Abstract
Although balanced translocations are among the most common human chromosomal aberrations, the constitutional t(11;22)(q23;q11) is the only known recurrent non-Robertsonian translocation. Evidence indicates that de novo formation of the t(11;22) occurs during meiosis. To test the hypothesis that spatial proximity of chromosomes 11 and 22 in meiotic prophase oocytes and spermatocytes plays a role in the rearrangement, the positions of the 11q23 and 22q11 translocation breakpoints were examined. Fluorescence in situ hybridization with use of DNA probes for these sites demonstrates that 11q23 is closer to 22q11 in meiosis than to a control at 6q26. Although chromosome 21p11, another control, often lies as close to 11q23 as does 22q11 during meiosis, chromosome 21 rarely rearranges with 11q23, and the DNA sequence of chromosome 21 appears to be less susceptible than 22q11 to double-strand breaks (DSBs). It has been suggested that the rearrangement recurs as a result of the palindromic AT-rich repeats at both 11q23 and 22q11, which extrude hairpin structures that are susceptible to DSBs. To determine whether the DSBs at these sites coincide with normal hotspots of meiotic recombination, immunocytochemical mapping of MLH1, a protein involved in crossing over, was employed. The results indicate that the translocation breakpoints do not coincide with recombination hotspots and therefore are unlikely to be the result of meiotic programmed DSBs, although MRE11 is likely to be involved. Previous analysis indicated that the DSBs appear to be repaired by a mechanism similar to nonhomologous end joining (NHEJ), although NHEJ is normally suppressed during meiosis. Taken together, these studies support the hypothesis that physical proximity between 11q23 and 22q11—but not typical meiotic recombinational activity in meiotic prophase—plays an important role in the generation of the constitutional t(11;22) rearrangement.
The t(11;22)(q23;q11) is the only known recurrent, non-Robertsonian constitutional translocation in humans.1,2 Factors that predispose to recurrence of this rearrangement have been difficult to identify. In general, 11;22-translocation carriers are phenotypically normal and are often identified only after the birth of abnormal offspring with an unbalanced form of the translocation. The supernumerary-der(22)t(11;22) syndrome (Emanuel syndrome [MIM #609029]) occurs as the result of 3:1 meiotic malsegregation of der(22).3 Individuals with Emanuel syndrome have a karyotype of 47,XX or XY,+der(22)t(11;22), and they present at birth with multiple congenital anomalies.1,4 The 11;22 translocation is rare, and its exact prevalence has not been determined. Clinical data indicate that there is a paucity of de novo translocation carriers in the population, and only a limited number of such occurrences have been identified in previous studies.5,6 Only one example was amenable to parent-of-origin determinations, and it was paternally derived. Translocation-specific der(11) and der(22) PCR products have been observed in DNA from sperm but not in somatic tissues isolated from karyotypically normal individuals, indicating that de novo translocations occur during male meiosis with a measurable frequency.7,8 Although the translocation has been identified in the sperm of normal males, it is not known whether it arises in female meiosis as well.
The genomic configuration of the breakpoint regions is likely to contribute to their propensity to rearrange. The 11q23- and 22q11-translocation breakpoint regions contain palindromic AT-rich repeats (PATRRs)9,10 consisting of a long AT-rich DNA sequence followed by its inverse complement. Palindromic sequences have the ability to self-pair, forming intrastrand hairpin or cruciform structures,11 and cloned DNA sequences derived from the 11q23 palindromic breakpoint form such structures in vitro.8,12 Further, polymorphisms of the PATRR11 sequence at the breakpoint alter the frequency of de novo translocations in gametes of normal males.8
Palindromic sequences represent a source of genetic instability in the genomes of many organisms through their potential to adopt these secondary structures, which can perturb a variety of biological processes. Hairpin structures can halt the progress of the replication fork and are also known to be intermediates in specialized mammalian recombination reactions such as V(D)J recombination.13 In mice, rearrangements of a palindromic transgene are consistent with a center-break mechanism where double-strand breaks (DSBs), created by hairpin nicking of an extruded cruciform, are imperfectly rejoined, creating microdeletions of a few nucleotides.14 In Escherichia coli, long palindromes are highly unstable and cannot be directly PCR amplified or sequenced because of their propensity to form intrastrand hairpins. The E. coli SbcCD-enzyme complex cuts near the tip of the hairpin, allowing homologous recombination to restore DNA replication.15 Thus, it has been suggested that cruciform extrusions of the palindromes at the 11q23 and 22q11 breakpoints might be similarly sensitive to enzymes that induce meiotic DSBs.7,9 In fact, when the 11;22 translocation forms, it sustains small symmetric central deletions at the site of the breakpoints, which supports the hypothesis that hairpin nicking of an extruded cruciform is a likely prelude to the repair that generates the translocation.7,9
Meiosis must provide the enzymatic machinery for translocation initiation and resolution, since the t(11;22) does not occur in somatic cells. Programmed DSBs, followed by DNA repair and recombination, are required for proper segregation of homologous chromosomes during meiosis. A break at the chromosome 11 or 22 PATRR hairpin extrusion might represent either (1) one of these programmed meiotic DSBs or (2) a break arising solely from the secondary structure of the palindrome. If the 11;22-translocation breakpoints are located at programmed DSBs, they should coincide with cytologically mapped hotspots of recombination. If they are rare palindrome-associated events and not programmed DSBs, the translocation breakpoints would be less likely to correspond to recombination hotspots. Whereas genetic analysis of chromosome 22 has shown that a peak of meiotic recombination coincides with the breakpoint region at 22q11,16 cytological examination of sites of crossing over has shown that distal events are twice as frequent as pericentromeric events in the vicinity of the breakpoint in males.17,18 In contrast, chromosome 11 has not yet been the subject of detailed cytological analysis, although this type of study has been performed for numerous other human chromosomes.19,20 Thus, the role of normal meiotic recombination in the genesis of the 11;22 translocation is unknown.
Programmed DSBs occur throughout the genome during meiotic prophase. This prompted us to seek a physical explanation for why the t(11;22) constitutional translocation recurs, whereas other balanced translocations, which occur in ∼0.1% of newborns,21 do not. There is evidence that individual chromosomes occupy compartmentalized domains and unique nuclear positions in the interphase nucleus, rather than being distributed in a random fashion.22–25 Studies of somatic cells have demonstrated that regions that preferentially rearrange with one another to produce nonrandom, tumor-associated rearrangements are more likely to be in closer proximity to one another in the interphase nucleus.26,27 Although analysis of sperm demonstrates the prevalence of chromosome 11 and 22 rearrangement in meiosis,7,8 little has been done to examine meiotic prophase chromosomal domains and recombinational behavior as a prelude to this rearrangement.
Molecular cytogenetics (i.e., FISH) and immunocytochemistry (i.e., fluorescent antibody localization [FAL]) provide powerful tools to address the question of meiotic chromosomal position and recombinational activity as a prelude to interchromosomal rearrangement. MLH1, a mismatch-repair protein, is one of several required for reciprocal recombination (crossing over).28,29 Antibodies to MLH1 can be used to identify sites of crossing over during meiosis.28,30,31 These combined FISH and FAL techniques have yet to be used to address the question of meiotic chromosomal position and recombinational activity. To our knowledge, the present study is the first to use FISH and FAL together to examine the location of chromosomes 11 and 22 during both male and female meiosis and the position of recombination events relative to the translocation breakpoint positions as determined by MLH1 foci. From these studies, it can be concluded that the position and secondary DNA structure—but not the programmed recombinational activity of 11q23 and 22q11 in the meiotic prophase nucleus—play an important role in the generation of the recurrent constitutional t(11;22) rearrangement.
Material and Methods
Ovarian and Testicular Samples
All samples were obtained with the appropriate Institutional Review Board approval. Fetal ovaries from first-trimester abortuses were obtained, by M.M.C., from Greater Baltimore Medical Center’s Cytogenetic Laboratory, and adult testicular samples were obtained either from the Cooperative Human Tissue Network or from biopsies performed by A.S. at University Hospitals of Cleveland or Cleveland VA Medical Center.
