Abstract
X-chromosome inactivation is widely believed to be random in early female development and to result in a mosaic distribution of cells, approximately half with the paternally derived X chromosome inactive and half with the maternally derived X chromosome inactive. Significant departures from such a random pattern are hallmarks of a variety of clinical states, including being carriers for severe X-linked diseases or X-chromosome cytogenetic abnormalities. To evaluate the significance of skewed patterns of X inactivation, we examined patterns of X inactivation in a population of >1,000 phenotypically unaffected females. The data demonstrate that only a very small proportion of unaffected females show significantly skewed inactivation, especially during the neonatal period. By comparison with this data set, the degree of skewed inactivation in a given individual can now be quantified and evaluated for its potential clinical significance.
In female mammals, most genes on all X chromosomes in excess of one per autosomal complement are silenced epigenetically early in development, thereby making female X-chromosome gene dosage largely equivalent to that of males.1,2 The choice of which X chromosome remains active in each cell is thought to be random, so that females have varying X-inactivation ratios, defined as the proportions of cells expressing alleles from one or the other X chromosome. The X-inactivation ratios of females can range from a highly skewed ratio of 0:100, where the same X chromosome is active for all cells, to a 50:50 ratio, where each X chromosome is active in an equal number of cells.3 In unaffected females, the X-inactivation ratio may be of no clinical importance, but a highly skewed X-inactivation ratio may be indicative of carrier status for many X-chromosome disorders, including both cytogenetic abnormalities and certain X-linked Mendelian conditions.4–6
Skewed X-inactivation patterns may occur either as a result of secondary cell selection during development or by primary stochastic or genetic processes.4,7,8 The most common reason for highly skewed X inactivation is postinactivation cell selection due to an X-chromosome abnormality that affects cell proliferation either in all cells in the embryo or in a tissue-specific manner. For example, females who carry balanced X;autosome translocations typically exhibit skewed X inactivation because of selection in favor of cells in which the normal X is inactivated.9,10
X-linked disorders are generally rare in females and are usually attributable to advantageous silencing of the X chromosome that carries the mutant allele.5 The presumption is that the initial X-inactivation choice is random but that, during proliferation, cells that have chosen the mutant X chromosome to be active have a significant or total growth disadvantage and are thus underrepresented in the adult carrier. This postinactivation cell selection has been documented in carriers for a number of X-linked diseases.11–15 In contrast to negative cell selection, the positive growth advantage of cells in which the mutant X is primarily active has been reported in a few conditions.16,17
It has long been recognized from a theoretical perspective that an additional explanation for nonrandom X-inactivation ratios might include mutations in the X-inactivation process itself, which causes one chromosome to be chosen over another at the time of X inactivation in the early embryo.8,18 Mutations in the X-inactivation pathway are thought to be rare, but studies of individuals with such mutations may provide much information regarding the regulation of the X-inactivation pathway. Studies of large families19,20 and females who express X-linked disorders18 have revealed that a locus or loci on the X chromosome can affect the X-inactivation ratio. Studies of females with Duchenne muscular dystrophy (MIM 300377) also suggest that other modifying loci exist that affect the X-inactivation ratio.21,22 These limited studies of human families are intriguing, since the X-inactivation ratio is known to be under primary genetic control in the mouse.23–25 The major locus controlling the preferential choice of which X chromosome is to remain active in the mouse is the X-linked X-controlling element (Xce) locus, although the mechanism responsible for this effect remains elusive. It has been suggested that similar XCE alleles might exist in the human population.19
Whereas the probability that highly skewed X-inactivation ratios occur by chance may be low,3 an accurate assessment of the X-inactivation variance is needed to quantify this probability. With a highly accurate measurement of X-inactivation in a large population, the number of cells present in the embryo when the choice of which X remains active can be determined.26 Previous studies to determine this number used distributions of X-inactivation ratios from apparently unaffected adult females.26–28 However, since skewing increases with age in unaffected adult and elderly females,29–32 use of these distributions to predict stem-cell number may not be appropriate, since the predicted stem-cell number is inversely proportional to the distribution variance.26 In addition, any errors in determining the X-inactivation ratio, likely dependent on the particular assay being used, would influence and likely increase the apparent variance.
