Abstract
Elevated levels of circulating low-density lipoprotein cholesterol (LDL-C) play a central role in the development of atherosclerosis. Mutations in proprotein convertase subtilisin/kexin type 9 (PCSK9) that are associated with lower plasma levels of LDL-C confer protection from coronary heart disease. Here, we show that four severe loss-of-function mutations prevent the secretion of PCSK9 by disrupting synthesis or trafficking of the protein. In contrast to recombinant wild-type PCSK9, which was secreted from cells into the medium within 2 hours, the severe loss-of-function mutations in PCSK9 largely abolished PCSK9 secretion. This finding predicted that circulating levels of PCSK9 would be lower in individuals with the loss-of-function mutations. Immunoprecipitation and immunoblotting of plasma for PCSK9 provided direct evidence that the serine protease is present in the circulation and identified the first known individual who has no immunodetectable circulating PCSK9. This healthy, fertile college graduate, who was a compound heterozygote for two inactivating mutations in PCSK9, had a strikingly low plasma level of LDL-C (14 mg/dL). The very low plasma level of LDL-C and apparent good health of this individual demonstrate that PCSK9 plays a major role in determining plasma levels of LDL-C and provides an attractive target for LDL-lowering therapy.
In 2003, Abifadel and colleagues1 reported that selected missense mutations in PCSK9 (proprotein convertase subtilisin/kexin type 9 [MIM 607786]), which encodes a cholesterol-regulated proprotein convertase,2,3 cause a new form of autosomal dominant hypercholesterolemia (MIM 603776). This discovery revealed a previously unrecognized mechanism that strongly influences the level of low-density lipoprotein cholesterol (LDL-C) in the circulation. PCSK9 comprises a signal sequence, a prodomain, a catalytic domain, and a cysteine-rich C-terminal domain (fig. 1A).7 Like other proprotein convertases, PCSK9 undergoes autocatalytic cleavage in the endoplasmic reticulum (ER) at residue 152, between the prodomain and the catalytic domain.8 The prodomain and the catalytic domain remain tightly associated and are secreted together from cultured cells.7,9 High-level expression of wild-type PCSK9 (PCSK9-WT) in the liver of mice results in a pronounced reduction in hepatic low-density lipoprotein receptor (LDLR) protein (but not mRNA).10,11 Since hepatic LDLR-mediated endocytosis is the major route of LDL clearance,12 the mice expressing high levels of PCSK9 in the liver are severely hypercholesterolemic. In contrast, mice expressing no PCSK9 have accelerated LDL clearance.13 These findings indicate that PCSK9 acts to limit the number of LDLRs at the cell surface. Thus, the PCSK9 mutations associated with hypercholesterolemia are presumably gain-of-function mutations.
Figure 1. .
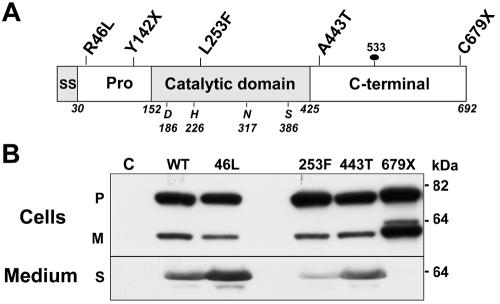
Effects of loss-of-function mutations on the synthesis and secretion of PCSK9. A, PCSK9, a protein of 692 aa that contains a signal sequence (SS), a 122-aa prodomain (Pro), a catalytic domain, and a C-terminal domain. The locations of the catalytic triad (D186, H226, and S386), oxyanion hole residue (N317), site of attachment of the N-linked sugar (533), and loss-of-function mutations4–6 are shown. B, Expression of recombinant PCSK9 in HEK-293 cells. Wild-type (WT) and mutant forms of PCSK9 were expressed in HEK-293 cells, and immunoblotting was performed on the cells and medium with use of an anti-FLAG M2 mAb, as described in the “Material and Methods” section. This experiment was repeated three times, with similar results. Lane C = control; P = precursor; M = mature; S = secreted.
Whereas gain-of-function mutations in PCSK9 in humans are apparently rare, a spectrum of more-frequent loss-of-function mutations associated with low LDL-C levels have been identified.4–6 Elsewhere, we demonstrated that 2%–2.6% of African Americans are heterozygous for one of two nonsense mutations in PCSK9 (Y142X and C679X).4,14 These mutations are associated with a 30%–40% reduction in plasma levels of LDL-C and an 88% reduction in coronary heart disease over a 15-year period.4,14 Other amino acid substitutions in PCSK9 reproducibly associated with significant reductions in plasma levels of LDL-C include R46L, L253F, and A443T; individuals heterozygous for these sequence variations have a 15%, 30%, and 2% reduction in plasma levels of LDL-C, respectively5,6 (fig. 1A). The effects of PCSK9 mutations on plasma levels of LDL-C and coronary heart disease suggest that PCSK9 is a major determinant of plasma levels of LDL-C and may be an attractive target for cholesterol-lowering therapy. However, the mechanism(s) by which these mutations affect PCSK9 function has not been fully defined. High-level expression of PCSK9 in cultured hepatocytes resulted in degradation of the LDLR in a post-ER compartment,15 but evidence supporting an extracellular effect of PCSK9 on LDLR number has also been reported.16 Furthermore, the phenotypic effects of total deficiency of PCSK9 have not been determined: to date, only heterozygotes for the severe loss-of-function mutations have been described. Here, we examined the effect of loss-of-function mutations on the synthesis and secretion of PCSK9. We found that the three mutations associated with the greatest reductions in plasma levels of LDL-C interfered with either the synthesis or the secretion of PCSK9. On the basis of these findings, we predicted that PCSK9 circulates in plasma and that individuals with two inactivating mutations in PCSK9 would have no circulating PCSK9. Immunoprecipitation and immunoblotting of plasma from family members of probands with PCSK9 mutations confirmed that the serine protease is present in the circulation and identified the first known individual with no immunodetectable circulating PCSK9.