Tissue Preparation
Two different protocols were used: “classic” 3:1 methanol:acetic acid fixation and “microspread” paraformaldehyde fixation. For the classic preparations, the tissue was minced in Hanks balanced salt solution. Following hypotonic treatment (0.075 M KCl solution), the meiotic cells were fixed in a 3:1 solution as described elsewhere,32 with minor modifications. The microspreads of spermatocytes were prepared as described elsewhere.17
FAL
Antibodies for the following proteins were used: SCP3, a component of the axial/lateral elements of the synaptonemal complex33; MLH1 (Pharmingen), a mismatch-repair protein that is required for meiotic recombination28; and human CREST, an autoimmune serum that detects kinetochores (centromeres) derived from individuals with CREST (calcinosis, Raynaud phenomenon, esophageal dysmotility, sclerodactyly, telangiectasia). The antibodies were: goat anti-SCP3 diluted 1:1,000,34 mouse monoclonal anti-MLH1 (Pharmingen) diluted 1:200, and CREST diluted 1:2,000. Secondary antibodies included: rhodamine (TRITC)-labeled anti-goat, FITC-labeled anti-mouse, and Cy5-labeled anti-human (all from Jackson ImmunoResearch). Application of the antibodies and their detection was as described elsewhere.35 After detection, the spermatocytes were counterstained with 4',6-diamidino-2-phenylindole (DAPI).
DNA Probes
Prelabeled chromosome-specific whole-chromosome paint (WCP) probes for chromosomes 6, 11, and 22 were purchased from Vysis. The locus-specific FISH probe for chromosome 21 was the prelabeled LSI 21 probe purchased from Vysis. The probe covers ∼200 kb in 21q22.13-22.2 and encompasses markers D21S342, D21S341, and D21S529. It is located at a distance ∼38 Mb from the centromere of chromosome 21 (UCSC Genome Browser). The chromosome 22 locus–specific FISH probe, c87f9, was a cosmid isolated from the LL22NCO3 cosmid library. The chromosome 11 locus–specific probe (BAC 442e11 [GenBank accession number AC007707]) and the 6q26 control locus-specific probe (BAC 849d12) are from the RPC11 human BAC library (Roswell Park Cancer Institute). The BAC and cosmid DNA were isolated with Qiagen Maxi Prep kits (Qiagen) and were labeled by nick translation with tetramethyl-rhodamine-5-dUTP or fluorescein-12-dUTP (Amersham).
FISH
FISH of the 3:1 fixed material was performed as described elsewhere,36 with minor modifications. After image acquisition of the FAL signals, the microspread spermatocyte preparations were denatured, and the probes were hybridized as described elsewhere,31 with the following exceptions: the probes were denatured at 85°C for 6 min and were preannealed for 15 min, and the hybridization step was extended to 72 h.
Image Analysis
All images were digitally captured using IP Lab Software (Applied Imaging). For the classic preparations, tools within IP Software were used to measure (in pixels) the distance between the FISH signals of the locus-specific probes. For chromosomes 11 and 21, the distance between the 11q BAC and the short arm of chromosome 21 was measured by specifying 21p as the end of the bivalent farthest from the LSI 21 signal. Measurements in pixels were converted to micrometers (1 μm=7.5 pixels) with use of a slide micrometer.
Collection of the FISH and FAL images from the microspread spermatocytes was a two-step procedure. The FAL data were first imaged on a Nikon Eclipse E800 fluorescence microscope equipped with a Photometrics-cooled charge-coupled device camera CH350, and the location of each cell was recorded electronically with IPLab software (version 3.6.3) (Scanalytics). Following completion of the FISH procedure, each nucleus previously imaged for the FAL experiment was electronically relocated and the FISH images captured. Although much of the antibody staining was lost during the denaturation step for FISH, enough remained to superimpose the images.
Chromosomes 11 and 22 were identified by the BAC and cosmid signals. MLH1 distribution and probe-separation distances along these synaptonemal complexes (SCs) were measured using MicroMeasure, a 32-bit Windows application,37 and the locations of each MLH1 focus and probe on the respective SCs were recorded as a relative position, with use of distance (fraction of SC arm length) from the centromere (identified by CREST).
Sequence Analysis
Ensembl (Build 35) was used to ascertain the available sequence for chromosome 21. Database searches were performed on the sequence from chromosome 21, to identify palindromes. The sequence for chromosome 21 was divided into two large contigs, 9,719,768–10,210,000 and 13,260,001–46,944,323. The sequence was examined using the EMBOSS palindrome-recognition program (PALINDROME). The following parameters were applied to the sequence contigs: arm size >60 bp, mismatch between arms <10 bp, and spacer <20 bp. Only AT-rich palindromes thus identified were subjected to further analysis with use of mfold,38 to calculate the free energy for each. This was determined by entering the obtained sequences into the mfold server.
Results
FISH Analysis of Chromosomes 11 and 22 in Oocytes (3:1 Preparations)
To determine whether regions that preferentially rearrange are located in proximity to one another in the meiotic prophase nucleus, the distance between 11q23 and 22q11 in oocytes and spermatocytes was assessed. Oocyte nuclei were first classified as to meiotic stage of meiotic prophase on the basis of microscopically observed cytologic characteristics: degree of chromosomal pairing and condensation. Of the 200 oocyte nuclei analyzed, 13.5% were in leptotenema, 24% in zygonema, 49% in pachynema, and 13.5% in diplonema. Since recombination occurs during pachynema, only oocytes in this stage were selected for further analysis after FISH analysis with DNA probes.
Dual-color FISH was performed on the fetal pachytene oocytes with use of four or six probes simultaneously. WCP probes labeled with either FITC or rhodamine were used to identify individual bivalents (11 with 22; 6 with 22; 11 with 21 and 22), and single-copy, locus-specific probes were used to identify the breakpoint regions on the test bivalents (11 and 22) or the selected regions on the control bivalents (6 or 21). The single-copy probes were labeled with the alternative color fluor to that of the WCP. The WCP- and probe-labeling combinations for the individual experiments are shown in table 1.
Table 1. .
Probes and Labeling Scheme for FISH Experiments
| WCP for Chromosome |
Probe (Chromosome) |
|||||||||
| Experiment | 11 | 22 | 6 | 21 | c87f9 (22) | B442e11 (11) | B849d12 (6) | LSI 21 (21) | No. of Cells | Sample Type |
| 1 | Rhodamine | FITC | … | … | Rhodamine | FITC | … | … | 50 | Oocytes (3:1) |
| 2 | Rhodamine | FITC | … | … | Rhodamine | FITC | … | … | 50 | Oocytes (3:1) |
| 3 | … | Rhodamine | FITC | … | FITC | … | Rhodamine | … | 50 | Oocytes (3:1) |
| 4 | … | Rhodamine | FITC | … | FITC | … | Rhodamine | … | 48 | Oocytes (3:1) |
| 5 | FITC | Rhodamine | … | FITC | FITC | Rhodamine | … | Rhodamine | 50 | Oocytes (3:1) |
| 6 | FITC | Rhodamine | … | … | FITC | Rhodamine | … | … | 50 | Spermatocytes (3:1) |
| 7 | … | Rhodamine | FITC | … | FITC | … | Rhodamine | … | 47 | Spermatocytes (3:1) |
| 8 | FITC | Rhodamine | … | FITC | FITC | Rhodamine | … | Rhodamine | 50 | Spermatocytes (3:1) |
| 9a | … | … | … | … | Rhodamine | FITC | … | … | 144 | Spermatocytes (microspread) |
In experiment 9, the marker for chromosome 21 was the CREST antibody.
To determine the distance between 11q23 and 22q11 in oocytes, two experiments were performed (experiments 1 and 2). The labeling scheme is described in table 1, and representative images are shown in figure 1A and 1B. See table 2 for data.
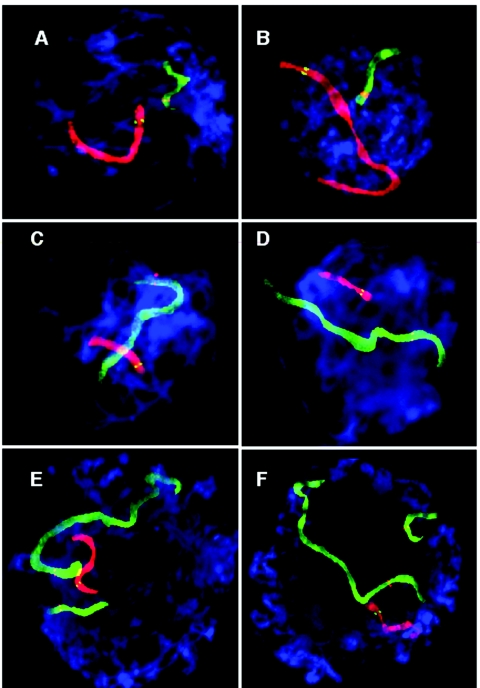
Figure 1. .