X-inactivation ratios have been determined with a variety of assays that rely on the differential expression of polymorphic X-linked genes or, less directly, on the differential methylation of sequences on the active and inactive chromosomes.26,28,29,33–35 Because of its high polymorphism content, the most commonly used assay examines differential methylation of the human androgen receptor (AR) gene.28 Although the assay has been used in many studies, a wide range of results has been reported with respect to the distribution of X-inactivation ratios in the population(s) under study,18,20,28,32,36–39 with as many as 20%–30% of the population reported to show highly skewed ratios. Although this presumably reflects, in part, the relatively small control groups reported, it may also reflect assay variation; reproducibility, in particular, is a key parameter for an assay that is highly sensitive to partial restriction-enzyme digestion and to potential PCR quantitation or electrophoresis artifacts.
A definitive data set of X-inactivation ratios in a large sample of phenotypically unaffected females from the general population would be useful as a baseline for studies of X inactivation in various patient populations and/or genetically defined cohorts. Toward this goal, we have determined the distribution of X-inactivation patterns in blood samples from >1,000 newborn infants and adult women, using the AR X-inactivation assay. The results provide a comprehensive estimate of the frequency of individuals in the general population with highly skewed X-inactivation ratios and suggest that screening of the newborn female population may be an effective means of ascertaining individuals at increased risk of carrying some deleterious X-linked defects.
Material and Methods
Study Population
A total of 1,284 anonymized DNA samples from either adult peripheral or newborn cord blood were obtained from several sources. All samples were obtained from phenotypically unaffected individuals from the general control population or from family studies in which it was determined that the individuals did not carry the specific defect in question. The racial and ethnic composition of the sample population was mixed. The following female samples were tested: 127 from the general adult population of Cleveland (provided by Drs. Stuart Schwartz and Georgia Wiesner, Case Western Reserve University), 77 from Botnia, Finland40 (provided by Drs. Melanie Mahtani and Eric Lander), 106 from a Mennonite group in western Pennsylvania (provided by Dr. Eric Puffenberger), 105 from Ashkenazi Jewish mothers with normal pregnancies (provided by Dr. David Gurwitz, National Laboratory for the Genetics of Israeli Populations, Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv). The remaining blood samples were from newborn cord or from mothers of newborns from a six-county area in the upper Savannah region of South Carolina. All samples were obtained with approval from the respective institutions and were fully anonymized before transfer to Case Western Reserve University for X-inactivation analysis.
Determination of X-Inactivation Patterns
To determine the X-inactivation pattern, we examined the methylation status of the AR locus, as described elsewhere.18 Briefly, ∼50–150 ng of DNA was digested in duplicate for 16 h either with HpaII plus RsaI or with RsaI alone as a control. In addition, digests with HhaI and RsaI, as well as CfoI and RsaI, were run on a select number of samples to test the accuracy of the assay. After digestion, the enzymes were inactivated at 80°C for 20 min, and one-tenth of the resulting digest was used for PCR amplification as described elsewhere.18 The resulting products were run on an ABI 373A automated sequencer or an ABI 3100 genetic analyzer and were analyzed by GeneScan software (Applied Biosystems). The raw peak height values of the digested samples were corrected for amplification efficiency by using the average of the two samples digested with RsaI alone (a restriction enzyme with sites outside the PCR product, which is not sensitive to methylation and thus digests active and inactive X alleles equally). The X-inactivation pattern (expressed arbitrarily as a ratio of the smaller:larger allele) is the resulting corrected average of the duplicate samples digested with HhaI and RsaI. Samples were scored only if the two alleles were separated by more than two CAG repeats, to avoid uncertain contributions of one peak to the other. In addition, one control male sample and one sample from a female known from an earlier study18 to have a completely nonrandom pattern of X inactivation (>98:2) were included in every batch of samples, to control for complete digestion and amplification efficiency.
Expression-Based X-Inactivation Assay
Transformed lymphoblast cell lines were grown and harvested for DNA and RNA, as described elsewhere.35 RNA was isolated with the Stratagene Strataprep Total RNA Miniprep Kit. cDNA was prepared by using plus and minus reverse-transcriptase reactions, to control for DNA contamination. The resulting cDNA was PCR-amplified with primers designed to detect an HinfI restriction-enzyme polymorphism in exon 6 of the XIST gene, followed by a single extension reaction with a fluorescently labeled primer.35 The resulting product was digested with HinfI for at least 4 h at 37°C, and the products were run on an ABI 373A automated sequencer. Peak height values of the resulting raw data were analyzed with GeneScan software, as described above. Control samples for complete digestion of the amplified products were included for every gel.