Material and Methods
Material
Rabbit polyclonal antibodies against full-length recombinant human PCSK9 (6389) and the catalytic domain of human PCSK9 (295A) were generated and purified. A polyclonal antibody IgG purified from serum of a nonimmune rabbit was provided by Russell DeBose-Boyd (UT Southwestern). Monoclonal antibody (15A6) was generated by fusion of Sp2/mIL-6 (ATCC catalog number CRL-2016) mouse myeloma cells with splenic B-lymphocytes derived from a female BALB/c mouse that was injected with full-length human PCSK9 protein by use of techniques described elsewhere.17 The antibody belongs to the IgG subclass 1 and recognizes epitopes in the C-terminal region of PCSK9. Mouse anti-FLAG M2 monoclonal antibody was purchased from Sigma. Unless otherwise specified, all other reagents were obtained from Sigma.
Expression Constructs for PCSK9-WT and Mutant Forms of PCSK9
An expression vector encoding amino acids 1–692 of human PCSK9 followed by a FLAG epitope tag (DYKDDDDK) under the control of the cytomegalovirus promoter-enhancer (pCMV-PCSK9-FLAG) was constructed in pcDNA3.1. Constructs expressing the truncation mutations (Y142X and C679X) were generated using the following primers: 5′ primer (5′-GCTCTAGAATGGGCACCGTCAGCTCCAG-3′) and 3′ primer (5′-GCGAAGCTTCTAGTCGACATGGGGCAACT-3′) for Y142X and 3′ primer (5′-GCGAAGCTTTCAGCAGATGGCAACGGCTG-3′) for C679X. The 5′ and 3′ primers contained XbaI and HindIII restriction sites, respectively. The PCR-amplified cDNA was then digested with XbaI and HindIII before ligation into pACCMVpLpA(1)loxP-SSP (provided by Robert Gerard [UT Southwestern]).
Single base-pair changes were introduced into the human PCSK9 cDNA expression constructs by use of QuikChange (Stratagene) as described elsewhere.18 Sequences of the oligonucleotides used to generate the expression constructs are available on request. The presence of the desired mutation and the fidelity of each construct were confirmed by DNA sequencing.
Transfections and Cell Lysates
Human embryonic kidney (HEK)–293 cells were seeded (1×105 cells/well) in 6-well plates. Expression plasmids (1 μg/well) were transiently transfected into HEK-293 cells with Fugene (6 μl of Fugene/μg of DNA) according to the manufacturer’s protocol (Roche Diagnostics). After a 48-h transfection, the medium was replaced with supplemental medium (Dulbecco's modified Eagle medium with 1% insulin-transferrin-selenium [Cellgro; Mediatech]). After 5 h, the medium was collected, and the protein was precipitated with trichloroacetic acid. Cells were washed twice in PBS (pH 7.4), were incubated in 0.2 ml of Triton lysis buffer (50 mM Tris, 80 mM NaCl, 2 mM CaCl2, 1% [v/v] Triton X-100, and complete mini EDTA-free protease inhibitor cocktail [Roche]) for 30 min at 4°C, and were harvested by scraping with a rubber policeman. Cells and lysis buffer were transferred to 1.5-ml tubes and were centrifuged for 15 min (15,000 g at 4°C). Supernatants were subjected to SDS-PAGE and immunoblot analysis.
Analysis of Glycosylation
Cell lysates and medium were collected from HEK-293 cells 48 h after transfection with PCSK9-expression constructs. The lysates and medium were denatured by heating (at 95°C for 10 min) in 0.5% SDS and 1 mM β-mercaptoethanol (β-ME) and were incubated overnight in the presence or absence of 10 units of peptide—N-glycosidase F (PNGaseF) or Endo H (New England Biolabs)—according to the manufacturer’s protocol. Samples were then diluted with sample loading buffer, were heated to 95°C for 5 min, were subjected to SDS-PAGE at 20 V for 20 h, and were immunoblotted using an anti-FLAG M2 monoclonal antibody (mAb).
SDS-PAGE and Immunoblot Analysis of PCSK9
Protein concentrations were determined using the Bio-Rad bicinchoninic acid assay, according to the manufacturer’s protocol. Sample loading buffer was added to a final concentration of 1×, and samples were heated to 95°C for 5 min. Proteins were size-fractionated on 8% or 10% SDS-polyacrylamide gels at 100 V and were subsequently transferred to nitrocellulose membranes at 100 V for 1 h. Membranes were incubated in PBST buffer (1× PBS [pH 7.4] and 0.5% Tween 20) with 5% dry milk for 60 min at room temperature (RT) before addition of the primary antibodies. Primary antibodies were diluted in PBST buffer with 5% dry milk and were incubated with membranes for 60 min. Membranes were washed three times for 5 min in PBST buffer. Horseradish peroxidase–conjugated donkey anti-rabbit IgG or goat anti-mouse IgG (Pierce) was diluted (1:10,000) in PBST buffer with 5% dry milk and was incubated with membranes for 60 min. Membranes were washed three times for 5 min in PBST and were visualized using SuperSignal-enhanced chemiluminescence (Pierce). Protein loading was assessed by visual inspection of Ponceau S-stained membranes.
For the dithiothreitol (DTT) treatment, recombinant PCSK9-WT and PCSK9-679X were expressed in HEK-293 cells after transfection with Lipofectamine 2000 (Invitrogen) by use of the protocol provided by the manufacturer. After 48 h at 37°C, the cells were collected in PBS and were isolated by centrifugation (1,000 g for 15 min). A total of 100 μl of buffer (50 mM Tris [pH 6.4], 2 mM CaCl2, and 150 mM NaCl) was added to the cell pellet. The cells were mechanically disrupted by 30 passages through a 23-gauge needle. A total of 7.3 μg of cellular protein was incubated with 4 μl of β-ME–free sample buffer (100 mM Tris [pH 6.5], 2% SDS, 1 mM EDTA, 20% glycerol, 0.001% bromophenol blue, and 0.05–20 μM DTT) at RT for 1 h. Immunoblotting was performed as described above, except that the proteins were electroblotted onto PVDF membrane (Amersham Biosciences).