FISH-labeled human pachytene oocytes (3:1 preparation). Chromosomes were cohybridized with WCPs and locus-specific probes. For details of the probes and labeling scheme, see table 1. A and B, WCP11 is shown in red, with the 11q23 locus-specific probe shown in green; WCP22 is shown in green, with the 22q11 locus-specific probe in red (table 1 [experiments 1 and 2]). C and D, WCP6 is shown in green, with the 6q26 locus-specific probe shown in red; WCP22 is shown in red, with the 22q11 locus-specific probe shown in green (table 1 [experiments 3 and 4]). E and F, WCP11 is shown in green, with the 11q23 probe in red; WCP22 is shown in red, with the 22q11 probe shown in green; WCP21 is shown in green, with the 21q11 probe in red (table 1 [experiment 5]).
Table 2. .
Distances between Probes for Three Females and Three Males[Note]
| Distancebetween Probes(μm) |
|||||
| Experiment and Sample |
Probe Combinations | No. of Cells Observed | Mean (SD) | Range | Pa |
| 1: | |||||
| Female 1 | 11q23+22q11 | 50 | 14.5 (8.5) | 0–33.2 | |
| 3: | |||||
| Female 1 | 6q26+22q11 | 50 | 18.1 (6.7) | 5.2–32.1 | .02 |
| 2: | |||||
| Female 2 | 11q23+22q11 | 50 | 11.0 (8.8) | 0–39.6 | |
| 4: | |||||
| Female 2 | 6q26+22q11 | 48 | 35.2 (11.3) | 4.1–65.0 | <.001 |
| 6: | |||||
| Male 1 | 11q23+22q11 | 50 | 8.5 (7.2) | 0–28.0 | |
| 7: | |||||
| M1 | 6q26+22q11 | 47 | 35.6 (10.0) | 14.6–55.5 | <.001 |
| 5: | |||||
| Female 3 | 11q23+22q11 | 50 | 14.5 (9.4) | 0–35.2 | |
| Female 3 | 11q23+21p11 | 50 | 26.2 (12.1) | 7.1–52.2 | <.001 |
| 8: | |||||
| Male 2 | 11q23+22q11 | 50 | 20.5 (11.7) | 0–47.8 | |
| Male 2 | 11q23+21p11 | 50 | 24.4 (10.8) | 3.9–52.2 | >.05b |
| 9: | |||||
| Male 3 | 11q23+22q11 | 145 | 21.1 (9.3) | 1.1–44.9 | |
| Male 3 | 11q23+21q11 | 144 | 23.4 (9.8) | 3.8–43.0 | .04 |
Note.— All measurements were taken from 3:1 fixed cells, except male 3, which was from microspread preparations.
Between probes in same individual (t tests).
Not significant.
As a control, the distance between 6q26 and 22q11 was compared with the distance between 11q23 and 22q11. Chromosome 6 was selected as a control for three reasons. Chromosome 6 is similar in size to chromosome 11, 6q26 is approximately the same fractional distance from the long arm telomere as is 11q23, and chromosome 6 has not been involved in recurrent rearrangements with 22q11. A BAC that maps to 6q26 (849d12) was selected as the control probe. The labeling scheme is described in table 1 (experiments 3 and 4). Representative images are shown in figure 1C and 1D). See table 2 for data.
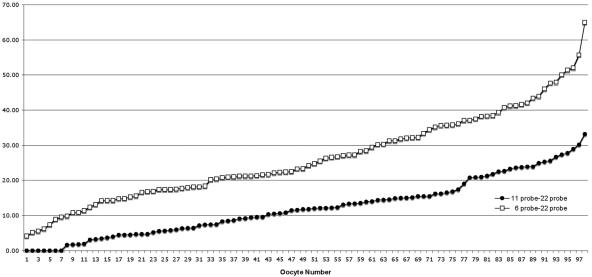
In these experiments, the distances between the relevant 11q23 and 22q11 or 6q26 and 22q11 single-copy probes were measured, and the two experiments for each bivalent pair were combined. The 98 oocytes were ranked by increasing probe-to-probe distance, and the data were plotted. Figure 2 compares the distance between chromosome 22 and either (1) chromosome 6 (open squares) or (2) chromosome 11 (filled circles) from the combined experiments (experiments 1–4). The Y-axis indicates the distance between probes from individual oocytes, which are enumerated on the X-axis to indicate the trend. Overall, it appears that 11q23 is closer to 22q11 than is 6q26, with average separation distances of 11–14.5 μm and 18–35 μm, respectively (table 2). These differences were statistically significant (P⩽.02).
Figure 2. .
Comparison of distances between 11q23 and 22q11 and between 6q26 and 22q11. Probe-to-probe distance is shown in micrometers on the Y-axis. Distances between 11q23 and 22q11 locus-specific probes are shown as filled circles and between 6q26 and 22q11 as open squares.
The distance of chromosome 21 (short arm) to 11q23 was selected as another control, to compare with the distance between the breakpoints on 22 and 11. Chromosome 21 was selected as a control for chromosome 22, because it is similar in size and morphology to 22, 21p is adjacent to the centromere of a small acrocentric chromosome, and chromosome 21 has not been involved in recurrent rearrangements with 11q23. For these experiments, all three bivalents (11, 21, and 22) were painted with WCPs, and the locus-specific probes were labeled in the fluor color alternative to that of the WCP (fig. 1E and 1F). Although there were several instances where chromosome 21 was closer to 11q23 than was 22q11, 22q11 was usually closer to 11q23 than was chromosome 21 in the 50 oocytes evaluated. The average distance (±SD) between 22q11 and 11q23 was 14.5±9.4 μm, whereas that between 21 and 11q23 was 26.2±12.1 μm. This difference is significant (P<.001), although less striking than that observed for the distances between 11 and 22 versus between 6 and 22.
FISH Analysis of Chromosomes 11 and 22 in Spermatocytes (3:1 Preparations)
Similar experiments were performed on spermatocytes. As can be seen from the representative images in figure 3A and 3B, 11q23 and 22q11 were often in very close proximity to one another, even occasionally overlapping in the vicinity of the breakpoint probe. The distance between 11q23 and 22q11 or 6q26 and 22q11 (fig. 3C and 3D) was measured in 50 (11q23) or 47 (6q26) spermatocytes (table 2 [experiments 6 and 7]). The average distance between 6q26 and 22q11 was more than four times greater than between 11q23 and 22q11 (table 2), a value that was highly significant (P<.001).
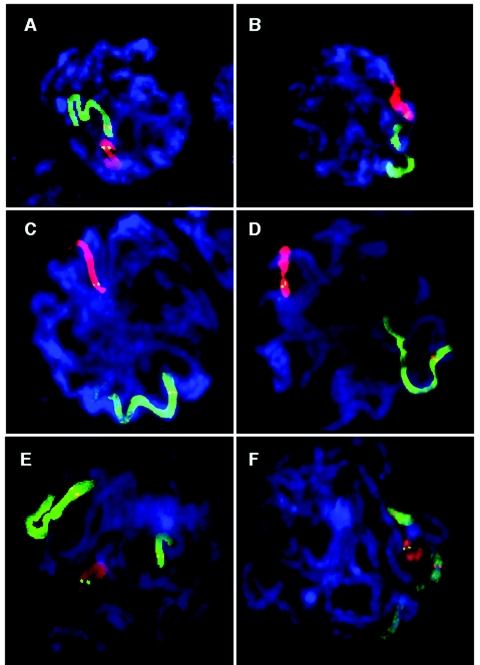
Figure 3. .
FISH-labeled human pachytene spermatocytes (3:1 preparation). Chromosomes were cohybridized with WCPs and locus-specific probes. For details of the probes and labeling scheme, see table 1. A and B, WCP11 is shown in green, with the 11q23 locus-specific probe shown in red; WCP22 is shown in red, with the 22q11 probe shown in green (table 1 [experiment 6]). C and D, WCP6 is shown in green, with the 6q26 probe shown in red; WCP22 is shown in red, with the 22q11 probe shown in green (table 1 [experiment 7]). E and F, WCP11 is shown in green, with the 11q23 probe shown in red; WCP22 is shown in red, with the 22q11 probe shown in green; WCP21 is shown in green, with the 21q11 probe shown in red (table 1 [experiment 8]).