Results
Technical Considerations
Because of the variability in the reports of distribution of X-inactivation patterns determined by use of the AR assay, we examined some of the variables that might influence the apparent results. We began by addressing which methylation-sensitive restriction enzyme yielded the most-consistent and reproducible results for us. Triplicate samples were assayed by using the restriction enzyme RsaI in conjunction with a methylation-sensitive restriction enzyme—HpaII, HhaI, or CfoI (an isoschizomer of HhaI), each of which has sites within the AR PCR product—as well as a mock-digested control with use of RsaI alone. HpaII and HhaI results were highly correlated with each other, but CfoI consistently generated more variable results (>10% variance). On further investigation, CfoI was found to be only partially heat-inactivated (Boehringer Mannheim and Gibco BRL) and could be completely inactivated only after phenol/chloroform extraction. Presumably, the resulting digested product continued to be digested during PCR amplification and yielded much more variable results between duplicates. To increase the level of precision and accuracy in this data set, we therefore used only HpaII as the methylation-sensitive restriction enzyme, in conjunction with RsaI for data generation. Reproducibility of the data set was confirmed by assaying all samples in duplicate and by reassaying 120 samples chosen at random with HhaI as the methylation-sensitive enzyme (for paired HpaII- and HhaI-digested samples, P>.1 [Student's t test]; r=0.97 [Pearson correlation coefficient]).
To determine, in a quantitative manner, whether the X-inactivation patterns measured indirectly at the AR locus correlate with X-inactivation patterns determined directly by gene expression, we also examined a transcribed polymorphism at the XIST locus. Since XIST is expressed exclusively from the inactive X chromosome in female somatic cells,41 this provides a direct estimate of the proportion of cells carrying a particular XIST allele on the inactive X chromosome.35 We determined the X-inactivation patterns of 18 polyclonal lymphoblast cell lines informative at both the AR and XIST loci, using both assays. Because the phase of the AR and XIST alleles was not known, we used a best-fit model to correlate the data. Whereas such a model is insensitive to allelic phase at X-inactivation patterns near 50:50, it is highly sensitive at more-extreme patterns, as frequently observed in clonal or oligoclonal lymphoblast lines.35 On the basis of this model, X-inactivation patterns determined by the AR methylation assay were highly correlated with those determined directly by XIST expression (R2=0.969), which indicates that the AR assay is an accurate measure of X inactivation (data not shown).
Notwithstanding the apparent robustness of the assay, we do note what appears to be a modest shift toward the left in the X-inactivation distributions (see, e.g., fig. 1), consistent with a slight bias toward amplification of the smaller AR allele. This may reflect a slight overcorrection when the RsaI digest is used; nonetheless, this shift is not statistically significant and has not been explored further.
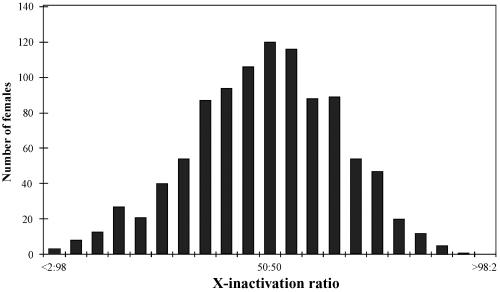
Figure 1. .
The X-inactivation patterns of 1,005 females were assigned to 21 “bins” with a range of <2:98 to >98:2, with increments of 5%. These are normally distributed, with the mean of the distribution residing at 49:51 and the median at 50:50 (SD of the mean = 17).
Distribution of X-Inactivation Patterns in 1,005 Samples from Female Subjects
The principal advantage of the AR assay for measuring X-inactivation patterns is the high heterozygosity of a (CAG)n polymorphism near the 5′ end.28 Indeed, only ∼8% of the 1,284 samples were uninformative for this polymorphism and were thus excluded from the study. However, because of the stutter bands frequently observed in minisatellite repeats and to avoid potential artifacts that might interfere with quantitation, we also rejected any heterozygous samples in which the stutter band from the larger allele overlapped the primary band of the lower allele. This resulted in the elimination of an additional 13% of the original samples.
