Abstract
Anopheles darlingi is the most important malaria vector in the Amazon basin of South America, and is capable of transmitting both Plasmodium falciparum and P. vivax. To understand the genetic structure of this vector in the Amazonian region of Peru, a simple polymerase chain reaction (PCR)-based test to identify this species of mosquito was used. A random amplified polymorphic DNA-PCR was used to study genetic variation at the micro-geographic level in nine geographically separate populations of An. darlingi collected in areas with different degrees of deforestation surrounding the city of Iquitos. Within-population genetic diversity in nine populations, as quantified by the expected heterozygosity (HE), ranged from 0.27 to 0.32. Average genetic distance (FST) among these populations was 0.017. These results show that the nine studied populations are highly homogeneous, suggesting that strategies can be developed to combat this malaria vector as a single epidemiologic unit.
INTRODUCTION
Peru has the second highest number of malaria cases in South America. Most of these cases are reported in the Peruvian Amazon, where malaria is a re-emerging disease.1 This high prevalence is attributed to the presence of Anopheles darlingi, a highly competitive and anthropophilic vector that colonized the Amazon Region at the start of the 1990s.2 Anopheles darlingi is the most significant vector of malaria in South America and is capable of transmitting both Plasmodium falciparum and P. vivax.3,4
In tropical zones, anthropogenic activities involving deforestation, such as the construction of highways, agriculture, and fish farming, produce ecologic changes and can alter the community of disease vectors.5,6 For instance, deforestation has caused partial or total substitution of Anopheles species in the Himalayas7 and Colombia.8 In the Peruvian Amazon, the population of An. darlingi has increased over the last 10 years and is now the major vector of malaria.1 Before 1991, An. darlingi was not found in the areas around Iquitos, the major city of the Peruvian jungle.9
The recent spread of An. darlingi in this zone of the Amazon is a public health problem. As such, we need cost-effective methodologies to not only identify this species but to understand the patterns of genetic diversity for this and other vectors. Identification of An. darlingi can be performed either by using specific primers (as was done in this study) or the use of internal transcribed spacers as described by Marrelli and others.10
Genetic diversity of Anopheles has been studied using random amplified polymorphic DNA-polymerase chain reaction (RAPD-PCR),11-15 internal transcribed spacer 2 (ITS2) markers,13,16 microsatellite loci,17 mitochondrial DNA,17,18 single nucleotide polymorphism genotyping,19 and isoenzymatic analysis.13,20 RAPD-PCR is based on the amplification of random DNA using short sequence primers (approximately 10 bases) at low annealing temperatures. It has been used to analyze the genetic variation of An. darlingi13 and other species of mosquitoes21 and as a tool to construct genetic maps.22
In this study, we present a new PCR-based assay that permits a simple and low cost identification of An. darlingi. Additionally, we used RAPD-PCR to analyze the genetic variation of natural populations of An. darlingi collected from areas with different levels of deforestation in Iquitos (Loreto, Peru), in the Amazonian region of Peru.
MATERIALS AND METHODS
Collection and manipulation of Anopheles. Female Anopheles adults were collected near the city of Iquitos (3°45′18″S and 73°14′40″W). Iquitos is the principal city of the Peruvian Amazon and is located on the western bank of the Amazon River in the northeast region of Peru at an altitude of 110-125 meters above sea-level in the department of Loreto, an administrative unit that borders with Ecuador, Colombia, and Brazil. The area in the study is tropical and humid with an annual average temperature that ranges between 24°C and 28°C, an annual relative humidity that varies between 80% and 90%, and an annual average precipitation of 1,500 mm.5 We defined the level of deforestation of the areas sampled according to a previously used classification: rural villages (Vil); shrub areas (Shrub), with young vegetation returning approximately five years after deforestation; secondary forest (SecFor), deforested approximately 15 years before the study; and primary forest (PriFor), referring to closed-canopy, tall forest.2
The areas sampled include five rural villages: Zungarococha, Santa Clara, Varillal, San Gerardo, and El Dorado; two shrub areas: San Gerardo and San Jose; one secondary forest: Monte Calvario; and one primary forest: Tiberias Pintuyacu I. The villages and shrub areas are located along the Iquitos-Nauta highway, which was completed in 2005, and the forests are located to the west of Iquitos (Figure 1). Samples were obtained by human bite collections (previously described) from December 1998 to June 1999 and from September 2000 to August 2001.2 The study was reviewed and approved by the ethics committee of the Johns Hopkins Bloomberg School of Public Health and Asociación Benéfica Prisma.
FIGURE 1.
Map of the study “population” areas’ relation to the city of Iquitos, showing the approximate locations of Anopheles darlingi collections with different levels of deforestation.