In experiments corresponding to those for oocytes, the distance between 21p11 and 11q23 was compared with the distance between the breakpoints on 22 and 11 in the spermatocytes. For these experiments, see table 1 for the labeling scheme, table 2 (experiment 8) for the data, and figure 3E and 3F for examples. On average, 22q11 is closer to 11q23 (20.5±11.7 μm) than is chromosome 21 (24.4±10.8 μm) in the 50 spermatocytes evaluated, but this difference was not statistically significant. This is in contrast to the oocyte experiments, in which the average separation distances between these two probes were significantly different. Whereas there is less separation between the 22q11 breakpoint region and the 11q23 breakpoint region, as compared with the same measurement for the 21p11 control region, the difference between the measurements was less striking than it was in the experiments comparing 6q26 and 22q11 versus 11q23 and 22q11. This suggests that proximity is not the only factor that plays a role in generating the translocation.
FISH Analysis of Chromosomes 11 and 22 in Spermatocytes (Microspread Preparations)
To determine whether preparative technique might affect proximity results, the distances between 11q23 and 22q11 versus 11q23 and 21p11 were evaluated in microspread spermatocytes. For these experiments, locus-specific probes were used to identify chromosomes 11 and 22. No probe was necessary to identify chromosome 21, since it is the smallest autosomal bivalent. The CREST antibody signal, rather than a locus-specific probe, was used for chromosome 21, and measurements were made from the distal border of the CREST signal. The average distances between either the 11q23 and 22q11 probes or 11q23 and 21q11 probes were not significantly different from those observed using 3:1 fixation (table 2 [experiment 9]). Unlike the 3:1 fixation in spermatocytes, the distances between chromosomes 11 and 22 and between chromosomes 11 and 21 were different for the microspread spermatocytes (P=.04). However, the level of significance is not high, and the large number of observations (n=145 and n=144) presumably contributed to the ability to detect the slight difference between the two probe combinations.
Comparison between Oocyte and Spermatocyte Preparations
To begin to address the question of whether there are differences in chromosomal position between oocytes and spermatocytes, the data sets for the two sexes were compared. For both oocyte and spermatocyte 3:1 preparations, there was at least one instance of zero distance between the 11 and 22 single-copy probes, whereas control probes (6–22 or 11–21) always had at least some distance between them (table 2). The distance between 6 and 22 is similar in oocytes and spermatocytes for experiments 4 and 7. The two data sets indicate that there is little effect of possible size differences between the two cell types. This is not the case when experiment 3 is compared with experiment 7. The distances between 6 and 22 are greater than the distances between 11 and 22 in both oocytes and spermatocytes. In addition, there appears to be a trend for 11 and 22 to lie closer to one another in oocyte 3:1 preparations than in spermatocytes prepared the same way (except for male 1). The data suggest a potential interindividual difference, as well as differences between oocytes and spermatocytes.
PATRRs and Chromosome 21
Since chromosomes 21 and 22 seem to lie at similar distances from chromosome 11q23 in meiotic prophase nuclei, most notably in spermatocytes, nuclear position alone is insufficient to explain the recurrent t(11;22). The presence of palindromic sequences on any given chromosome might be predicted to influence translocation permissiveness.39 To determine whether there exists a PATRR on chromosome 21 that is as conducive to translocation with 11q23 as is the PATRR on chromosome 22, a sequence search with the DNA sequence for chromosome 21 was performed with the PALINDROME software. The minimum length of a palindrome was set at 60 nt, the gap length was set at 20 nt, and mismatch was set at 10 nt. The search identified 160 palindromic sequences within the two large sequence contigs that exist for chromosomes 21. Formation of hairpin secondary structures requires denaturation of double-stranded chromosomal DNA containing an inverse complement such that the single-stranded DNA can self-anneal into a stem loop. The stability of a particular strand of DNA in its double-stranded configuration or in a secondary structure formed within a single strand can be described by separate Gibbs free energy (G) values. Because a maximum amount of base pairing occurs in double-stranded DNA, it is inherently more stable and always has a free energy value (GDS) that is more negative than that of a single strand folding within itself (GSTRUC). The free energy of a single strand of sequence forming a stem loop depends on the position and number of complementary base pairs, as well as the GC content of these nucleotides.
The palindromic sequences were submitted to the mfold server that provided free energy values (GSTRUC) as part of the output for the PATRRs. Of the 160 palindromes studied, 25 sequences were identified with a GSTRUC lower than −50. For these 25 sequences, the GDS was calculated (table 3). This was accomplished by “annealing” the sequence to its reverse complement in silico, reanalyzing the DS sequence, and halving the output. Free energy for the formation of a secondary structure (ΔG) is the GDS-GSTRUC difference. Small ΔGs are predicted to be more susceptible to hairpin extrusion and breakage. When the ΔGs of the 25 chromosome 21 PATRR-like sequences were compared with those for PATRR17, -11, and -22, it was determined that all of the chromosome 21 PATRR ΔG values were greater than those calculated for PATRR17, -11, and -22. These data indicate that there is no PATRR on chromosome 21 that is likely to engage in translocation with 11q23 at a frequency similar to what has been determined for the translocation-permissive chromosome 17 and the chromosome 22 PATRRs. This suggests that proximity and presence of PATRRs alone do not predispose to translocation; rather, palindromes with specific characteristics are required.
Table 3. .
Palindromic Sequences Identified on Human Chromosome 21
| Locationin Chromosome 21 |
|||||||
| Palindromea | Start | End | Length (nt) |
GSTRUC (kcal/mol) |
GDS (kcal/mol) |
ΔG (kcal/mol) |
ΔG/nt (kcal/mol) |
| 5 | 16108261 | 16108407 | 147 | −62.69 | −94.97 | 32.28 | .2196 |
| 10 | 17041640 | 17041792 | 153 | −55.27 | −71.21 | 15.94 | .1042 |
| 22 | 20574998 | 20575241 | 244 | −119.23 | −135.13 | 15.90 | .0651 |
| 27 | 20951787 | 20951942 | 156 | −65.74 | −87.45 | 21.71 | .1391 |
| 35 | 24454851 | 24455058 | 208 | −81.69 | −129.21 | 47.52 | .2285 |
| 36 | 24615664 | 24615817 | 154 | −5.40 | −82.27 | 31.87 | .2069 |
| 38 | 24927548 | 24927717 | 170 | −51.76 | −81.01 | 29.25 | .1720 |
| 39 | 25121832 | 25122002 | 171 | −61.11 | −102.27 | 41.16 | .2407 |
| 48 | 28030093 | 28030220 | 128 | −54.32 | −73.10 | 18.78 | .1467 |
| 55 | 29263048 | 29263183 | 136 | −54.58 | −98.61 | 44.03 | .3238 |
| 83 | 33617249 | 33617425 | 177 | −56.89 | −89.28 | 32.39 | .1830 |
| 84 | 33617282 | 33617448 | 167 | −61.12 | −82.33 | 21.21 | .1270 |
| 86 | 33617284 | 33617461 | 178 | −65.93 | −88.45 | 22.52 | .1265 |
| 89 | 33617296 | 33617477 | 182 | −67.52 | −9.31 | 22.79 | .1252 |
| 91 | 33617305 | 33617496 | 192 | −77.01 | −97.06 | 2.05 | .1044 |
| 93 | 33617312 | 33617489 | 178 | −74.11 | −87.74 | 13.63 | .0765 |
| 94 | 33617314 | 33617515 | 202 | −8.63 | −10.71 | 2.08 | .0994 |
| 96 | 33617342 | 33617515 | 174 | −68.09 | −86.28 | 18.19 | .1045 |
| 99 | 33758611 | 33758766 | 156 | −72.62 | −113.39 | 4.77 | .2613 |
| 104 | 37508486 | 37508642 | 157 | −7.80 | −11.29 | 39.49 | .2515 |
| 147 | 42468805 | 42468949 | 145 | −63.73 | −96.84 | 33.11 | .2283 |
| 149 | 43404786 | 43404925 | 140 | −67.39 | −96.42 | 29.03 | .2073 |
| 152 | 44600367 | 44600499 | 133 | −7.47 | −99.51 | 29.04 | .2183 |
| 153 | 45475447 | 45475586 | 140 | −6.17 | −104.79 | 44.62 | .3187 |
| 154 | 46446323 | 46446567 | 245 | −142.41 | −18.51 | 38.10 | .1555 |
| PATRR11b | 445 | −178.40 | −196.25 | 17.85 | .0401 | ||
| PATRR17c | 187 | −89.47 | −92.11 | 2.64 | .0141 | ||
| PATRR22d,e | 582 | −313.60 | −324.75 | 11.15 | .0192 | ||
Palindromes with GSTRUC less than −50 kcal/mol were analyzed.