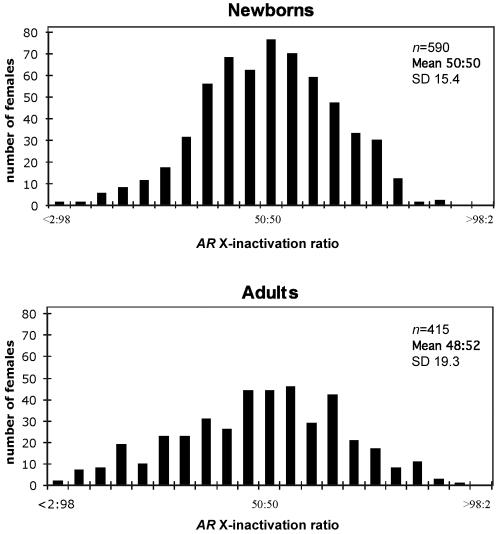
On the basis of these criteria, DNA samples from 1,005 females who were informative for the AR polymorphism were analyzed for their X-inactivation patterns (fig. 1). In the resulting data set, 25% of the samples had X-inactivation patterns skewed to the extent of >70:30 or <30:70; only 8% of the samples were skewed to the extent of >80:20 or <20:80 (table 1). We further examined the distribution by dividing the data set by population groups (see the “Material and Methods” section); there were no statistical differences between the means of any of the distributions. However, there was a highly significant difference between variances (P=.0008 [F test]) when the newborn population was compared with the remaining group of samples (mostly adult women of child-bearing age and a small number of young adults aged >13 years) (fig. 2). This finding is consistent with previous suggestions based on a limited number of samples that indicated a higher proportion of skewed samples in older females.30,32,37,42,43 This effect may reflect random fluctuation in the hematopoietic cell population over time or the possible ascertainment of a small proportion of phenotypically unaffected females in whom postinactivation cell selection has skewed the cell population in favor of one allele on the active X.
Table 1. .
Comparison of Skewed X-Inactivation Ratios in Newborn and Adult Populations
| Percentage of Population withX-Inactivation Ratio of |
||||||
| Population | n | Mean | SD | <20:80/>80:20 | <10:90/>90:10 | <5:95/>95:5 |
| Newborns | 590 | 50:50 | 15.4 | 4.9 | .5 | .2 |
| Adults | 415 |
48:52 | 19.3 | 14.2 | 3.6 | 1.7 |
| Total | 1,005 | 49:51 | 17.2 | 8.8 | 1.8 | .8 |
Figure 2. .
Distributions of X-inactivation ratios of both newborn samples (n=590) and unaffected adult females (n=415).
Estimate of the Number of Stem Cells at the Time of X Inactivation
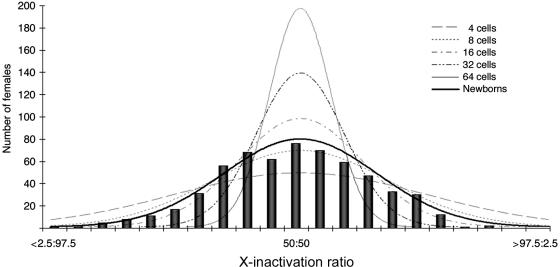
Previous studies determined the number of hematopoietic stem cells present at the time of X-inactivation choice by use of the variance of the X inactivation–pattern distribution.26 To estimate the primordial stem-cell pool size (N), the formula for the variance of a binomial distribution was used (variance=pq/N) and setting the probability that either allele (p or q) is active to 0.5. Normal distributions were drawn by determining the variance (σ2) for each of the values of N and drawing the curve with use of the equation for the normal distribution (fig. 3). To limit the potential effect that somatic mutations and environmental factors might impose on hematopoietic stem cells, we performed these calculations using only the 590 newborn samples. With the SD of 15.3%, the number of stem cells is predicted to be between 8 and 16, which is consistent with previous estimates26,27,42 (fig. 3).
Figure 3. .
Comparison of the newborn distribution with predicted normal distributions of varying stem-cell pool size. The solid black curve follows a normal distribution with use of the actual SD of the newborn samples (15.4 [fig. 2]). The estimated number of precursor stem cells predicted by the newborn SD is between 10 and 12 cells.