Identification of Anopheles darlingi. The adult females of An. darlingi were identified with the diagnostic characteristics described by Linthicum and others,23 individually preserved in 96% alcohol, and stored at -20°C. We developed a PCR-based test to specifically identify An. darlingi individuals. First, we searched Genbank for the An. darlingi sequences showing lowest similarity with other homologous sequences from other Anopheles species. We selected a sequence in the internal spacers of subunits 5.8S and 28S of the genes coding ribosomal RNA (Genbank accession no. AF462389) and designed the primers 5′-CCC GTG TGT GGT CAA GCA TT-3′ and 5′-TTG AGG CCC ACT TGA GAT CC-3′. We then tested the specificity of these primers by performing a PCR on genomic DNA extracted following the same protocol from An. darlingi and the following related species: Aedes aegypti, Culex quinquefasciatus, An. oswaldoi, An. rangeli, An. benarrochi, An. nuneztovari, An. punctimacula, An. albimanus, An. triannulatus, An. mediopunctatus, An. mattogrossensis, and An. forattini. The conditions for the PCRs were the following: 2 mM MgCl2, 0.2 mM dNTPs, 0.5 μM of each primer, 0.5 units of Taq polymerase (Invitrogen, Carlsbad, CA), and 2 ng of genomic DNA in a final volume of 12 μL per tube. The reaction mixture was incubated for one cycle at 94°C for five minutes; 35 cycles at 94°C for one minute, 57°C for one minute, and 72°C for one minute, and at 72°C for five minutes. Electrophoresis was conducted on a 2% agarose gel at 75 V for two hours.
Extraction of genomic DNA. DNA was isolated according to the method of Snounou and others,24 and the quantification of the DNA was carried out by comparison with bacteriophage λ genomic DNA (Invitrogen) of known concentration.25
RAPD-PCR. RAPD analyses were carried out on 270 DNA samples from 9 zones (30 per population) according to the protocol described by Wilkerson and others14 with some modifications, such as the use of Taq (Biolase, Santa Clara, CA); four oligonucleotides: A09, A05, A08 and B04;12 and oligonucleotide 1283, which provided high rates of differentiation in studies on genetic variation of Helicobacter pylori (Table 1).26
TABLE 1.
RAPD-PCR oligonucleotides, their sequences, number of bands, and margins of analysis*
| Oligonucleotides | Sequence (5′ to 3′ ) | No. of polymorphic bands | Margins of analysis (basepairs) |
|---|---|---|---|
| OPA09 | GGGTAACGCC | 15 | 400-1570 |
| OPA08 | GTGACGTAGG | 12 | 436-1100 |
| OPA05 | AGGGGTCTTG | 14 | 387-1433 |
| OPB04 | GGACTGGAGT | 11 | 400-1277 |
| 1283 | GCGATCCCCA | 13 | 434-1249 |
RAPD-PCR = random amplified polymorphic DNA-polymerase chain reaction.
The specific reaction conditions were 2.5 mM MgCl2, 0.3 mM dNTPs, 0.8 μM of each oligonucleotide (Table 1), 1 unit of Taq polymerase (Biolase), and 0.5-1.0 ng of genomic DNA in a final volume of 25 μL per tube. The reaction mixtures were incubated for one cycle at 94°C for five minutes; 44 cycles at (94°C for one minute, 36°C for one minute, and 72°C for two minutes, and 72°C for five minutes. Electrophoresis was conducted on a 2% agarose gel at 60 V for four hours with a 100-basepair molecular mass marker (Invitrogen).
Genetic and statistical analysis. DNA ProScan version 2.39 (DNA Proscan, Inc., Nashville, TN) was used for analysis of polymorphic patterns from the populations of An. darlingi. This program calculates the length of the DNA fragments based on the migration coefficient (RF) normalized to the molecular weight of the 100-basepair molecular mass marker (Invitrogen). The molecular weights of the bands from each polymorphic pattern are stored in a database (and compared with 5% error to new polymorphic patterns). A polymorphic pattern was considered new for An. darlingi when we found a minimum of three different bands for each primer. For the final analysis, a binary matrix was constructed based on the presence or absence of the bands amplified, which considered bands with frequencies within the margins of > 5% and < 96%. Unless differently specified, polymorphic alleles from the RAPD-PCR were analyzed assuming that RAPD-PCR products segregate as dominant traits in a Mendelian fashion, the DNA sequences with equal molecular weight are homologs, the different loci segregate independently, and populations are in Hardy-Weinberg equilibrium.