PATRR22 was inferred from the two junction-fragment sequences on der(11) and der(22).
FAL Analysis to Determine Crossover Distribution on Chromosomes 11 and 22
Reciprocal exchange between chromosomes 11 and 22 requires DSBs and DNA repair. To determine whether the position of the breakpoints coincides with normal hotspots of meiotic recombination during pachynema, MLH1 was used to identify crossover sites. In addition, an antibody to SCP3, a component of the axial/lateral elements of the SC, was used to visualize the SCs, and CREST, a human autoimmune serum that localizes to kinetochores, was used to identify the centromere regions and distinguish between the long and short arms (fig. 4).
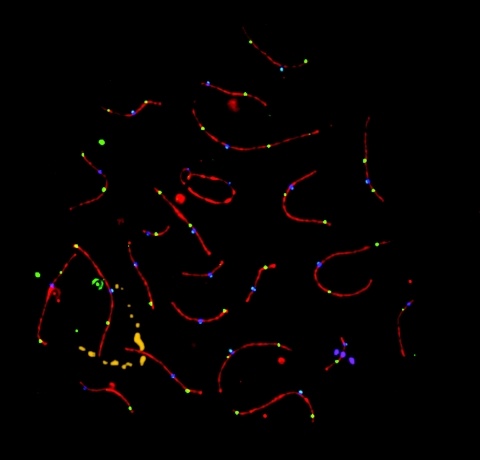
Figure 4. .
Immunostained and FISH-labeled human pachytene spermatocyte (microspread preparation). SCP3 is shown in red, MLH1 in green, and CREST in blue. In addition, the 11q23 probe is shown in gold, and the 22q11 probe is shown in magenta. The entire nucleus is stained with DAPI (cyan). MLH1 foci mark crossover sites.
For chromosome 11, FISH with the BAC 442e11 probe provided both a positive identification of the bivalent and a marker for the breakpoint region. Several previous human MLH1 mapping studies have divided the chromosomal arms into fourths and have assigned each focus to quarter segments.17,19 To better compare the distribution patterns for chromosomes 11 and 22 at a higher resolution, we used the procedure of Froenicke et al.31 to map MLH1 foci in 0.2-μm segments for the relevant chromosomal bivalents. The distribution graphs for MLH1 are shown in figures 5 and 6.
Figure 5. .
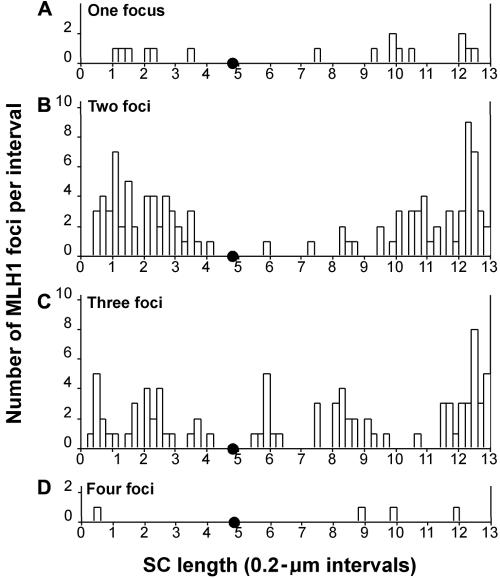
Histograms of the distribution of MLH1 on chromosomal bivalent 11. The X-axis indicates the position (in μm) of MLH1 foci along the length of the bivalent. The position of the centromere is indicated by a small blackened circle; the p arm lies to the left, and the q arm lies to the right. The Y-axis indicates the number of MLH1 foci mapped to each interval. A, Distribution of MLH1 on bivalents with only one MLH1 focus. B, Distribution of MLH1 on bivalents with two foci. C, Distribution of MLH1 on bivalents with three foci. D, Distribution of MLH1 on the single bivalent with four foci.
Figure 6. .
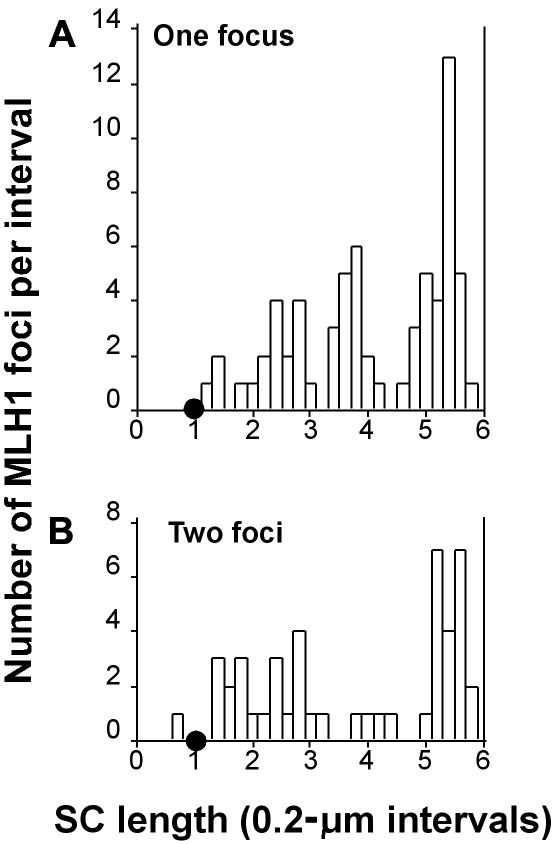
Histograms of the distribution of MLH1 on chromosomal bivalent 22. The X-axis indicates the position (in μm) of MLH1 foci along the length of the bivalent. The position of the centromere is indicated by a small blackened circle; the p arm lies to the left, and the q arm lies to the right. The Y-axis indicates the number of MLH1 foci mapped to each interval. A, Distribution of MLH1 on bivalents with only one focus. B, Distribution of MLH1 on bivalents with two foci.
The number of MLH1 foci on the chromosome 11 bivalent varied between one and four, with a mean of 2.17 (SD 0.70) (n=101). Of these pachytene nuclei, 16% had a single MLH1 focus, 52% had two foci, and 31% had three foci. In addition, one chromosome 11 bivalent had four MLH1 foci. The distribution of MLH1 foci along the SC bivalent in each category is presented in figure 5. No MLH1 foci localized near the centromere (located at ∼4.8 μm on the histograms), consistent with previous observations on biarmed chromosomes in human spermatocytes.17,19 Consistent with reported high rates of recombination in human males, the largest MLH1 peak on chromosomal bivalent 11 was near but not at 11qter. However, there were also a few MLH1 foci in the terminal 0.2 μm of 11q.
It has long been recognized that when two crossovers occur on the same chromosome, they do not occur close together, a phenomenon called “positive crossover interference.”40 Therefore, it is not surprising that a clear bimodal distribution pattern exists in those chromosome 11 bivalents with two foci (figs. 5 and 7). Within this group, one MLH1 focus was generally located on each arm, which suggests that the crossover interference signal can be transmitted across the centromere. In the middle of each of these “peaks,” there was a distinct trough—a 0.2-μm region that had no MLH1 foci.
Figure 7. .
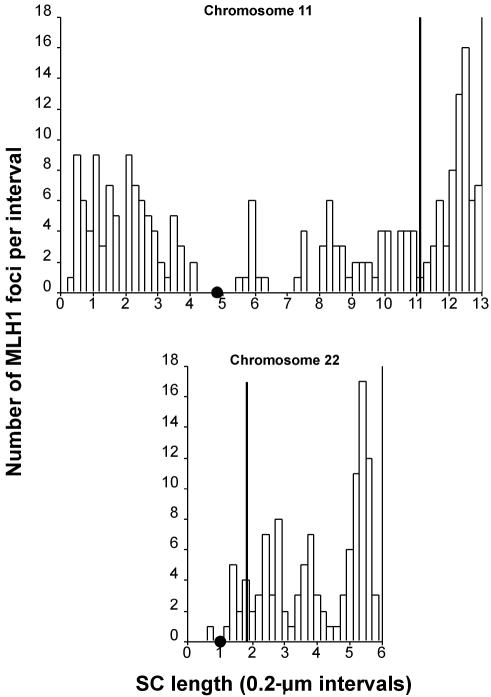
Histograms of the distribution of all MLH1 foci relative to the position of the probe for chromosomal bivalents 11 and 22. The X-axis indicates the position (in μm) of MLH1 foci (unblackened bars) and the probe (solid line) along the length of the bivalent. The chromosomes are positioned relative to one another on the X-axis on the basis of centromere position, which is indicated by a small blackened circle; the p arm lies to the left, and the q arm lies to the right. The Y-axis indicates the number of MLH1 foci mapped to each interval, as well as the location of the probe.