Discussion
The distribution of X-inactivation patterns in phenotypically unaffected females has been reported in a number of previous studies that used the AR polymorphism assay.14,15,18,20,28,32,37,39,44 We believe that our data set, which was based on a large number of individuals and for which we used a robust assay subjected to rigorous validation, provides an accurate profile of X-inactivation patterns in the general female population. The distribution reported here thus establishes a set of baseline data for comparing patterns in various patient populations and/or genetically stratified study populations. We attempted to limit over- or underestimating the proportion of individuals who exhibit highly skewed patterns of X inactivation (i.e., more extreme than 80:20 or 90:10, which are the threshold values adopted in many studies) by controlling for a number of potential technical complications, such as incomplete enzyme digestion or scoring of alleles that differ by only a single repeat unit. The reasonable accuracy of our distribution is supported by the fact that the X-inactivation patterns determined by the AR differential methylation assay are highly correlated with those determined more directly by assay of X-linked gene expression (data not shown). Any systematic problems with the AR assay would presumably have reduced the correlation to an unacceptable level.
Nonetheless, it is important to acknowledge that the current study has several limitations. First, the reported distribution is based on methylation analysis, an indirect assessment of X-inactivation status. Although AR methylation does correlate with allele-specific expression of at least one X-linked locus (XIST), an extension of this study could examine the increasing number of other expressed polymorphisms now amenable to analysis.2 Second, the current analysis was performed on only cord or peripheral blood samples. Although this is also true of most other X-inactivation studies performed elsewhere, it is conceivable that X-inactivation patterns will vary somewhat in different clinically relevant tissues.
These limitations notwithstanding, our data support previous observations that, with advancing age, there is greater variation in X inactivation–ratio distribution. The relative paucity of skewed ratios (table 1), however, argues that skewing, when present, may be more clinically informative than previously suspected, especially in newborns. Because <5% of newborn samples show patterns of skewing of <20% or >80%, any newborn who exhibits skewing at birth may warrant further genetic evaluation, on the basis of statistical considerations alone. Because of the blinding of the samples, we were unable to go back and examine the highly skewed samples for chromosomal abnormalities and/or history of X-linked disease; however, such a prospective study might now be appropriate.
The most striking revelation is that very few samples in our study were highly skewed, with only a few newborn samples exhibiting >95:5% X-inactivation ratios. However, we believe we have ruled out partial digestion as the cause of the paucity of skewed samples, because we can digest with a different enzyme or with double digests and get highly reproducible results. Further, the number of skewed samples we observed is similar to that predicted by using the determined variance in the normal distribution equation. On the basis of this evidence, we believe it is highly noteworthy that so few females exhibit skewed X-inactivation ratios, at least in the tissue sampled.
Also noteworthy is the fact that the distribution was platykurtic, with a kurtosis value of −0.26. If there were extensive partial digestion, the distribution would have a decreased variance about the mean, with more of the samples residing at or near 50:50, which would result in positive kurtosis. The negative value that we observe suggests that what we are observing is not partial digestion of the DNA but, rather, that there are multiple overlapping distributions with smaller SDs with different means. This is statistical evidence for the suggestion that there may be a genetic locus (or loci) that affects the determination of which X chromosome will remain active in the cell. This locus (loci) is not likely to be closely linked to the AR locus, since we do not see a directionality to the skewing (reflected in a change of the mean); rather, we see a broadening of the spread of the distribution or a decrease in the kurtosis of the curve.
The data in table 1 provide a basis for evaluating the presence of skewed X-inactivation patterns in various clinical populations. Highly skewed inactivation patterns have been described in carriers of several X-linked disorders5,6,8—most notably, various X-linked mental retardation conditions15 and X-linked immune disorders.45 The unusual nature of skewing in carriers of such defects can now be quantified, in both newborn and adult age groups.
Given the rarity of highly skewed X inactivation in both populations, particularly among newborns, a finding of skewed inactivation as extreme as 90:10 or 95:5 deserves increased scrutiny, since it is, a priori, such an unlikely event (table 1). The application of Bayesian statistics may allow quantification of the level of suspicion that might be appropriate. For example, in a family with an X-linked condition known to be characterized by highly skewed patterns of inactivation, the finding of such a pattern in a newborn potential carrier would be much more likely to be due to her being a carrier than to chance alone (posterior odds ratio ∼100:1). A corollary of this would be that, even in the general population (in which one might estimate the overall frequency of X-linked disease to be, at most, 1%), a finding of highly skewed inactivation is at least as likely to be explained by being a carrier for an X-linked trait that affects X-inactivation ratios as by chance alone. Thus, even in the general population (and, again, especially in the newborn population), a finding of skewed inactivation should prompt questions about possible family histories consistent with a potential X-linked condition (e.g., a history of miscarriage, a deficit of live male births, or idiopathic mental retardation in a male family member).