Within-population genetic diversity was quantified by the method of expected heterozygosity (HE).27 The 95% credibility intervals of HE were calculated using the program Hickory version 1.3.27 Pairwise FST (the between-population component of genetic variance) were estimated using two estimators: Wright’s F-coefficient28 and the θ statistic.29 Assuming an island model of population structure, we used FST to estimate the number of migrants per generation (NM) using the formula FST + 1/(1 + 4NM).30 Genetic distances (FST) and the NM estimators were calculated using the RAPDFST computer program.29 Genetic-distance matrices were graphically summarized by non-metric multidimensional scaling (MDS),31 through use of the software NewMDSX (Sigma Essex Research and Consultancy, Argyll, United Kingdom). MDS uses an iterative process to transform a similarity/dissimilarity matrix into distances represented in a Euclidean n-dimensional space. We assessed the correlation among matrices of genetic and geographic distances by the Mantel test,32,33 as implemented in Arlequin version 2.0 (Schneider S, Roessli D, Excoffier L, Genetics and Biometry Laboratory, University of Geneva, Geneva, Switzerland). To control for the effects of deforestation, we created a matrix of ecologic distances. For this purpose, we coded populations as 0 if they were primary forest, 1 if secondary forest, 2 if shrub area, and 3 if rural village, For each pair of populations, we defined the ecologic distance as the absolute value of the subtraction of their codes. We then calculated a partial correlation between genetic and geographic distance matrices, controlling for the ecologic distances, using the Mantel test as suggested by Smouse and others.34
Analysis of molecular variance (AMOVA) was used to quantify the proportion of the total genetic variance within population, between populations, and among groups of populations (i.e. rural villages, shrub areas, secondary and primary forest), using a framework similar to the analysis of variance adapted for genetic data.35 The AMOVA calculations were performed from the band frequencies using the program Arlequin version 2.0. Significance of the AMOVA results was assessed by a randomization test (10,000 repetitions). To verify that AMOVA calculations were not affected by deviations from Hardy-Weinberg equilibrium, we also used the Bayesian approach developed by Holsinger and others to calculate θβ, an estimator of FST among the nine populations.36 This probabilistic approach uses the full data set and incorporates the uncertainty regarding Hardy-Weinberg equilibrium, while the previously mentioned analyses assume Harding-Weinberg equilibrium.
RESULTS
Population and identification of An. darlingi. Approximately 1,000 mosquitoes were collected and identified morphologically as An. darlingi from the nine study areas: five rural villages, Zungarocoha, Santa Clara, San Gerardo, Varillal, and El Dorado; two shrub areas, San Jose and San Gerardo; one secondary forest, Monte Calvario; and one primary forest, Tiberias Pintuyacu I.2 Thirty mosquitoes were randomly selected from each of the populations, from which DNA was extracted and the species verified by PCR. The result of the PCR with different species of mosquito is shown in Figure 2. The expected result for An. darlingi was 300 basepairs. There was no amplification in the other species of mosquito tested.
FIGURE 2.

Specific polymerase chain reaction products for Anopheles darlingi (300 basepairs [bp]). Lane 1, Culex quinquefasciatus; Lane 2, An. albimanus; Lane 3, An. mattogrossensis; Lane 4, An. mediopuntactus; Lane 5, Aedes aegypti; Lane 6, An. rangeli; Lanes 7, 8, 14, 15, 18, and 19, An. darlingi; Lane 9, An. oswaldoi; Lane 10, 100-bp DNA ladder; Lane 11, An. nuneztovari; Lane 12, An. benarrochi; Lane 13, An. triannulatus; Lane 16, An. forattini; Lane 17, An. puntimacula; Lane 20, negative control.
Descriptive analysis of the RAPD-PCR loci. We identified 65 polymorphic bands with molecular weights in the range of 387-1,570 basepairs in the 270 samples of An. darlingi (Figure 3). Seventy-five percent of the total samples were analyzed twice; none of the repeats differed by three or more bands. The number of polymorphic bands for each type of oligonucleotide is shown in Table 1.
FIGURE 3.
Results of random amplified polymorphic DNA amplification patterns of An. darlingi samples (lanes 2-7) with the oligonucleotides specified in Table 1. Lane 1, 100 basepair DNA ladder, horizontal lines indicate the limits of analysis (negative control not shown).
Within-population genetic diversity. Within-population genetic diversity was low (Table 2). This genetic diversity was quantified by the average expected heterozygosity across the 65 loci (for each locus, HE + 1-p2, where p is the frequency of the presence of the band). The average values across the 65 RAPD loci varied from 0.27 to 0.32. In general, there was no association among the environment of mosquito populations and level of within-population variability.
TABLE 2.
Expected Heterozygosity (He) values of the “populations” of Anopheles darlingi
| Population | Expected heterozygosity | 95% confidence intervals |
|---|---|---|
| Vil-Zungarococha | 0.28 | 0.27-0.30 |
| Vil-Santa Clara | 0.27 | 0.27-0.30 |
| Vil-San Gerardo | 0.29 | 0.28-0.30 |
| Shrub-San Jose | 0.29 | 0.28-0.30 |
| Vil-Varillal | 0.30 | 0.28-0.31 |
| SecFor-Monte Calvario | 0.28 | 0.28-0.30 |
| Vil-El Dorado | 0.30 | 0.28-0.31 |
| Shrub-San Gerardo | 0.28 | 0.28-0.30 |
| PriFor-Tiberias Pintuyacu I | 0.29 | 0.28-0.30 |
| All populations | 0.30 | 0.29-0.30 |
Between-population genetic diversity. The correlation between the Wright’s FST (Table 3) and the θ matrices of genetic distances was very high (R2 + 0.95, P < 0.0001, by Mantel test). Therefore, hereafter we used the Wright’sFST estimator to compare genetic diversity between populations. The average genetic distance among the nine populations was 0.017. In general, genetic distances were very low. For each population, Table 3 also shows, across the diagonal, the average genetic distance with the other eight populations.