Given the inhibitory effect of a second crossover occurring in close proximity to a first, one would predict the widest distribution spread in those bivalents with the highest number of crossovers—that is, very proximal and very distal crossovers on these bivalents. Consistent with this expectation, both the two-focus group and the three-focus group have MLH1 foci nearer the telomeres than any observed in the one-focus group. Since there was only one chromosome 11 bivalent with four foci, the sample size was too small to draw any further conclusions.
Chromosome 22 is the second-shortest in the human complement, and all chromosome 22 bivalents had either one or two foci, with the majority (73% [n=92]) having only one (mean 1.26; SD 0.44 [n=90]). The distribution of MLH1 foci along the SC of chromosomes 22 is presented in figure 6. Although only three MLH1 foci mapped to the terminal 0.2 μm of 22q, there was a major MLH1 peak near the end of 22q. Only one MLH1 focus mapped to the short arm of chromosome 22. When the fact that 22p consists of highly repetitive ribosomal sequences—in which recombination is known to be repressed—is considered, the paucity of MLH1 foci in this region is hardly surprising. However, unlike chromosome 11, there were a few crossovers very close to the centromere on chromosome 22.
When there were two MLH1 foci on the 22 bivalent, the distribution was bimodal. The distal peak on 22q in the group with two foci coincides with the main peak in the single-focus group. Rather than a second sharp peak of MLH1 foci on proximal 22q, there was a broad distribution of foci in the two-focus group. Contrary to the expectation of suppression of crossover near the centromere, in both the one- and two-focus groups, there were a few foci in the intervals adjacent to the centromere.
Location of the Breakpoints Relative to the Peaks of Recombination
The 11;22 translocation could arise as a result of an error in a normal meiotic recombination event (repair of a meiotically programmed DSB) or as an attempt to repair an unprogrammed DSB that arose as a result of the secondary structure of the PATRRs. To discriminate between these two possibilities, probes from the breakpoint regions on chromosomes 11 and 22 were hybridized and FISH mapped to microspread spermatocytes. The microspread preparative technique preserves the chromatin structure better than does the 3:1 fixation. Consequently, both the chromosome 11 BAC and the chromosome 22 cosmid produce a FISH signal that is more like that seen in fiber-FISH. Consistent with the differences in size, the chromosome 11 BAC (180 kb) produces a more elongated signal than does the cosmid on 22 (fig. 4). These signals represent the chromatin loops extending from the SC. The position where the signals from the two homologues crossed the SC was measured and mapped to a region 11.2 μm from the distal end of the chromosome 11 bivalent. As seen in figure 7, the location of the BAC 442e11 does not coincide with a cytological region of high recombination activity but with a trough of MLH1 on 11q. On the basis of the combined MLH1 and probe map, the exchange event that leads to the t(11;22) cannot be attributed to a hotspot of normal meiotic recombinational activity.
The 87f9 cosmid from the breakpoint region of chromosome 22 mapped ∼1.8 μm from 22pter and ∼0.8 μm distal to the centromere (fig. 7). Again, on the basis of the MLH1 map, this region is one of low recombinational activity, which argues against typical meiotic recombination as the mechanism of DSB repair leading to the t(11;22).
Discussion
The remarkable similarity between numerous constitutional t(11;22) breakpoint junctions derived from unrelated individuals suggested that, in addition to the presence of PATRRs at both breakpoints, proximity between these two chromosomal regions in the meiotic prophase nucleus is likely to facilitate the rearrangement. In recent years, studies on the spatial organization of chromosomes within the mitotic interphase nucleus have indicated that the arrangement of chromosomes and genes is nonrandom (for reviews, see Misteli41 and Parada et al.42). Gene-dense, euchromatic chromosomes are located more toward the center of the nucleus, and gene-poor, G-band–positive chromosomes are more peripherally positioned.24 Nonrandom proximity between nonhomologous chromosomes has been observed and proposed as playing a role in recurrent chromosomal rearrangements.43 Moreover, there seem to be changes in the position of specific loci and regions in different cell types.43 However, few such studies have attempted to examine chromosomal position during mammalian meiosis, and most of these have focused on alignment of homologues prior to synapsis.44 Only a few have looked at the spatial disposition of specific chromosomes during meiotic prophase.45,46 Thus, the current studies offer a first glimpse at comparative chromosomal organization of pachytene nuclei and indicate that chromosomal domains may not be randomly distributed and that relative positions may differ between the sexes.
These studies demonstrate that both chromosomes 21 and 22 reside close to 11q23 in the meiotic prophase nucleus, especially in males. It is not surprising that 21 and 22 reside close to one another, since they are both small chromosomes and previous ultrastructural studies found that the nucleolar organizer regions of human acrocentric bivalents associate with the nucleolus during meiotic prophase.47 However, despite the fact that they are close to one another and are both in proximity to 11q23, chromosome 21 does not engage in recurrent translocation with 11q23. The results reiterate the idea that proximity alone is insufficient to promote chromosomal rearrangement.
Specific DNA-sequence content and genomic configuration undoubtedly play significant roles in translocation permissiveness. For example, even though Robertsonian translocations take place between acrocentric chromosomes with homologous DNA sequence on their short arms, the prevalence of specific translocations among the potential chromosomal combinations differs. Thus, the more common recurrent Robertsonian translocations—t(13;14) and t(14;21)—have translocation breakpoints in the same chromosomal region with respect to specific satellite DNA subfamilies that are shared between them. These translocations arise primarily during oogenesis, when the chromosomes are in close proximity, perhaps through a common sequence-based mechanism.48 Similarly, the common breakpoints for the t(11;22) suggest that the recurrence of the translocation is based not only on the proximity of the two chromosomes but also on the presence of the rearrangement-permissive PATRRs.
The role of polymorphic variation in PATRR11 DNA sequence in generating the t(11;22) has recently been demonstrated.8 Polymorphisms of the PATRR11 in normal males results in a greater than threefold variation in susceptibility for generating the recurrent translocation in male gametes. Although DNA sequence variation influences translocation frequency, it is important to note that the chromosome 11 and 22 PATRRs appear to have limited cell-type susceptibility to interchromosomal rearrangement. Previous studies of DNA samples from normal cell lines from a variety of tissues—as well as those derived from individuals with Bloom syndrome or ataxia-telangiectasia, which are DNA-instability syndromes—did not identify translocation-specific PCR products.9 This indicates that production of the recurrent t(11;22) requires the conditions that exist during meiosis.
Since the t(11;22) rearrangement occurs during meiosis and obviously requires DNA breakage and repair, it is important to consider the likely enzymes and their availability during meiotic prophase. The MRE11 complex (MRE11/RAD50/NBS1 in mammals) is probably a key player. This complex has been implicated in both major DNA-repair pathways: nonhomologous end joining (NHEJ) and homologous recombination (see Assenmacher and Hopfner49 for review). The Mre11 complex recognizes and cleaves hairpin structures such as those adopted by the PATRRs on chromosomes 11 and 2250; thus, it is logical to predict binding under the current circumstances. Intriguingly, the Mre11 complex is also required for meiotic synapsis and recombination.51 Another prerequisite for meiotic recombination is programmed DSBs executed by SPO11, a topoisomerase.52
The Mre11 complex is not only a key component of DSB repair, but it is often prepositioned at sites vulnerable to a variety of DSBs. In addition to hairpin binding,53 the mammalian Mre11 complex binds to transcription factors at replication origins54 and continues to colocalize with replication forks as DNA synthesis proceeds.55 In meiosis in Saccharomyces cerevisiae, the Mre11 complex is part of a large prerecombination complex that includes SPO11 and is required for programmed meiotic DSBs.51 A similar role for the Mre11 complex has been proposed in mammals.56
Therefore, in humans, association of the Mre11 complex with the PATRRs during early stages of meiosis might theoretically assure their inclusion in a prerecombination complex. Alternatively, the Mre11 complex might associate with the PATRR hairpins and cleave them independent of SPO11. If binding of the Mre11 complex to the PATRR hairpins on chromosomes 11 and 22 were to assure inclusion of these sequences in a prerecombination complex, it would be logical to expect that most DSBs would be repaired via homologous recombination between homologues. If this scenario were correct, the breakpoints on chromosomes 11 and 22 should coincide with hotspots of meiotic recombination. As shown in the current study, this is not the case. The lack of such a correlation suggests that the DSB(s) giving rise to the translocation are not programmed meiotic DSBs but occur independently, as a result of the hairpin structure itself. Although programmed meiotic DSBs are thought to occur during leptonema or early zygonema, the time of breakage as a result of extruded hairpins remains unresolved.