Acknowledgments
The authors thank Drs. Stuart Schwartz, Georgia Wiesner, Eric Puffenberger, David Gurwitz, and Melanie Mahtani, for providing anonymous DNA samples. This work was supported in part by National Institutes of Health research grant GM45441.
Web Resource
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Duchenne muscular dystrophy)
References
- 1.Lyon MF (1961) Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190:372–373 10.1038/190372a0 [DOI] [PubMed] [Google Scholar]
- 2.Carrel L, Willard HF (2005) X inactivation profile reveals extensive variability in X-linked gene expression. Nature 434:400–404 10.1038/nature03479 [DOI] [PubMed] [Google Scholar]
- 3.Nance WE (1964) Genetic tests with a sex-linked marker: glucose-6-phosphate dehydrogenase. Cold Spring Harbor Symp Quant Biol 29:415–425 [DOI] [PubMed] [Google Scholar]
- 4.Brown CJ, Robinson WP (2000) The causes and consequences of random and non-random X chromosome inactivation in humans. Clin Genet 58:353–363 10.1034/j.1399-0004.2000.580504.x [DOI] [PubMed] [Google Scholar]
- 5.Willard HF (2000) The sex chromosomes and X chromosome inactivation. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Vogelstein B (eds) The metabolic and molecular bases of inherited disease, 8th ed. McGraw-Hill, New York, pp 1191–1221 [Google Scholar]
- 6.Van den Veyver IB (2001) Skewed X inactivation in X-linked disorders. Semin Reprod Med 19:183–191 10.1055/s-2001-15398 [DOI] [PubMed] [Google Scholar]
- 7.Belmont JW (1996) Genetic control of X inactivation and processes leading to X-inactivation skewing. Am J Hum Genet 58:1101–1108 [PMC free article] [PubMed] [Google Scholar]
- 8.Puck JM, Willard HF (1998) X inactivation in females with X-linked disease. N Engl J Med 338:325–328 10.1056/NEJM199801293380511 [DOI] [PubMed] [Google Scholar]
- 9.Mattei MG, Mattei JF, Vidal I, Giraud F (1981) Structural anomalies of the X chromosome and inactivation center. Hum Genet 56:401–408 10.1007/BF00274702 [DOI] [PubMed] [Google Scholar]
- 10.Schmidt M, Du Sart D (1992) Functional disomies of the X chromosome influence the cell selection and hence the X inactivation pattern in females with balanced X-autosome translocations: a review of 122 cases. Am J Med Genet 42:161–169 10.1002/ajmg.1320420205 [DOI] [PubMed] [Google Scholar]
- 11.Nyhan WL, Bakay B, Connor JD, Marks JF, Keele DK (1970) Hemizygous expression of glucose-6-phosphate dehydrogenase in erythrocytes of heterozygotes for the Lesch-Nyhan syndrome. Proc Natl Acad Sci USA 65:214–218 10.1073/pnas.65.1.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conley ME, Brown P, Pickard AR, Buckley RH, Miller DS, Raskind WH, Singer JW, Fialkow JJ (1986) Expression of the gene defect in X-Linked agammaglobulinemia. N Engl J Med 315:564–567 [DOI] [PubMed] [Google Scholar]
- 13.Goodship J, Carter J, Espanol T, Boyd Y, Malcolm S, Levinsky R (1991) Carrier detection in Wiskott-Aldrich syndrome: combined use of M27 β for X inactivation studies and as a linked probe. Blood 77:2677–2681 [PubMed] [Google Scholar]
- 14.Plenge RM, Tranebjaerg L, Jensen PKA, Schwartz CE, Willard HF (1999) Evidence that mutations in the X-linked DDP gene cause incompletely penetrant and variable skewed X inactivation. Am J Hum Genet 64:759–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plenge RM, Stevenson RA, Lubs HA, Schwartz CE, Willard HF (2002) Skewed X-chromosome inactivation is a common feature of X-linked mental retardation disorders. Am J Hum Genet 71:168–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Migeon BR, Moser H, Moser A, Axelman J, Sillence D, Norum R (1981) Adrenoleukodystrophy: evidence for X linkage, inactivation, and selection favoring the mutant allele in heterozygous cells. Proc Natl Acad Sci USA 78:5066–5070 10.1073/pnas.78.8.5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Josten KM, Tooze JA, Borthwick-Clarke C, Gordon-Smith EC, Rutherford TR (1991) Acquired aplastic anemia and paroxysmal nocturnal hemoglobinuria: studies on clonality. Blood 78:3162–3167 [PubMed] [Google Scholar]