TABLE 3.
Estimation of FST statistics (above the diagonal) in population of Anopheles darlingi from nine areas around Iquitos*
| Population | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| 1 Vil-Zungarococha | 0.017 | 0.019 | 0.021 | 0.018 | 0.020 | 0.017 | 0.010 | 0.017 | 0.014 |
| 2 Vil-Santa Clara | 12.8 | 0.019 | 0.011 | 0.016 | 0.022 | 0.021 | 0.018 | 0.022 | 0.023 |
| 3 Vil-San Gerardo | 11.6 | 22.2 | 0.018 | 0.013 | 0.026 | 0.030 | 0.015 | 0.019 | 0.011 |
| 4 Shrub-San Jose | 13.7 | 15.5 | 18.6 | 0.015 | 0.022 | 0.015 | 0.011 | 0.010 | 0.017 |
| 5 Vil-Varillal | 12.0 | 11.0 | 9.2 | 11.4 | 0.019 | 0.015 | 0.017 | 0.017 | 0.016 |
| 6 SecFor-Monte Calvario | 14.6 | 11.5 | 8.0 | 16.1 | 16.8 | 0.018 | 0.008 | 0.018 | 0.019 |
| 7 Vil-El Dorado | 24.6 | 13.5 | 16.5 | 21.7 | 14.2 | 29.7 | 0.013 | 0.013 | 0.014 |
| 8 Shrub-San Gerardo | 14.8 | 11.0 | 13.2 | 24.4 | 14.2 | 13.4 | 18.3 | 0.016 | 0.011 |
| 9 PriFor-Tiberias Pintuyacu I | 17.8 | 10.6 | 22.7 | 14.4 | 14.9 | 13.2 | 18.3 | 22.7 | 0.016 |
NM values (the number of migrants per generation) are below the diagonal and the average genetic distance are on the diagonal.
Assuming an island model of population structure, we estimated NM (i.e., the number of migrants per generation) from the FST values (Table 3). All NM values are greater than 8, which suggests that gene flow, rather than genetic drift, is the predominant evolutionary force shaping the genetic structure of these mosquito populations.37 Alternatively, all populations studied derived from a unique ancestral population that has expanded very recently, without time to differentiate.
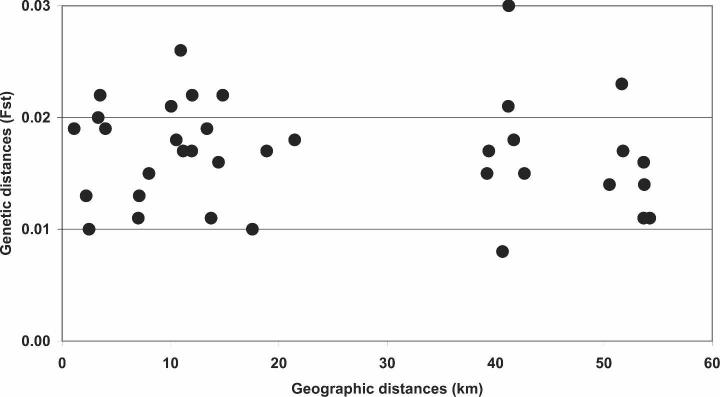
The MDS of the FST genetic distance matrix is shown in Figure 4. Consistent with the genetic distance matrix, the populations of Vil-Varillal and Vil-Santa Clara, which show the largest average genetic distances with the other populations (0.19 in both cases), are the more peripheral in the graphic. El Dorado and San Jose, which show the smallest average genetic distances (0.13 and 0.15, respectively), are central in the graphic. In general, the distribution of the populations in the MDS does not match their geographic distances. The scatterplot of genetic distances (FST) against geographic distances (Figure 5) confirms this result. In this figure, two sets of observations are evident. On the left of the scatterplot, a set of points represents comparisons among geographically close populations (i.e, those rural villages and shrub areas located along the Iquitos-Nauta highway near Iquitos) and between SecFor-Monte Calvario and PriFor-Tiberias Pintuyacu I. On the right of the scatterplot, we observe comparisons among each of the populations along the highway and the distant forest populations, and it is clear that the genetic distances for the former, geographically close populations are not smaller than for the latter, geographically distant populations. The Mantel test does not show any correlation between the FST and geographic distances (R2 = -0.06, P = 0.62), even when the effect of ecologic distances was controlled (partial correlation = 0.01, P = 0.63). These results suggest that, despite the different ecological environments, at the micro-geographic level of this study, there is high homogeneity among populations.