Once the breaks occur, they must be repaired, a process that can leave “footprints” that offer clues related to the repair pathway used. Reconstruction of the original genomic DNA configuration that is based on end products of the translocation event for both derivative chromosomes indicates that the rearrangement resembles a process seen in mice and other eukaryotes called “center break palindrome modification.” The palindromic DNA sustains small central deletions that create junction fragments, a hallmark of NHEJ.57 Despite their AT-richness, no substantial homology has been observed between the PATRR11 and the PATRR22. The breakpoints on the two chromosomes possess only a small number of identical nucleotides. Since the DNA sequences that reside on 22 and 11 are not homologous to one another, it is unlikely that a typical homologous recombination pathway is responsible. In contrast, little homology is required for NHEJ. Thus, it appears that the mechanism for this recurrent chromosomal translocation involves DSBs at the two PATRRs, followed by their repair through a pathway that resembles NHEJ.
However, the meiotic process has evolved to assure homologous recombination. The rarity of ectopic recombination or exchange between homologous or homeologous sequences on nonhomologous chromosomes in mammals is testimony to the success of this strategy. One component of this meiotic tactic is the suppression of NHEJ during meiotic prophase. In somatic cells, NHEJ repairs hairpin lesions with the ku70/ku80/DNA-PK(cs) complex.58 The ku proteins are not present during early meiotic prophase in spermatocytes when programmed meiotic DSBs occur, even though DNA-PK is expressed throughout meiotic prophase. In fact, ku70 and ku80 do not reappear until midpachynema,56,59 approximately three to four days after meiotic programmed DSBs are presumed to occur. There are at least two explanations for this conundrum: (1) the palindromic breaks occur long after meiotic recombination events have been initiated and after the ku proteins are again available or (2) repair is accomplished without ku70/ku80 via a mechanism currently indistinguishable from ku-assisted NHEJ.
Variation in the regional rates of recombination has led to the identification of recombination hot- and coldspots on individual human chromosomes.60 In a detailed analysis of the genomic sequence of chromosome 22, a significant correlation between long-tandem GT repeats and recombination hotspots on human chromosome 22 was observed.16 One of these hotspots appears to reside in the general vicinity of the PATRR in 22q11. However, since the 22q11 PATRR represents one of the unclonable gaps in the sequence of chromosome 22, it is likely to have eluded this analysis. In addition, although the known PATRR sequence does not contain the GT repeat–motif characteristic of such a meiotic hotspot, the PATRR itself likely leads to a DNA structure susceptible to DSBs. This would occur as a result of cruciform extrusion promoting the DSBs that initiate stabilizing rearrangements or recombination events.61
Disease-related recombination on proximal 22 seems to contradict expectations of typical meiotic behavior. Haplotype reconstruction of 22q11-deletion cases demonstrates proximal interchromosomal exchanges between homologs giving rise to de novo deletions in 95% of those studied.18 By contrast, the normal chromosome 22 in the same deleted probands showed interchromosomal exchanges in <14% of informative meioses, a rate more in keeping with the genetic distance and cytological observations. Recombination, visualized as MLH1 foci, localizes to the distal long arm of chromosome 22 in the majority of human spermatocytes examined, also reflecting the genetic map.17,18 The MLH1 data presented here support the previous cytological studies indicating that more crossover events take place on the distal long arm than in 22q11.2. Thus, it is interesting that this chromosome is extremely susceptible to rearrangements of the proximal long arm but that these rearrangements do not appear to have been repaired via homologous recombination.
In fact, proximal chromosome 22q demonstrates a propensity to undergo aberrant meiotic homologous or nonhomologous repair events that result in translocations, interstitial deletions, and small marker chromosomes. The 11;22 translocation breakpoint at 22q11 has been localized within one of the low-copy repeats (LCRs) on 22q that produce other human chromosomal disorders, most notably the 22q11 deletion syndrome.3,10,62,63 Each of the LCRs on 22q11 extends over several hundred kilobases, and they share >95% sequence homology over short stretches. Synapsis of homologous chromosomes requires a molecular check for homology that involves single-strand DNA (ssDNA).64,65 If pairing problems resulted in sequence similarities but not identities between the LCRs, extensive unwinding of the DNA can be predicted to follow.64,65 This would permit LCR-B and the PATRR22 contained within it to persist as ssDNA, rendering it susceptible to cruciform extrusion and creating sites for Mre11 binding. In fact, the potential for unwinding longer stretches of DNA than would normally occur during replication in somatic cells might provide an explanation for the preferential occurrence of the 11;22 translocation during meiosis.
In addition to the t(11;22), other 22q11 translocation breakpoints cluster within the chromosome-specific LCR region that encompasses the PATRR22.12,39,66–71 The PATRR22 itself has been implicated in the etiology of these rare 22q11-related translocations. The data presented herein suggest that physical proximity between 11q23 and 22q11, in addition to the genomic instability introduced by the PATRRs, plays a role in facilitating the t(11;22). Greater physical separation in the prophase nucleus may account for the infrequent occurrence of these other PATRR22 single-translocation events. Verification of this hypothesis awaits further investigation. Thus, additional analysis of chromosomal domains and proximities in meiotic prophase may be warranted. The recurrence of the t(11;22) translocation reminds us of the potential for nonhomologous exchange during meiosis. Given the minimal estimate of ∼150 DSBs during meiotic prophase in human spermatocytes72 and the fact that a significant proportion of the human genome is repetitive DNA, such detailed analysis might provide us with an understanding of why there are so few constitutional chromosomal rearrangements.