Pulse-Chase Analysis of PCSK9 Processing and Secretion
Wild-type PCSK9-253F and PCSK9-679X were expressed in HEK-293 cells with use of Fugene 6 transfection reagent. After 2 d in culture, the cells were washed twice with cold PBS and were then incubated with Cys/Met-free medium for 1 h at 37°C. Cells were pulsed with 0.1 mCi/ml S35-Cys/Met for 10 min at 37°C and then were chased for the time intervals indicated in figure 2. PCSK9 was immunoprecipitated from cell lysates and medium with use of the polyclonal antibody 6389, and the precipitates were subjected to 8% SDS-PAGE and were visualized by exposure to Biomax MS film.
Figure 2. .
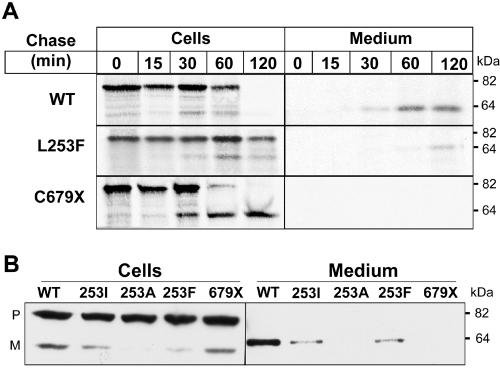
Pulse-chase analysis of the synthesis, processing, and secretion of PCSK9-253F and PCSK9-679X. A, Cells expressing PCSK9-WT (WT) or mutant PCSK9 were incubated with 0.1 mCi/ml S35-Cys/Met for 10 min and then were chased for the indicated time periods. PCSK9 was immunoprecipitated from cell lysates and medium with use of antibody 6389 and then was subjected to 8% SDS-PAGE. Proteins were visualized by exposure to XOMAT film. B, PCSK9-WT (WT) and mutant forms of PCSK9 were expressed in HEK-293 cells, and immunoblotting was performed on the cells and medium as described in the figure 1 legend. P = precursor; M = mature.
Immunoprecipitation of PCSK9 from Plasma
After written consent to participate in the institutional review board–approved study was obtained, fasting blood was collected from a Dallas Heart Study participant identified elsewhere to be heterozygous for the Y142X mutation and from selected family members.4,19 Plasma was isolated from the blood as described elsewhere19 and was aliquoted and maintained at −80°C. A total of 500 μl of plasma was adjusted to a final concentration of 50 mM HEPES [pH 7.4], 2.5 mM MgCl2, 1% Triton X-100, 0.5% sodium deoxycholate, 1 mM phenylmethylsulphonylfluoride, and protease inhibitor cocktail (Roche) in a final volume of 1 ml. Samples were rotated at 4°C for 30 min and then were centrifuged at 21,000 g for 15 min. Supernatants were then divided into two 500-μl aliquots, and 20 μg of rabbit anti-human PCSK9 antibody (295A) or 20 μg of purified IgG from a nonimmunized rabbit (4752) was added to each aliquot. A total of 50 μl of PBS-equilibrated protein A agarose beads (RepliGEN) was added to each sample. The samples were rotated at 4°C overnight before being washed four times with 1× IP buffer (50 mM Hepes [pH 7.4], 100 mM NaCl, 1.5 mM MgCl2, 1% Triton X-100, and 0.5% sodium deoxycholate). The beads were resuspended in 100 μl SDS-PAGE sample buffer, were boiled at 96°C for 5 min, and were centrifuged at 21,000 g for 5 min. A total of 30 μl of supernatant was subjected to SDS-PAGE (8%), and immunoblotting was performed using a monoclonal antibody to PCSK9 (15A6).
Structural Model of PCSK9
A BLAST search20 with use of the PCSK9 sequence gi|31317307 identified the fervidolysin sequence gi|56553650 (residue range 25–446, E value=10-9) and established an initial alignment of the two sequences. The sequence of PCSK9 was aligned with the N-terminal and catalytic domains of the fervidolysin structure (PDB ID 1r6v) with use of the fold-recognition program Meta-BASIC,21 which incorporates evolutionary information and secondary structure predictions (significant score 144.9). The resulting PCSK9 model structure was generated on the basis of the 1r6v coordinates with the program MolScript.22 The alignment produced by PSI-BLAST and the alignment produced by Meta-BASIC differ in some loop regions and in the extreme N-terminus (residues 1–67 of PCSK9) and C-terminus (residue 420 to the end of PCSK9); for mutations falling within these regions (R46L, F216L, and A443T), the range bounded by the BLAST alignment position and the BASIC alignment position were estimated. Alignments were in agreement for the other mutations in the prodomain (ΔR97 and S127R) and in the catalytic domain (L253F and D374Y).
Results
Expression of Loss-of-Function Mutations in HEK-293 Cells
PCSK9 is synthesized as an ∼72-kDa soluble zymogen (precursor) and undergoes autocatalytic cleavage to form an ∼16-kDa prodomain and an ∼62-kDa catalytic and C-terminal domain (mature protein).7,9 To determine the effect of loss-of-function mutations on PCSK9 synthesis, processing, and secretion, we expressed recombinant forms of the protein in HEK-293 cells and analyzed the cell lysates and medium for the presence of PCSK9 (fig. 1B). The two mutations associated with more-modest reductions in plasma LDL-C levels (R46L and A443T) had no detectable effect on the processing or secretion of PCSK9 (fig. 1B). The Y142X mutation occurs early in the transcript and is predicted to initiate nonsense-mediated mRNA decay24; expression of a cDNA construct containing this mutation resulted in no immunodetectable protein with use of either an antibody to the prodomain or the epitope tag (data not shown).
In cells expressing PCSK9-253F, the amount of mature protein was reduced compared with that in cells expressing PCSK9-WT, which suggests that the mutation inhibits autocatalytic cleavage. In contrast to the L253F substitution, the C679X mutation did not interfere with protein cleavage. Rather, the mature protein accumulated in the cells, and none was detected in the medium. Thus, both the L253F and the C679X mutations interfere with the secretion of PCSK9.