FIGURE 4.
Synthetic representation of the FST genetic distance matrix, obtained by the non-metric Multidimensional Scaling (MDS).
FIGURE 5.
Scatter plot of genetic distances (FST) on geographic distance in kilometers.
We also performed a hierarchical analysis of variance developed for molecular data (AMOVA), which confirms the very low level of differentiation among the nine populations (FST = 0.015, P < 0.0001). The differentiation among populations within the ecologic groups (rural villages and shrub areas) is also low (FSC = 0.017, P < 0.0001), but no significant differentiation was observed among the ecologic groups (rural villages, shrub areas, secondary forest, and primary forest; FCT = 0.000, P = 0.84).
DISCUSSION
The migration of humans into previously unpopulated areas of Iquitos and the subsequent deforestation have been associated with a new distribution of An. darlingi in these areas.2 In the more rural areas of Iquitos, An. darlingi was notable for its absence in surveys prior to 1991, but it is now the most important malaria vector. We aimed to determine whether there was an association between the variability of invading An. darlingi populations and habitat. Particularly, we tested the hypothesis that mosquito populations in recently cleared forest habitats were more uniform than populations in secondary and primary forest. Using RAPD-PCR, we found no significant population structure and, therefore, no evidence that cleared forest has facilitated the differentiation of any specific An. darlingi subpopulation.
This study also used newly developed primers to identity An. darlingi to confirm that microscopy correctly identified this mosquito species. The identities of 270 adult females of the species An. darlingi, as determined by morphologic analysis, were reconfirmed with a PCR test. The PCR had a 100% sensitivity and specificity compared with the morphologic analysis, which is the gold standard for species identification. The availability of our highly efficient and cheap test, based on a single PCR reaction, permits the identification of An. darlingi individuals even when specimens are damaged.
The design of the reported primers was based on the sequence of the ribosomal DNA regions 5.8S and 28S (between which the ITS2 are encountered). We confirmed that our primers were good candidates for the identification of An. darlingi. Marrelli and others used this same region for the identification of multiple anopheline mosquito species from the Brazilian Amazon from groups of morphologically similar species.10 Also, Garros and others demonstrated that the ITS2 fragment is able to successfully discriminate between different Anopheles species. This assay has the advantage of permitting the identification of at least 13 Anopheles species within the subgenus Cellia from Africa and Asia;38 however, it cannot be done like our assay, as a simple PCR test.
In this study, the RAPD-PCR identified 65 RAPD loci, a comparable number to those identified in previous studies: (46-65).10,12,19,39 The within-population diversity (i.e., expected average heterozygosity across loci) observed in our samples was comparable to previous studies on various Anopheles species in different South and Central American populations.17 Using various molecular markers, other investigators found similar values of heterozygosity. In three populations of An. nuneztovari from Colombia, the within-population diversity was 0.343,11 and for the same species in four populations from Brazil and two from Colombia the within-population diversity ranged from 0.087 to 0.116.21 In Brazil, a within-population diversity for An. darlingi of 0.171 was found,20 and in the same species in seven countries of Central and South America a range of 0.063 to 0.122 was found.13
The level of differentiation among populations in the studied area was very low (θβ, an FST estimator, is 0.0125). In particular, there was no evidence of a higher differentiation among the cluster of populations of deforested areas along the Iquitos-Nauta highway and An. darlingi populations from the primary forest (Tiberias Pintuyacu I) and the secondary forest of Monte Calvario, which are located at more than 50 km with respect to the Iquitos-Nauta highway. These results suggest that enough gene flow exists among these populations to maintain homogeneity across the area. These results were consistent with other studies in different parts of the world that did not show a strong population structure among mosquitoes populations for different species, even at distances as great as 100 and 200 km.39,40
Posso and others11 evaluated the degree of genetic differentiation between populations of An. nuneztovari in Colombia using RAPD-PCR.11 They found that populations separated by 325 km show higher migration rates than populations separated by only 250 km and divided by a geographic barrier. At a continental level, Maguin and others13 found that RAPD-PCR isolated populations of An. darlingi geographically and detected a small genetic distance between them, whereas ITS2 grouped all of them together as identical. Malafronte and others also found by means of ITS2 that the populations of An. darlingi from different states of Brazil were identical.16 However, Conn and others demonstrated that between the populations of An. darlingi from different countries of South America there exists a significant correlation between the genetic and geographic distances, and this is reflected in the different phenotypes.18
If the studied populations of An. darlingi maintain a high level of gene flow across an area such as that considered in this study, it implies that genetic effects of human intervention for the control of mosquito populations will spread across areas encompassing deforested environments as well as the adjacent forest. For instance, if use of insecticides in villages raises frequencies of alleles conferring insecticide resistance, our study suggests that these alleles can spread even to the near (i.e. 50-60 km) primary and secondary forest. Conversely, if populations are highly homogeneous, they likely show the same level of genetic susceptibility to insecticides, and the same eradication program can be applied to different villages and the surrounding forested areas.
