stability to modulate synaptic structure and function. Curr Biol 14:1135–1147 10.1016/j.cub.2004.06.058 [DOI] [PubMed] [Google Scholar]
- 11.Evans KJ, Gomes ER, Reisenweber SM, Gundersen GG, Lauring BP (2005) Linking axonal degeneration to microtubule remodeling by Spastin-mediated microtubule severing. J Cell Biol 168:599–606 10.1083/jcb.200409058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reid E, Connell J, Edwards TL, Duley S, Brown SE, Sanderson CM (2005) The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum Mol Genet 14:19–38 10.1093/hmg/ddi003 [DOI] [PubMed] [Google Scholar]
- 13.Sanderson CM, Connell JW, Edwards TL, Bright NA, Duley S, Thompson A, Luzio JP, Reid E (2006) Spastin and atlastin, two proteins mutated in autosomal-dominant hereditary spastic paraplegia, are binding partners. Hum Mol Genet 15:307–318 10.1093/hmg/ddi447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciccarelli FD, Proukakis C, Patel H, Cross H, Azam S, Patton MA, Bork P, Crosby AH (2003) The identification of a conserved domain in both spartin and spastin, mutated in hereditary spastic paraplegia. Genomics 81:437–441 10.1016/S0888-7543(03)00011-9 [DOI] [PubMed] [Google Scholar]
- 15.Finken-Eigen M, Rohricht RA, Kohrer K (1997) The VPS4 gene is involved in protein transport out of a yeast pre-vacuolar endosome-like compartment. Curr Genet 31:469–480 10.1007/s002940050232 [DOI] [PubMed] [Google Scholar]
- 16.Yoshimori T, Yamagata F, Yamamoto A, Mizushima N, Kabeya Y, Nara A, Miwako I, Ohashi M, Ohsumi M, Ohsumi Y (2000) The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol Biol Cell 11:747–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi S, Okada T, Igarashi N, Fujita T, Jahangeer S, Nakamura S (2002) Identification and characterization of RPK118, a novel sphingosine kinase-1-binding protein. J Biol Chem 277:33319–33324 10.1074/jbc.M201442200 [DOI] [PubMed] [Google Scholar]
- 18.Phillips SA, Barr VA, Haft DH, Taylor SI, Haft CR (2001) Identification and characterization of SNX15, a novel sorting nexin involved in protein trafficking. J Biol Chem 276:5074–5084 10.1074/jbc.M004671200 [DOI] [PubMed] [Google Scholar]
- 19.Svenson IK, Ashley-Koch AE, Gaskell PC, Riney TJ, Cumming WJ, Kingston HM, Hogan EL, Boustany RM, Vance JM, Nance MA, Pericak-Vance MA, Marchuk DA (2001) Identification and expression analysis of spastin gene mutations in hereditary spastic paraplegia. Am J Hum Genet 68:1077–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stenmark H, Aasland R (1999) FYVE-finger proteins-effectors of an inositol lipid. J Cell Sci 112:4175–4183 [DOI] [PubMed] [Google Scholar]
- 21.Stenmark H, Aasland R, Driscoll PC (2002) The phosphatidylinositol 3-phosphate-binding FYVE finger. FEBS Lett 513:77–84 10.1016/S0014-5793(01)03308-7 [DOI] [PubMed] [Google Scholar]
- 22.Mills IG, Jones AT, Clague MJ (1998) Involvement of the endosomal autoantigen EEA1 in homotypic fusion of early endosomes. Curr Biol 8:881–884 10.1016/S0960-9822(07)00351-X [DOI] [PubMed] [Google Scholar]
- 23.Simonsen A, Lippé R, Christoforidis S, Gaullier J-M, Brech A, Callaghan J, Toh B-H, Murphy C, Zerial M, Stenmark H (1998) EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 394:494–498 10.1038/28879 [DOI] [PubMed] [Google Scholar]
- 24.Raiborg C, Bremnes B, Mehlum A, Gillooly DJ, D’Arrigo A, Stang E, Stenmark H (2001) FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J Cell Sci 114:2255–2263 [DOI] [PubMed] [Google Scholar]
- 25.Van de Velde HJ, Roebroek AJ, Senden NH, Ramaekers FC, Van de Ven WJ (1994) NSP-encoded reticulons, neuroendocrine proteins of a novel gene family associated with membranes of the endoplasmic reticulum. J Cell Sci 107:2403–2416 [DOI] [PubMed] [Google Scholar]
- 26.Senden NH, Timmer ED, Boers JE, van de Velde HJ, Roebroek AJ, Van de Ven WJ, Broers JL, Ramaekers FC (1996) Neuroendocrine-specific protein C (NSP-C): subcellular localization and differential expression in relation to NSP-A. Eur J Cell Biol 69:197–213 [PubMed] [Google Scholar]
- 27.Errico A, Claudiani P, D’Addio M, Rugarli EI (2004) Spastin interacts with the centrosomal protein NA14, and is enriched in the spindle pole, the midbody and the distal axon. Hum Mol Genet 13:2121–232 10.1093/hmg/ddh223 [DOI] [PubMed] [Google Scholar]
- 28.Meijer IA, Cossette P, Roussel J, Benard M, Toupin S, Rouleau GA (2004) A novel locus for pure recessive hereditary spastic paraplegia maps to 10q22.1-10q24.1. Ann Neurol 56:579–582 10.1002/ana.20239 [DOI] [PubMed] [Google Scholar]
- 29.Lo Nigro C, Cusano R, Scaranari M, Cinti R, Forabosco P, Morra VB, De Michele G, Santoro L, Davies S, Hurst J, Devoto M, Ravazzolo R, Seri M (2000) A refined physical and transcriptional map of the SPG9 locus on 10q23.3-q24.2. Eur J Hum Genet 8:777–782 10.1038/sj.ejhg.5200546 [DOI] [PubMed] [Google Scholar]