Interference by PCSK9 Loss-of-Function Mutations with Protein Processing and Secretion
The effects of the L253F and C679X mutations on PCSK9 processing were confirmed by pulse-chase studies (fig. 2A). No differences in the synthesis or secretion of the A443T or the R46L mutations were seen (data not shown). Neither mutation affected the amount of precursor protein observed at the 15-min time point. In cells expressing PCSK9-WT, the precursor protein was fully converted to the mature form of the protein and was secreted within 2 h. Only a small amount of mature wild-type protein was detected in cells at any time point, which suggests that the protein was rapidly secreted after processing. In cells expressing PCSK9-253F, the precursor was almost completely retained at the 2-h time point, and only a small quantity of mature protein was detected in the cells or in the medium. Substitution of an isoleucine or valine for leucine at this position also significantly reduced the rate of processing and secretion of PCSK9 (fig. 2B), which suggests that leucine is required at this position and that the adverse effect of the substitution of phenylalanine was not attributable simply to the introduction of a bulky aromatic side chain. Thus, the L253F mutation interferes with autocatalytic cleavage of PCSK9, which presumably reflects decreased catalytic activity of the mutant protein.
PCSK9-679X Is Retained in the ER
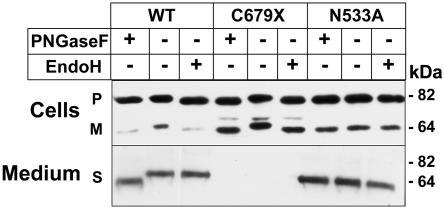
In contrast to PCSK9-253F, recombinant PCSK9-679X underwent rapid cleavage, but the mature protein accumulated in the cells and was not secreted (fig. 2). To determine the site of accumulation of the mature PCSK9-679X, we examined the sensitivity of the N-linked sugar attached to residue 533 to PNGaseF and endoglycosidase H (EndoH) (fig. 3). In the cells expressing PCSK9-WT, the precursor and mature forms of the protein remained sensitive to both glycosidases. In contrast to these results, the secreted form of the wild-type protein was resistant to EndoH, as would be expected of a protein that had transitted through the Golgi. This result is consistent with rapid secretion of PCSK9 after autocatalytic cleavage. In cells expressing PCSK9-679X, both the precursor and mature forms of PCSK9 remained sensitive to EndoH, which indicates that these forms of the protein reside in the ER (fig. 3). Substitution of an alanine for asparagine at residue 533 did not interfere with either the processing or the secretion of PCSK9. These data suggest that the C679X mutation spares catalytic activity but prevents normal folding of the protein, which is thus retained in the ER.
Figure 3. .
Effect of the C679X mutation on the maturation of N-linked sugars in PCSK9. Cell lysate and medium were collected from HEK-293 cells expressing PCSK9-WT (WT) and mutant PCSK9 and were treated with PNGaseF or EndoH as described in the “Material and Methods” section. Analysis of recombinant PCSK9 containing an alanine rather than asparagine at the site of attachment of the N-linked sugar was also performed. Deglycosylated samples were subjected to 10% SDS-PAGE, and immunoblotting was performed using anti-FLAG M2 mAb. P = precursor; M = mature; S = secreted.
C679X Is Retained in the ER Because of Aberrant Folding
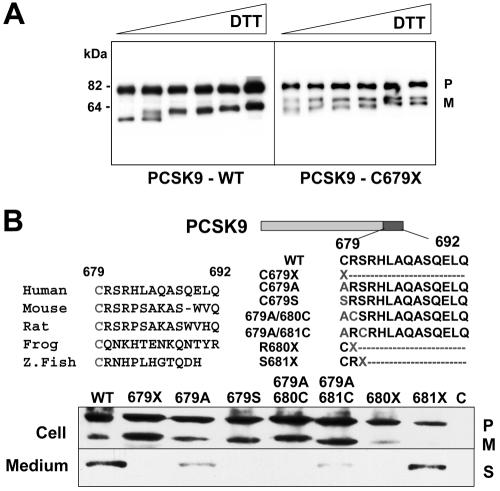
The cysteine at residue 679 is the last of 18 cysteines in the 269-aa C-terminal domain. The failure of PCSK9-679X to transit out of the ER is presumably due to abnormal disulfide bond formation, which results in improper protein folding. Aberrant folding of C679X was confirmed by inspection of the migration of the mutant protein on a nonreducing gel after treatment with increasing concentrations of DTT (fig. 4A). Substitution of either serine or alanine for cysteine at residue 679 resulted in retention of the mature protein in the ER. Introduction of cysteine at position 680 or 681 failed to rescue trafficking of the protein (fig. 4B). These results suggest a specific requirement for cysteine at residue 679. Introduction of a stop mutation at residue 680 (but not at 681) also prevented secretion of the protein. Thus, proper folding of the C-terminal domain is not required for catalytic cleavage but is required for secretion.
Figure 4. .
DTT reduction of PCSK9-679X and secretion of mutant forms of PCSK9. A, Aliquots of the cell lysates and medium from HEK-293 cells expressing wild-type (WT) or PCSK9-679X were treated with increasing concentrations of DTT at RT for 1 h. Samples were then subjected to 8% SDS-PAGE before immunoblotting. B, The highly conserved cysteine C679 (top left) was mutated (top right), and the constructs were expressed in HEK-293 cells (bottom). Immunoblotting was performed on the cells and medium as described in the figure 1 legend. Z. Fish = zebrafish; P = precursor; M = mature; S = secreted.