In this study, we showed high homogeneity among nine populations of An. darlingi separated by 20-60 km located in different habitats from the areas surrounding the city of Iquitos. This result suggests implementation of strategies to combat these populations because one epidemiologic unit would permit the control of the principal malaria vector in this study area. In this context, this study is a contribution to malaria programs that are striving to eradicate the vector in these areas.
Acknowledgments
Acknowledgments: We appreciate the helpful work of Drs. Jan E. Conn (Wadsworth Center, New York State Department of Health, Slingerlands, NY) and Ranulfo Gonzáles (Department of Biology, Valle University, Cali, Colombia). We also thank Dr. Gregory J. Devine (Rothamsted Research, Harpenden, United Kingdom) for his helpful comments and Lizardo Fachin (Unidad de Información Geográfica y Teledetección-IIAP, Iquitos) for designing the map. We are grateful to numerous collaborators for assistance in collecting samples for this research, particularly the field team in Peru.
Footnotes
Financial support: This study was supported by the Doris Duke Charitable Foundation, Infectious Diseases Training Program, Charitable RG-ER funds (3D43TW006581); and the Fogarty Global Research and Tutorial in Tropical Health at Johns Hopkins University/Peru Overseas (T35AI07646 National Institutes of Health/National Institute of Allergy and Infectious Diseases).
Disclaimer: The opinions and assertions made by the authors do not reflect the official position of any organization listed.
Disclosure: All authors declare that they have no conflict of interest in relation to this work.
Authors’ addresses: Viviana Vanessa Pinedo-Cancino, Patricia Sheen, and Cesar Jeri, Laboratory of Infectious Diseases, Department of Microbiology, Faculty of Sciences, Peruvian University Cayetano Heredia, Lima, Peru Avenida Honorio Delgado No. 430 Urbanización Ingeniería, San Martín de Porras, Peru, Telephone: 1-483-2942, Fax: 1-483-2942. Eduardo Tarazona-Santos, Section of Genomic Variation, Pediatric Oncology Branch, National Cancer Institute; Department of General Biology, Federal University of Minas Gerais, Belo Horizonte, Brazil Av. Antônio Carlos, 6627, Caixa-Postal: 486 Pampulha, 31270910, Belo Horizonte, Minas Gerais, Brazil. William E. Oswald, Biomedical Research Unit, Asociación Benéfica PRISMA (Projects in Informatics, Health, Medicine and Agriculture), Lima, Peru Avenida Carlos Gonzales 251, Urbanización Maranga, San Miguel, Peru, Telephone: 1-464-0221, Fax: 1-464-0781. Amy Yomiko Vittor, School of Medicine, Stanford University, Stanford, CA 94305. Jonathan A. Patz, Nelson Institute for Environmental Studies, University of Wisconsin, 1710 University Avenue, Madison, WI 53726, Telephone: 608-262-4775, Fax: 608-265-4113. Robert H. Gilman, Department of Molecular Microbiology and Immunology, Johns Hopkins Bloomberg School of Public Health, 615 North Wolfe Street, Room 5515, Baltimore, MD 21205, Telephone: 410-614-3959, E-mail: rgilman@jhsph.edu.
REFERENCES
- 1.Aramburu Guarda J, Ramal Asayag C, Witzig R. Malaria reemergence in the Peruvian Amazon region. Emerg Infect Dis. 1999;5:209–215. doi: 10.3201/eid0502.990204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vittor AY, Gilman RH, Tielsch J, Glass G, Shields T, Sanchez Lozano W, Pinedo Cancino V, Patz JA. The effect of deforestation on the human-biting rate of Anopheles darlingi, the primary vector of falciparum malaria in the Peruvian Amazon. Am J Trop Med Hyg. 2006;74:3–11. [PubMed] [Google Scholar]
- 3.Lourenço de Oliveira R, Da Gama A, Arle M, Fernández da Silva T. Anopheline species, some of their habits and relation to malaria in endemic areas of Rondonia state, Amazon region of Brazil. Mem Inst Oswaldo Cruz. 1989;84:501–514. doi: 10.1590/s0074-02761989000400008. [DOI] [PubMed] [Google Scholar]
- 4.Oliveira FJ, De Oliveira R, Teva A, Deane L, Ribeiro C. Natural infections of Anophelines in Rondonia State, Brazilian Amazon. Am J Trop Med Hyg. 1990;43:6–10. [PubMed] [Google Scholar]
- 5.Gómez-Romero E, Tamariz-Ortiz T. Uso de la tierra y patrones de deforestación en la zona de Iquitos. In: Kalliola R, Flores-Paitan S, editors. Geoecologia y Desarrollo Amazónico. Finnreklama Oy; Sulkava: 1998. [Google Scholar]
- 6.Harlem G. Informe Sobre las Enfermedades Infecciosas, Datos Epidemiológicos 1998-1999. http://www.who.int/infectious-disease-report/idr99-spanish/index .htm. Organización Mundial de la Salud. 2000 Accessed February 9, 2006.