- 18.Plenge RM, Hendrich BD, Schwartz C, Arena JF, Naumova A, Sapienza C, Winter RM, Willard HF (1997) A promoter mutation in the XIST gene in two unrelated families with skewed X-chromosome inactivation. Nat Genet 17:353–356 10.1038/ng1197-353 [DOI] [PubMed] [Google Scholar]
- 19.Naumova AK, Olien L, Bird LM, Smith M, Verner AE, Leppert M, Morgan K, Sapienza C (1998) Genetic mapping of X-linked loci involved in skewing of X chromosome inactivation in the human. Eur J Hum Genet 6:552–562 10.1038/sj.ejhg.5200255 [DOI] [PubMed] [Google Scholar]
- 20.Naumova AK, Plenge RM, Bird LM, Leppert M, Morgan K, Willard HF, Sapienza C (1996) Heritability of X chromosome–inactivation phenotype in a large family. Am J Hum Genet 58:1111–1119 [PMC free article] [PubMed] [Google Scholar]
- 21.Azofeifa J, Voit T, Hubner C, Cremer M (1995) X-chromosome methylation in manifesting and healthy carriers of dystrophinopathies: concordance of activation ratios among first degree female relatives and skewed inactivation as a cause of the affected phenotypes. Hum Genet 96:167–176 10.1007/BF00207374 [DOI] [PubMed] [Google Scholar]
- 22.Pegoraro E, Whitaker J, Mowery-Rushton P, Surti U, Lanasa M, Hoffman EP (1997) Familial skewed X inactivation: a molecular trait associated with high spontaneous-abortion rate maps to Xq28. Am J Hum Genet 61:160–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cattanach BM, Isaacson JH (1967) Controlling elements in the mouse X chromsome. Genetics 57:331–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Percec I, Plenge RM, Nadeau JH, Bartolomei MS, Willard HF (2002) Autosomal dominant mutations affecting X inactivation choice in the mouse. Science 296:1136–1139 10.1126/science.1070087 [DOI] [PubMed] [Google Scholar]
- 25.Chadwick LH, Willard HF (2005) Genetic and parent-of-origin influences on X chromosome choice in Xce heterozygous mice. Mammal Genome 16:691–699 10.1007/s00335-005-0059-2 [DOI] [PubMed] [Google Scholar]
- 26.Fialkow PJ (1973) Primordial cell pool size and lineage relationships of five human cell types. Ann Hum Genet 37:39–48 [DOI] [PubMed] [Google Scholar]
- 27.Puck JM, Stewart CC, Nussbaum RL (1992) Maximum-likelihood analysis of human T-cell X chromosome inactivation patterns: normal women versus carriers of X-linked severe combined immunodeficiency. Am J Hum Genet 50:742–748 [PMC free article] [PubMed] [Google Scholar]
- 28.Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW (1992) Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 51:1229–1239 [PMC free article] [PubMed] [Google Scholar]
- 29.Fey MF, Liechti-Gallati S, von Rohr A, Borisch B, Theilkas L, Schneider V, Oestreicher M, Nagel S, Ziemiecki A, Tobler A (1994) Clonality and X-inactivation patterns in hematopoietic cell populations detected by the highly informative M27β DNA probe. Blood 83:931–938 [PubMed] [Google Scholar]
- 30.Busque L, Mio R, Mattioli J, Brais E, Blais N, Lalonde Y, Maragh M, Gilliland DG (1996) Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood 88:59–65 [PubMed] [Google Scholar]
- 31.Gale RE, Fielding AK, Harrison CN, Linch DC (1997) Acquired skewing of X-chromosome inactivation patterns in myeloid cells of the elderly suggests stochastic clonal loss with age. Br J Haematol 98:512–519 10.1046/j.1365-2141.1997.2573078.x [DOI] [PubMed] [Google Scholar]