Identification of a Compound Heterozygote with No Circulating PCSK9
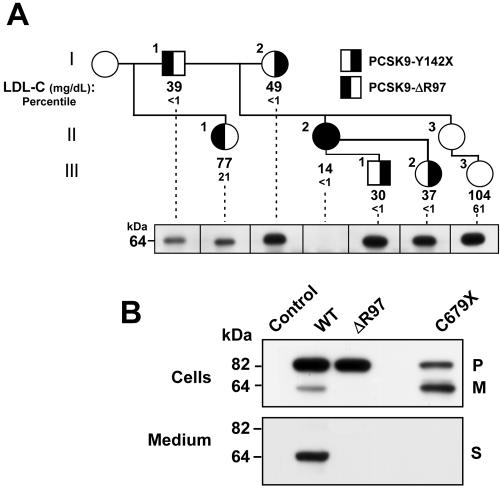
The three mutations in PCSK9 associated with the greatest reductions in plasma levels of LDL-C prevent the secretion of mature PCSK9 by disrupting the synthesis (Y142X), cleavage (L253F), or folding (C679X) of the protein. On the basis of these findings, we predicted that individuals with these mutations would have reduced circulating levels of PCSK9. To test this hypothesis, we examined the relationship between the amount of circulating PCSK9 and the segregation of the PCSK9 nonsense alleles in families. In one such family, we identified an individual with no immunodetectable circulating PCSK9 (fig. 5A and table 1). The proband of this family was a 53-year-old woman who was a participant in the Dallas Heart Study.19 This subject was heterozygous for the Y142X allele and had an LDL-C level of 49 mg/dL (<1st percentile when compared with age- and sex-matched controls). Genetic analysis of her family revealed that her daughter and two of her granddaughters were also heterozygous for the nonsense mutation. All four of the family members who were heterozygous for the Y142X allele had LDL-C levels <1st percentile when compared with age- and sex-matched controls. However, the proband’s daughter (II.2) had a lower plasma level of LDL-C (14 mg/dL) than that of either her mother (49 mg/dL) and her children (30 mg/dL and 27 mg/dL); since the daughter's father was also hypocholesterolemic (LDL-C of 39 mg/dL), we screened her PCSK9 gene for a mutation on the paternal allele. DNA sequencing revealed that II.2 was heterozygous for an in-frame 3-bp deletion (c.290_292delGCC) that deletes an arginine at codon 97. Expression of the mutant protein in HEK-293 cells revealed that the mutation prevented autocatalytic cleavage and secretion of PCSK9 (fig. 5B).
Figure 5. .
Pedigree of a 32-year-old African American woman (II.2) who is compound heterozygous for loss-of-function mutations in PCSK9. A, The proband (I.2) of the family is a participant in the Dallas Heart Study19 who was found to be heterozygous for the Y142X allele in PCSK9. Fasting blood samples were obtained from additional family members. Plasma and serum were isolated, and the lipids and lipoprotein levels were measured using commercial reagents (table 1). The LDL-C and age- and sex-adjusted percentiles are provided for each family member. PCSK9 was immunoprecipitated from the plasma of selected family members with use of a polyclonal anti-PCSK9 antibody (295A), was size-fractionated by SDS-PAGE, and then was immunoblotted as described in the “Material and Methods” section. Individual II.3 was sampled, but the analysis of circulating PCSK9 was not performed on this subject. B, PCSK9-WT (WT) and mutant PCSK9 (ΔR97) were expressed in HEK-293 cells. After 2 d, the cell lysates and the medium were collected and subjected to immunoblotting as described in the figure 1 legend. NA = not available; P = precursor; M = mature; S = secreted.
Table 1. .
Plasma Lipid and Lipoprotein Levels in Family Members of an Individual Who Is a Compound Heterozygote for Loss-of-Function Mutations in PCSK9[Note]
| Fasting Plasma Levels, in mg/dL (Percentile) |
PCSK9 Allelea |
|||||||
| Individual | Age (years) |
Sex | Cholesterol | LDL-C | HDL-Cb | TGc | 1 | 2 |
| I.1 | 51 | M | 96 (<1st) | 39 (<1st) | 44 (55th) | 88 (25th) | ΔR97 | WT |
| I.2 | 53 | F | 144 (<1st) | 49 (<1st) | 88 (90th) | 51 (<1st) | Y142X | WT |
| II.1 | 10 | F | 137 (15th) | 77 (20th) | 49 (45th) | 78 (50th) | ΔR97 | WT |
| II.2 | 32 | F | 96 (<1st) | 14 (<1st) | 65 (80th) | 119 (70th) | ΔR97 | Y142X |
| II.3 | 28 | F | 152 (20th) | 80 (15th) | 59 (65th) | 92 (55th) | WT | WT |
| III.1 | 6 | M | 106 (<1st) | 30 (<1st) | 65 (75th) | 77 (75th) | Y142X | WT |
| III.2 | 3 | F | 106 (<1st) | 37 (<1st) | 59 (70th) | 71 (55th) | Y142X | WT |
| III.3 | 13 | F | 174 (60th) | 104 (60th) | 55 (65th) | 110 (75th) | WT | WT |
Note.— Plasma lipid and lipoprotein levels were measured after a 12-h fast. The percentile of the lipid and lipoprotein levels are compared with age- and sex-matched African American control individuals.
WT = wild type.
HDL-C = high-density lipoprotein cholesterol.
TG = triglycerides.
Thus, II.2 is a compound heterozygote for mutations that disrupt synthesis (maternal allele) and processing/secretion (paternal allele) of PCSK9. If PCSK9 is secreted from hepatocytes in vivo, II.2 would be predicted to have no circulating PCSK9. To test this possibility, we examined the plasma of each family member for the presence of PCSK9. Immunoprecipitation of PCSK9 from 500 μl of fasting plasma revealed a single band of the expected size (64 kDa) in all family members except the proband’s daughter (II.2).
Despite having no immunodetectable circulating PCSK9 and an LDL-C of only 14 mg/dL, II.2 is an apparently healthy, fertile, normotensive, college-educated woman with normal liver and renal function tests (including urinalysis) who works as an aerobics instructor.