- 7.Sharma V, Malhotra M, Mani T. Facets of Environmental Problems, Five Case Studies. Indian National Science Academy Press; New Delhi: 1984. Entomological and epidemiological study of malaria in Terai; p. 198. [Google Scholar]
- 8.Kroeger A, Mancheno M, Peese K. Métodos para Mejorar el Control de la Malaria en Ecuador y Colombia. Abya Yala Press; Quito: 1995. p. 230. [Google Scholar]
- 9.Need J, Rogers E, Phillips I. Mosquitoes (Diptera: Culicidae) captured in the Iquitos area of Peru. J Med Entomol. 1993;30:634–638. doi: 10.1093/jmedent/30.3.634. [DOI] [PubMed] [Google Scholar]
- 10.Marrelli MT, Floeter-Winter LM, Malafronte RS, Tadei WP, Lourenço de Oliveira R, Flores-Mendoza C, Marinotti O. Amazonian malaria vector anopheline relationships interpreted from ITS2 rDNA sequences. Med Vet Entomol. 2005;19:208–218. doi: 10.1111/j.0269-283X.2005.00558.x. [DOI] [PubMed] [Google Scholar]
- 11.Posso C, Gonzalez R, Cardenas H, Gallego G, Duque M, Suarez M. Random amplified polymorphic DNA analysis of Anopheles nuneztovari (Diptera: Culicidae) from western and northeastern Colombia. Mem Inst Oswaldo Cruz. 2003;98:469–476. doi: 10.1590/s0074-02762003000400007. [DOI] [PubMed] [Google Scholar]
- 12.Lounibos L, Wilkerson R, Conn J, Hribar L, Fritz G, Danoff-Burg J. Morphological, molecular, and chromosomal discrimination of cryptic Anopheles (Nysorhynchus) (Diptera: Culicidae) from South America. J Med Entomol. 1998;35:830–838. doi: 10.1093/jmedent/35.5.830. [DOI] [PubMed] [Google Scholar]
- 13.Manguin S, Wilkerson RC, Conn JE, Rubio-Palis Y, Danoff-Burg JA, Roberts DR. Population structure of the primary malaria vector in South America, Anopheles darlingi, using isozyme, random amplified polymorphic DNA, internal transcribed spacer 2, and morphologic markers. Am J Trop Med Hyg. 1999;60:364–376. doi: 10.4269/ajtmh.1999.60.364. [DOI] [PubMed] [Google Scholar]
- 14.Wilkerson RC, Parsons TJ, Klein TA, Gaffigan TV, Bergo E, Consolim J. Diagnosis by random amplified polymorphic DNA polymerase chain reaction of four cryptic species related to Anopheles (Nyssorhynchus) albitarsis (Diptera: Culicidae) from Paraguay, Argentina, and Brazil. J Med Entomol. 1995;32:697–704. doi: 10.1093/jmedent/32.5.697. [DOI] [PubMed] [Google Scholar]
- 15.Sucharit S, Komalamisra N. Differentiation of Anopheles minimus species complex by RAPD-PCR, technique. J Med Assoc Thai. 1997;80:598–602. [PubMed] [Google Scholar]
- 16.Malafronte RS, Marrelli MT, Marinotti O. Analysis of ITS2 DNA sequences from Brazilian Anopheles darlingi (Diptera: Culicidae) J Med Entomol. 1999;36:631–634. doi: 10.1093/jmedent/36.5.631. [DOI] [PubMed] [Google Scholar]
- 17.Molina-Cruz AP, De Merida AM, Mills K, Rodriguez F, Schoua C, Yurrita MM, Molina E, Palmieri M, Black WC., IV Gene flow among Anopheles albimanus populations in Central America, South America, and the Caribbean assessed by microsatellites and mitochondrial DNA. Am J Trop Med Hyg. 2004;71:350–359. [PubMed] [Google Scholar]
- 18.Conn JE, Rosa-Freitas MG, Luz SL, Momen H. Molecular population genetics of the primary neotropical malaria vector Anopheles darlingi using mtDNA. J Am Mosq Control Assoc. 1999;15:468–474. [PubMed] [Google Scholar]
- 19.Morlais I, Ponçon N, Simard F, Cohuet A, Fontenille D. Intraspecific nucleotide variation in Anopheles gambiae: New insights into the biology of malaria vectors. Am J Trop Med Hyg. 2004;71:795–802. [PubMed] [Google Scholar]
- 20.Dos Santos J, Maia J, Tadei W, Rodriguez G. Izoenzimatic variability among five Anopheles species belonging to the Nysorhynchus and Anopheles subgenera of the Amazon region, Brazil. Mem Inst Oswaldo Cruz. 2003;98:247–253. doi: 10.1590/s0074-02762003000200014. [DOI] [PubMed] [Google Scholar]
- 21.Scarpassa VM, Tadei WP, Suarez MF. Population structure and genetic divergence in de Anopheles nuneztovari (Diptera: Culicidae) from Brazil and Colombia. Am J Trop Med Hyg. 1999;60:1010–1018. doi: 10.4269/ajtmh.1999.60.1010. [DOI] [PubMed] [Google Scholar]