- 32.Sandovici I, Naumova AK, Leppert M, Linares Y, Sapienza C (2004) A longitudinal study of X inactivation ratio in human females. Hum Genet 115:387–392 [DOI] [PubMed] [Google Scholar]
- 33.Gandini E, Gartler SM (1969) Glucose-6-phosphate dehydrogenase mosaicism for studying the development of blood cell precursors. Nature 224:599–600 10.1038/224599a0 [DOI] [PubMed] [Google Scholar]
- 34.Vogelstein B, Fearon ER, Hamilton SR, Preisinger AC, Willard HF, Michelson AM, Riggs AD, Orkin SH (1987) Clonal analysis using recombinant DNA probes from the X-chromosome. Cancer Res 47:4806–4813 [PubMed] [Google Scholar]
- 35.Rupert JL, Brown CJ, Willard HF (1995) Direct detection of non-random X chromosome inactivation by use of a transcribed polymorphism in the XIST gene. Eur J Hum Genet 3:333–343 [DOI] [PubMed] [Google Scholar]
- 36.Gale RE, Mein CA, Linch DC (1996) Quantification of X-chromosome inactivation patterns in haematological samples using the DNA PCR-based HUMARA assay. Leukemia 10:362–367 [PubMed] [Google Scholar]
- 37.Sharp A, Robinson D, Jacobs P (2000) Age- and tissue-specific variation of X chromosome inactivation ratios in normal women. Hum Genet 107:343–349 10.1007/s004390000382 [DOI] [PubMed] [Google Scholar]
- 38.Karasawa M, Tsukamoto N, Yamane A, Okamoto K, Maehara T, Yokohama A, Nojima Y, Omine M (2001) Analysis of the distribution of CAG repeats and X-chromosome inactivation status of HUMARA gene in healthy female subjects using improved fluorescence-based assay. Int J Hematol 74:281–286 [DOI] [PubMed] [Google Scholar]
- 39.Beever CL, Stephenson MD, Peñaherrera MS, Jiang RH, Kalousek DK, Hayden M, Field L, Brown CJ, Robinson WP (2003) Skewed X-chromosome inactivation is associated with trisomy in women ascertained on the basis of recurrent spontaneous abortion or chromosomally abnormal pregnancies. Am J Hum Genet 72:399–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahtani MM, Widen E, Lehto M, Thomas J, McCarthy M, Brayer J, Bryant B, Chan G, Daly M, Forsblom C, Kanninen T, Kirby A, Kruglyak L, Munnelly K, Parkkonen M, Reeve-Daly MP, Weaver A, Brettin T, Duyk G, Lander ES, Groop LC (1996) Mapping of a gene for type 2 diabetes associated with an insulin secretion defect by a genome scan in Finnish families. Nat Genet 14:90–94 10.1038/ng0996-90 [DOI] [PubMed] [Google Scholar]
- 41.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF (1991) A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349:38–44 10.1038/349038a0 [DOI] [PubMed] [Google Scholar]
- 42.Tonon L, Bergamaschi G, Dellavecchia C, Rosti V, Lucotti C, Malabarba L, Novella A, Vercesi E, Frassoni F, Cazzola M (1998) Unbalanced X-chromosome inactivation in haemopoietic cells from normal women. Br J Haematol 102:996–1003 10.1046/j.1365-2141.1998.00867.x [DOI] [PubMed] [Google Scholar]
- 43.Hatakeyama C, Anderson CL, Beever CL, Penaherrera MS, Brown CJ, Robinson WP (2004) The dynamics of X inactivation skewing as women age. Clin Genet 66:327–332 10.1111/j.1399-0004.2004.00310.x [DOI] [PubMed] [Google Scholar]
- 44.Orstavik KH, Orstavik RE, Schwartz M (1999) Skewed X chromosome inactivation in a female with haemophilia B and in her non-carrier daughter: a genetic influence on X chromosome inactivation? J Med Genet 36:865–866 [PMC free article] [PubMed] [Google Scholar]
- 45.Belmont JW (1995) Insights into lymphocyte development from X-linked immune deficiencies. Trends Genet 11:112–116 10.1016/S0168-9525(00)89012-5 [DOI] [PubMed] [Google Scholar]