Structural Model of PCSK9
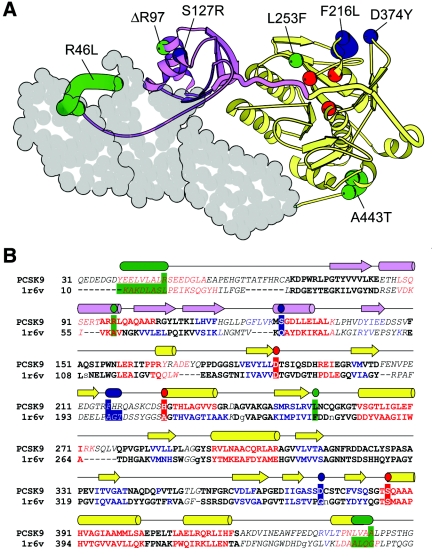
To relate the functional characterization of the loss-of-function variants to protein structure, we developed a structural model of PCSK9 on the basis of homology with fervidolysin, a keratin-degrading enzyme from the bacterium Fervidobacterium pennivorans.23 Significant sequence similarity between PCSK9 and fervidolysin extends from the subtilisin-like N-terminal prodomain (fig. 6A [purple]) to the peptidase S8 catalytic domain (fig. 6A [yellow]); within these two domains, the two sequences can be aligned with high confidence (fig. 6B). Additionally, both proteins include a C-terminal extension from the catalytic domain that is similar in length and secondary structure. In fervidolysin, the C-terminus contains two β-sandwich domain repeats that interact with both the prodomain and the catalytic domain.23 In PCSK9, the C-terminus contains a conserved cysteine-rich pattern (Cys × 6) repeated three times within the context of predicted β-strands, suggesting the presence of three—rather than two—C-terminal domains. Thus, despite secondary structural similarities between PCSK9 and fervidolysin, the topology of the PCSK9 C-terminus remains ambiguous; accordingly, it is represented as a space-filled model without a corresponding alignment (fig. 6).
Figure 6. .
Structural model (A) and alignment of residues 31–450 of PCSK9 with fervidolysin (B). A, A model of PCSK9, generated using coordinates from the crystal structure of fervidolysin (Protein Database ID 1r6v).23 The N-terminal prodomain (purple) is connected to the catalytic domain (yellow) by a long, thickened coil. The prodomain cleavage site (color boundary) is near the catalytic triad (red). PCSK9 mutations that map to the prodomain or catalytic domain are labeled and numbered according to the human sequence and are depicted in blue (gain of function) or green (loss of function). Those mutations that reside in less accurately mapped regions of the protein are represented by a range of residues (see the “Material and Methods” section). Three predicted C-terminal domains of PCSK9 are represented as a space-fill that is based on the two fervidolysin C-terminal domains. B, An alignment between PCSK9 and the 1r6v N-terminal and catalytic domain sequences, generated with a fold-recognition program (Meta-BASIC). Compared with an alignment made by BLAST (not shown), the residues whose positions in the alignment were in agreement with those obtained using BLAST are in bold type, whereas those that differ are italicized. Noncapitalized residues in 1r6v represent positions where sequence was deleted. Residues with predicted secondary structure are red (helix), blue (strand), and black (coil) and are compared with the observed 1r6v secondary structure (cylinders and arrows shown above the alignment and colored as in panel A). Residues corresponding to the catalytic triad, the gain-of-function mutations, and the loss-of-function mutations are highlighted with the colors and ranges described for panel A.
In the PCSK9 model, the R46L substitution is located within an N-terminal extension of the prodomain. In fervidolysin, this N-terminal coil makes extensive contacts with the C-terminal domain and is thought to contribute to the association of the prodomain with the catalytic/C-terminal domains, which is maintained after autocatalytic cleavage.23 The substitution of a hydrophobic leucine for a polar arginine within the corresponding region of PCSK9 may alter the association between the prodomain and the catalytic domain/C-terminal domains after autocatalytic cleavage. The arginine deleted at residue 97 in II.2 (family DHS20) is predicted to disrupt an α-helix in the prodomain; the finding that this mutation prevents autocatalytic cleavage (fig. 5B) is consistent with the prediction that it would interfere with proper folding of the protein. One of the gain-of-function mutations (S127R) is also located in the prodomain and also reduces the rate of protein processing.11 The remaining two gain-of-function mutations (F216L and D374Y) are predicted to reside in close proximity to each other on an external face of the catalytic domain. Although the molecular consequences of these mutations remain unknown, this mapping might point to a role for these residues in interaction with another protein. The loss-of-function mutation L253F that impairs processing (fig. 2A) resides in close proximity to the catalytic triad. The corresponding residue in fervidolysin points toward the proteolytic cleavage site and is located within 4 angstroms of the aspartate of the catalytic triad. Leucine at this location in PCSK9 appears to be essential and cannot be replaced by isoleucine, alanine, or phenylalanine (fig. 2B), which suggests that this residue helps position the PCSK9 peptide backbone for autocatalytic cleavage. Finally, the substitution of an alanine for a threonine at amino acid 443 has only a modest effect on plasma levels of LDL-C (homozygosity is associated with a 3.5% reduction in plasma LDL-C level);5 it is not clear whether the LDL-lowering effect of this substitution is a direct effect of the amino acid substitution or is due to a sequence variation that is in linkage disequilibrium.5
Discussion
A major finding of this study is that PCSK9 circulates in plasma and that those mutations associated with the greatest reductions in plasma levels of LDL-C interfere with either the synthesis or the secretion of PCSK9. These observations led to the identification of the first known compound heterozygote with two inactivating mutations in PCSK9. Although PCSK9 is expressed in the brain, liver, intestine, and kidneys,7 the compound heterozygote was apparently healthy. Therefore, it appears that secretion of PCSK9 is not essential for normal development and function. The normal fertility and development of the PCSK9-knockout mice are consistent with this conclusion.13 We cannot exclude the possibility that the mutant PCSK9 expressed from the ΔR97 allele, which remains sequestered in the ER, serves some essential function; however, this seems unlikely, since the mutation prevents autocatalytic cleavage, which is required for the function of all other proprotein convertases.25
Nonsense mutations in PCSK9 are relatively common among individuals of African descent. In the United States, 1 in every 40–50 African Americans has at least one PCSK9 allele that contains a nonsense mutation.4,14 The less frequent nonsense mutation (Y142X) occurs early in the transcript in exon 3 and is located 51 nt upstream of the exon-intron junction. These properties predict that the mutant mRNA is recognized by the nonsense-mediated mRNA-decay machinery. We failed to detect PCSK9 mRNA in leukocytes and were therefore unable to directly confirm that this mutation results in degradation of the mRNA.5 To exclude the possibility that a peptide formed from the Y142X allele might adversely affect the function of the wild-type protein, we coexpressed the 14-kDa N-terminal peptide and the wild-type protein in HEK-293 cells. The mutant peptide did not affect the synthesis, cleavage, or secretion of the PCSK9-WT (data not shown). Furthermore, Y142X heterozygotes have circulating PCSK9, presumably expressed from the normal allele, which suggests that the nonsense allele does not prevent secretion of the normal protein. These data are consistent with the hypothesis that the Y142X mutation acts as a loss-of-function mutation rather than as a gain-of-function mutation.