- 22.Mutebi JP, Black WC, IV, Bosio CF, Sweeney WP, Craig GB. Linkage map for the Asian tiger mosquito Aedes (Stegomyia) albopictus based on SSCP analysis of RAPD markers. J Hered. 1997;88:489–494. doi: 10.1093/oxfordjournals.jhered.a023142. [DOI] [PubMed] [Google Scholar]
- 23.Linthicum KJ. A revision of the Argyritarsis section of the subgenus Nyssorynchus of Anopheles (Diptera: Culicidae) Mosq Syst. 1988;20:98–271. [Google Scholar]
- 24.Snounou G, Pinheiro L, Goncalves A, Fonseca L, Dias F, Brown K, Do Rosario V. The importance of sensitive detection of malaria parasites in the human and insect hosts in epidemiological studies, as shown by the analysis of field samples from Guinea-Bissau. Trans R Soc Trop Med Hyg. 1993;87:649–653. doi: 10.1016/0035-9203(93)90274-t. [DOI] [PubMed] [Google Scholar]
- 25.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 26.Soto G, Bautista CT, Roth DE, Gilman RH, Velapatino B, Ogura M, Dailide G, Razuri M, Meza R, Katz U, Monath TP, Berg DE, Taylor DN. Helicobacter pylori reinfection is common in Peruvian adults after antibiotic eradication therapy. J Infect Dis. 2003;188:1263–1275. doi: 10.1086/379046. [DOI] [PubMed] [Google Scholar]
- 27.Holsinger KE, Lewis PO, Dey DK. A Bayesian approach to inferring population structure from dominant markers. Mol Ecol. 2002;11:1157–1164. doi: 10.1046/j.1365-294x.2002.01512.x. [DOI] [PubMed] [Google Scholar]
- 28.Wright S. The genetical structure of populations. Ann Eugen. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 29.Black WC., IV . Statistical analysis of arbitrary primed PCR patterns in molecular taxonomic studies. In: Clapp JP, editor. Methods in Molecular Biology: Species Diagnostic Protocol PCR and other Nucleic Acid Methods. Humana Press; Totowa, NJ: 1995. pp. 39–55. [DOI] [PubMed] [Google Scholar]
- 30.Nei M. Molecular Evolutionary Genetics. Columbia University Press; New York: 1987. [Google Scholar]
- 31.Kruskal J. Nonmetric multidimensional scaling: a numerical method. Psychometrika. 1964;29:28–42. [Google Scholar]
- 32.Mantel NA. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- 33.Sokal RR, Rohlf FJ. Biometry. W. H .Freeman and Company Press; New York: 1995. [Google Scholar]
- 34.Smouse PE, Long JC, Sokal RR. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst Zool. 1986;35:627–632. [Google Scholar]
- 35.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction sites. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holsinger KE, Wallace LE. Bayesian approaches for the analysis of population genetic structure: an example from Platanthera leucophaea (Orchidaceae) Mol Ecol. 2004;13:887–894. doi: 10.1111/j.1365-294x.2004.02052.x. [DOI] [PubMed] [Google Scholar]
- 37.Hartl DL, Clark AG. Principles in Population Genetics. Sinauer Associates, Inc; Sunderland, MA: [Google Scholar]
- 38.Garros C, Koekemoer L, Kamau L, Awolola S, Van Bortel W, Coetzee M, Coosemans M, Manguin S. Restriction fragment length polymorphism method for the identification of major African and Asian malaria vectors within the Anopheles funestus and An. minimus groups. Am J Trop Med Hyg. 2004;70:260–265. [PubMed] [Google Scholar]
- 39.Temu EA, Hunt RH, Coetzee M. Microsatellite DNA polymorphism and heterozygosity in the malaria vector mosquito Anopheles funestus (Diptera: Culicidae) in east and southern Africa. Acta Trop. 2004;90:39–49. doi: 10.1016/j.actatropica.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 40.Gorrochotegui-Escalante N, Gomez-Machorro C, Lozano-Fuentes S, Fernandez-Salas L, De Lourdes Munoz M, Farfan-Ale JA, Garcia-Rejon J, Beaty BJ, Black WC., IV Breeding structure of Aedes aegypti populations in Mexico varies by region. Am J Trop Med Hyg. 2002;66:213–222. doi: 10.4269/ajtmh.2002.66.213. [DOI] [PubMed] [Google Scholar]