The most common nonsense mutation of PCSK9 is predicted to truncate the protein by 14 aa (PCSK9-679X). This mutation did not interfere with the synthesis or autocatalytic cleavage of PCSK9 but did prevent secretion of the protein from cells (fig. 2). These results are consistent with the previous findings that recombinant PCSK9 containing truncations at the C-terminal underwent normal autocatalytic cleavage.8,9 Analysis of the N-linked sugar attached to residue 533 revealed that the mature, processed form PCSK9-679X remained EndoH sensitive, which is consistent with retention of the protein in the ER. Thus, a functional catalytic domain is not sufficient for the secretion of PCSK9 or for its effect on modulating LDLR levels.
The PCSK9-679X mutation eliminates the final cysteine in the C-terminal domain. This cysteine has been highly conserved through vertebrate evolution (fig. 4B). When either a serine or alanine was substituted for the cysteine, the protein was retained in the ER. Introduction of another cysteine downstream of the substituted cysteine failed to rescue the trafficking of the protein out of the ER, which indicates that the presence and the spacing of the cysteine residue in this region is critical for secretion. The aberrant migration of the C679X mutant protein on SDS-PAGE after DTT reduction suggests that the absence of the last cysteine residue interferes with proper disulfide formation and protein folding. The failure of the processed protein to transit out of the ER is consistent with PCSK9 acting in a post-ER compartment, as suggested elsewhere.15
The very low level of plasma LDL-C in the compound heterozygote reveals that circulating levels of LDL-C are almost totally depleted in the absence of PCSK9. In contrast to other Mendelian forms of severe hypocholesterolemia—abetalipoproteinemia (MIM 200100) and homozygous hypobetalipoproteinemia (MIM 107730), which are both associated with malnutrition, steatorrhea, hepatic steatosis, and manifestations of fat-soluble vitamin deficiency (night blindness and vibratory and proprioception defects)26—hypocholesterolemia due to deficiency of PCSK9 appears to be benign. Individuals heterozygous for a nonsense mutation in PCSK9 do not have any detectable increase in hepatic triglyceride content,5 which often accompanies heterozygous hypobetalipoproteinemia.27 The differences in outcome between these conditions presumably reflect the disparate mechanistic basis for the hypocholesterolemia. Genetic defects in apolipoprotein B or microsomal triglyceride transfer protein (MIM 157147) prevent formation of chylomicrons, thus limiting the absorption of fat-soluble vitamins and the secretion of the LDL precursor lipoprotein, very-low-density lipoprotein.26 Although it remains controversial,28,29 the hypocholesterolemia associated with PCSK9 deficiency appears to be caused primarily by increased LDL clearance and not by reduced lipoprotein production.12,13,30
The observation that PCSK9 deficiency is associated with very low LDL-C levels in an otherwise healthy individual makes PCSK9 an attractive therapeutic target. We have shown elsewhere that heterozygotes for loss-of-function mutations in PCSK9 enjoy significant protection from coronary heart disease.14 Taken together with the results of the present study, these findings are consistent with the notion that inhibition of PCSK9 should be a safe and effective strategy for the primary prevention of coronary heart disease.
Acknowledgments
We thank Y. K. Ho for generating the antibodies used in these experiments. This work was supported by grants from the National Institute of Health (P01 HL20948), the Donald W. Reynolds Foundation, and the Perot Family Fund. T.A.L. is supported by the Natural Sciences and Engineering Research Council of Canada. Y.T.-W. is supported by the Stanley J. Sarnoff Endowment.
Web Resource
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PCSK9, autosomal dominant hypercholesterolemia, abetalipoproteinemia, homozygous hypobetalipoproteinemia, and microsomal triglyceride transfer protein)
References
- 1.Abifadel M, Varret M, Rabes JP, Allard D, Ouguerram K, Devillers M, Cruaud C, et al (2003) Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet 34:154–156 10.1038/ng1161 [DOI] [PubMed] [Google Scholar]
- 2.Maxwell KN, Soccio RE, Duncan EM, Sehayek E, Breslow JL (2003) Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. J Lipid Res 44:2109–2119 10.1194/jlr.M300203-JLR200 [DOI] [PubMed] [Google Scholar]
- 3.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL (2003) Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA 100:12027–12032 10.1073/pnas.1534923100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH (2005) Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet 37:161–165 10.1038/ng1509 [DOI] [PubMed] [Google Scholar]
- 5.Kotowski IK, Pertsemlidis A, Luke A, Cooper RS, Vega GL, Cohen JC, Hobbs HH (2006) A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am J Hum Genet 78:410–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berge KE, Ose L, Leren TP (2006) Missense mutations in the PCSK9 gene are associated with hypocholesterolemia and possibly increased response to statin therapy. Arterioscler Thromb Vasc Biol 26:1094–1100 10.1161/01.ATV.0000204337.81286.1c [DOI] [PubMed] [Google Scholar]
- 7.Seidah NG, Benjannet S, Wickham L, Marcinkiewicz J, Jasmin SB, Stifani S, Basak A, Prat A, Chretien M (2003) The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc Natl Acad Sci USA 100:928–933 10.1073/pnas.0335507100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naureckiene S, Ma L, Sreekumar K, Purandare U, Lo CF, Huang Y, Chiang LW, Grenier JM, Ozenberger BA, Jacobsen JS, Kennedy JD, DiStefano PS, Wood A, Bingham B (2003) Functional characterization of Narc 1, a novel proteinase related to proteinase K. Arch Biochem Biophys 420:55–67 10.1016/j.abb.2003.09.011 [DOI] [PubMed] [Google Scholar]
- 9.Benjannet S, Rhainds D, Essalmani R, Mayne J, Wickham L, Jin W, Asselin MC, Hamelin J, Varret M, Allard D, Trillard M, Abifadel M, Tebon A, Attie AD, Rader DJ, Boileau C, Brissette L, Chretien M, Prat A, Seidah NG (2004) NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the LDLR and LDL-cholesterol. J Biol Chem 279:48865–48875 10.1074/jbc.M409699200 [DOI] [PubMed] [Google Scholar]
- 10.Maxwell KN, Breslow JL (2004) Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc Natl Acad Sci USA 101:7100–7105 10.1073/pnas.0402133101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park SW, Moon YA, Horton JD (2004) Post-transcriptional regulation of LDL receptor protein by proprotein convertase subtilisin/kexin type 9a (PCSK9) in mouse liver. J Biol Chem 279:50630–50638 10.1074/jbc.M410077200 [DOI] [PubMed] [Google Scholar]
- 12.Goldstein JL, Hobbs HH, Brown MS (2001) Familial hypercholesterolemia. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease, 8th ed. McGraw Hill, New York, pp 2863–2913 [Google Scholar]
- 13.Rashid S, Curtis DE, Garuti R, Anderson NN, Bashmakov Y, Ho YK, Hammer RE, Moon YA, Horton JD (2005) Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc Natl Acad Sci USA 102:5374–5379 10.1073/pnas.0501652102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen JC, Boerwinkle E, Mosley TH, Hobbs HH (2006) Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med 354:34–42 10.1056/NEJMoa052985 [DOI] [PubMed] [Google Scholar]
- 15.Maxwell KN, Fisher EA, Breslow JL (2005) Overexpression of PCSK9 accelerates the degradation of the LDLR in a post-endoplasmic reticulum compartment. Proc Natl Acad Sci USA 102:2069–2074 10.1073/pnas.0409736102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cameron J, Holla OL, Ranheim T, Kulseth MA, Berge KE, Leren TP (2006) Effect of mutations in the PCSK9 gene on the cell surface LDL receptors. Hum Mol Genet 15:1551–1558 10.1093/hmg/ddl077 [DOI] [PubMed] [Google Scholar]
- 17.Herz J, Kowal RC, Ho YK, Brown MS, Goldstein JL (1990) Low density lipoprotein receptor-related protein mediates endocytosis of monoclonal antibodies in cultured cells and rabbit liver. J Biol Chem 265:21355–21362 [PubMed] [Google Scholar]
- 18.Graf GA, Cohen JC, Hobbs HH (2004) Missense mutations in ABCG5 and ABCG8 disrupt heterodimerization and trafficking. J Biol Chem 279:24881–24888 10.1074/jbc.M402634200 [DOI] [PubMed] [Google Scholar]
- 19.Victor RG, Haley RW, Willett DL, Peshock RM, Vaeth PC, Leonard D, Basit M, Cooper RS, Iannacchione VG, Visscher WA, Staab JM, Hobbs HH (2004) The Dallas Heart Study: a population-based probability sample for the multidisciplinary study of ethnic differences in cardiovascular health. Am J Cardiol 93:1473–1480 10.1016/j.amjcard.2004.02.058 [DOI] [PubMed] [Google Scholar]
- 20.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ginalski K, von Grotthuss M, Grishin NV, Rychlewski L (2004) Detecting distant homology with Meta-BASIC. Nucleic Acids Res 32:W576–W581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esnouf RM (1999) Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr D Biol Crystallogr 55:938–940 10.1107/S0907444998017363 [DOI] [PubMed] [Google Scholar]
- 23.Kim JS, Kluskens LD, de Vos WM, Huber R, van der Oost J (2004) Crystal structure of fervidolysin from Fervidobacterium pennivorans, a keratinolytic enzyme related to subtilisin. J Mol Biol 335:787–797 10.1016/j.jmb.2003.11.006 [DOI] [PubMed] [Google Scholar]
- 24.Maquat LE (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5:89–99 10.1038/nrm1310 [DOI] [PubMed] [Google Scholar]
- 25.Seidah NG, Benjannet S, Hamelin J, Mamarbachi AM, Basak A, Marcinkiewicz J, Mbikay M, Chretien M, Marcinkiewicz M (1999) The subtilisin/kexin family of precursor convertases: emphasis on PC1, PC2/7B2, POMC and the novel enzyme SKI-1. Ann N Y Acad Sci 885:57–74 [DOI] [PubMed] [Google Scholar]
- 26.Kane JP (2001) Disorders of the biogenesis and secretion of lipoproteins containing the B apolipoproteins. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease, 8th ed. McGraw Hill, New York, pp 2717–2752 [Google Scholar]
- 27.Tarugi P, Lonardo A, Ballarini G, Grisendi A, Pulvirenti M, Bagni A, Calandra S (1996) Fatty liver in heterozygous hypobetalipoproteinemia caused by a novel truncated form of apolipoprotein B. Gastroenterology 111:1125–1133 10.1016/S0016-5085(96)70082-3 [DOI] [PubMed] [Google Scholar]
- 28.Sun XM, Eden ER, Tosi I, Neuwirth CK, Wile D, Naoumova RP, Soutar AK (2005) Evidence for effect of mutant PCSK9 on apolipoprotein B secretion as the cause of unusually severe dominant hypercholesterolaemia. Hum Mol Genet 14:1161–1169 10.1093/hmg/ddi128 [DOI] [PubMed] [Google Scholar]
- 29.Ouguerram K, Chetiveaux M, Zair Y, Costet P, Abifadel M, Varret M, Boileau C, Magot T, Krempf M (2004) Apolipoprotein B100 metabolism in autosomal-dominant hypercholesterolemia related to mutations in PCSK9. Arterioscler Thromb Vasc Biol 24:1448–1453 10.1161/01.ATV.0000133684.77013.88 [DOI] [PubMed] [Google Scholar]
- 30.Lalanne F, Lambert G, Amar MJ, Chetiveaux M, Zair Y, Jarnoux AL, Ouguerram K, Friburg J, Seidah NG, Brewer HB Jr, Krempf M, Costet P (2005) Wild-type PCSK9 inhibits LDL clearance but does not affect apoB-containing lipoprotein production in mouse and cultured cells. J Lipid Res 46:1312–1319 10.1194/jlr.M400396-JLR200 [DOI] [PubMed] [Google Scholar]