Acknowledgments
The authors acknowledge the excellent technical assistance of Danielle Conforto. These studies were supported in part by National Institutes of Health (NIH) National Cancer Institute grant CA39926 (to B.S.E.), the Charles E. H. Upham chair in Pediatrics (B.S.E.), NIH General Medical Institute grant GM67846 (to T.A.), and National Science Foundation grant MCB-0314644 (to L.K.A.).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for BAC 442e11 [accession number AC007707], PATRR11 [accession number AF391129], PATRR17 [accession number AB195814], and PATRR22 [accession numbers AC087065 and AC074203])
- mfold, http://www.bioinfo.rpi.edu/applications/mfold/dna/form1.cgi
- MicroMeasure, http://www.colostate.edu/Depts/Biology/MicroMeasure
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Emanuel syndrome)
- PALINDROME, http://bioweb.pasteur.fr/seqanal/interfaces/palindrome.html (for the EMBOSS palindrome recognition program)
- UCSC Genome Browser, http://genome.ucsc.edu/
References
- 1.Zackai EH, Emanuel, BS (1980) Site-specific reciprocal translocation, t(11;22))q23;q11), in several unrelated families with 3:1 meiotic disjunction. Am J Med Genet 7:507–521 10.1002/ajmg.1320070412 [DOI] [PubMed] [Google Scholar]
- 2.Fraccaro M, Lindsten J, Ford C, Iselius L (1980) The 11q;22q translocation: a European collaborative analysis of 43 cases. Hum Genet 56:21–51 [DOI] [PubMed] [Google Scholar]
- 3.Shaikh TH, Budarf M, Celle L, Zackai EH, Emanuel BS (1999) Clustered 11q23 and 22q11 breakpoints and 3:1 meiotic malsegragation in multiple unrelated t(11;22) families. Am J Hum Genet 65:1595–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin TY, Viswanathan S, Wood C, Wilson PG, Wolf N, Fuller MT (1996) Coordinate developmental control of the meiotic cell cycle and spermatid defferentiation in Drosophila males. Development 122:1331–1341 [DOI] [PubMed] [Google Scholar]
- 5.Kurahashi H, Shaikh TH, Hu P, Roe BA, Emanuel BS, Budarf M (2000) Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22). Hum Mol Genet 9:1665–1670 10.1093/hmg/9.11.1665 [DOI] [PubMed] [Google Scholar]
- 6.Kurahashi H, Shaikh TH, Zackai EH, Celle L, Driscoll DA, Budarf ML, Emanuel BS (2000) Tightly clustered 11q23 and 22q11 breakpoints permit PCR-based detection of the recurrent constitutional t(11;22). Am J Hum Genet 67:763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurahashi H, Emanuel BS (2001) Unexpectedly high rate of de novo constitutional t(11;22) translocations in sperm from normal males. Nat Genet 29:139–140 10.1038/ng1001-139 [DOI] [PubMed] [Google Scholar]
- 8.Kato T, Inagaki H, Yamada K, Kogo H, Ohye T, Kowa H, Nagaoka K, Taniguchi M, Emanuel BS, Kurahashi H (2006) Genetic variation affects de novo translocation frequency. Science 311:971 10.1126/science.1121452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurahashi H, Emanuel BS (2001) Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum Mol Genet 10:2605–2617 10.1093/hmg/10.23.2605 [DOI] [PubMed] [Google Scholar]
- 10.Edelmann L, Spiteri E, Koren K, Pulijaal V, Bialer M, Shanske A, Goldberg R, Morrow BE (2001) AT-rich palindromes mediate the consitutional t(11;22) translocation. Am J Hum Genet 68:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connelly JC, Leach DR (1996) The sbcC and sbcD genes of Escherichia coli encode a nuclease involved in palindrome inviability and genetic recombination. Genes Cells 1:285–291 10.1046/j.1365-2443.1996.23024.x [DOI] [PubMed] [Google Scholar]
- 12.Kurahashi H, Inagaki H, Yamada K, Ohye T, Taniguchi M, Emanuel BS, Toda T (2004) Cruciform DNA structure underlies the etiology for palindrome-mediated human chromosomal translocations. J Biol Chem 279:35377–35383 10.1074/jbc.M400354200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roth DB, Menetski JP, Nakajima PB, Bosma MJ, Gellert M (1992) V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell 70:983–991 10.1016/0092-8674(92)90248-B [DOI] [PubMed] [Google Scholar]
- 14.Cunningham LA, Cote AG, Cam-Ozdemir C, Lewis SM (2003) Rapid, stabilizing palindrome rearrangements in somatic cells by the center-break mechanism. Mol Cell Biol 23:8740–8750 10.1128/MCB.23.23.8740-8750.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connelly JC, Kirkham L, Leach D (1998) The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc Nat Acad Sci USA 95:7969–7974 10.1073/pnas.95.14.7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majewski J, Ott J (2000) GT repeats are associated with recombination on human chromosome 22. Genome Res 10:1108–1114 10.1101/gr.10.8.1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynn A, Koehler KE, Judis L, Chan ER, Cherry JP, Schwartz S, Seftel A, Hunt PA, Hassold TJ (2002) Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science 296:2222–2225 10.1126/science.1071220 [DOI] [PubMed] [Google Scholar]
- 18.Saitta SC, Harris SE, Gaeth AP, Driscoll DA, McDonald-McGinn DM, Maisenbacher MK, Yersak JM, Chakraborty PK, Hacker AM, Zackai EH, Ashley T, Emanuel BS (2004) Aberrant interchromosomal exchanges are the predominant cause of the 22q11.2 deletion. Hum Mol Genet 13:417–428 10.1093/hmg/ddh041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun F, Trpkov K, Rademaker A, Ko E, Martin R (2005) Variation in meiotic recombination frequencies among human males. Hum Genet 116:172–178 10.1007/s00439-004-1215-6 [DOI] [PubMed] [Google Scholar]
- 20.Tease C, Hartshorne GM, Hultén MA (2002) Patterns of meiotic recombination in human fetal oocytes. Am J Hum Genet 70:1469–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen J, Sillesen I (1975) Incidence of chromosome aberrations among 11148 newborn children. Humangenetik 30:1–12 10.1007/BF00273626 [DOI] [PubMed] [Google Scholar]
- 22.Nagele RG, Freeman T, Fazekas J, Lee KM, Thomson Z, Lee HY (1998) Chromosome spatial order in human cells: evidence of early origin and faithful propagation. Chromosoma 107:330–338 10.1007/s004120050315 [DOI] [PubMed] [Google Scholar]
- 23.Vourc’h C, Taruscio D, Boyle AL, Ward DC (1993) Cell cycle-dependent distribution of telomeres, centromeres and chromosome-specific subsatellite domains in the interphase nucleus of mouse lymphocytes. Exp Cell Res 205:142–151 10.1006/excr.1993.1068 [DOI] [PubMed] [Google Scholar]
- 24.Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore W (1999) Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 145:1119–1131 10.1083/jcb.145.6.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA (2001) The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet 10:211–219 10.1093/hmg/10.3.211 [DOI] [PubMed] [Google Scholar]
- 26.Kozubek S, Lukasova E, Mareckova A, Skalnikova M, Kozubek M, Bartova E, Kroha V, Krahulcova E, Slotova J (1999) The topological organization of chromosomes 9 and 22 in cell nuclei has a determinative role in the induction of t(9,22) translocations and in the pathogenesis of t(9,22) leukemias. Chromosoma 108:426–435 10.1007/s004120050394 [DOI] [PubMed] [Google Scholar]
- 27.Nikiforova MN, Stringer JR, Blough R, Medvedovic M, Fagin JA, Nikiforov YE (2000) Proximity of chromosomal loci that participate in radiation-induced rearrangements in human cells. Science 290:138–141 10.1126/science.290.5489.138 [DOI] [PubMed] [Google Scholar]
- 28.Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A, Ashley T, Liskay RM (1996) Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet 13:336–342 10.1038/ng0796-336 [DOI] [PubMed] [Google Scholar]
- 29.Edelmann W, Cohen PE, Kane M, Lau K, Morrow B, Bennett S, Umar A, Kunkel T, Cattoretti G, Chaganti R, Pollard JW, Kolodner RD, Kucherlapati R (1996) Meiotic pachytene arrest in Mlh1-deficient mice. Cell 85:1125–1134 10.1016/S0092-8674(00)81312-4 [DOI] [PubMed] [Google Scholar]
- 30.Anderson LK, Reeves A, Webb LM, Ashley T (1999) Distribution of crossovers on mouse chromosomes using immunofluorescent localization of MLH1 protein. Genetics 151:1569–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Froenicke L, Anderson LK, Wienberg J, Ashley T (2002) Male mouse recombination maps for each autosome identified by chromosome painting. Am J Hum Genet 71:1353–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans EP, Breckon G, Ford CE (1964) An air-drying method for meiotic preparations from mammalian testis. Cytogenetics 3:288–295 [DOI] [PubMed] [Google Scholar]
- 33.Dodson G, Shi L, Tibbetts RS (2004) DNA replication defects, spontaneous DNA damage, and ATM-dependent checkpoint activation in replication protein A-deficient cells. J Biol Chem 279:34010–34014 10.1074/jbc.C400242200 [DOI] [PubMed] [Google Scholar]
- 34.Walpita D, Plug AW, Neff N, German J, Ashley T (1999) Bloom’s syndrome protein (BLM) co-localizes with RPA in meiotic prophase nuclei of mammalian spermatocytes. Proc Natl Acad Sci USA 96:5622–5627 10.1073/pnas.96.10.5622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashley T, Plug AW, Xu J, Solari AJ, Reddy G, Golub EI, Ward DC (1995) Dynamic changes in Rad51 distribution on chromatin during meiosis in male and female vertebrates. Chromosoma 104:19–28 [DOI] [PubMed] [Google Scholar]
- 36.Lichter P, Tang CC, Call K, Hermanson G, Evans GA, Housman D, Ward DC (1990) High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science 247:64–69 [DOI] [PubMed] [Google Scholar]
- 37.Reeves A (2001) Micromeasure: a new computer program for the collection and analysis of cytogenetic data. Genome 44:439–443 10.1139/gen-44-3-439 [DOI] [PubMed] [Google Scholar]
- 38.Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gotter AL, Shaikh TH, Budarf ML, Rhodes CH, Emanuel BS (2004) A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum Mol Genet 13:103–115 10.1093/hmg/ddh004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lynn A, Ashley T, Hassold T (2004) Variation in human meiotic recombination. In: Lander E, Page D, Chakravarti A (eds) Annual review of genomics and human genetics. Annual Reviews, Palo Alto, pp 317–349 [DOI] [PubMed] [Google Scholar]
- 41.Misteli T (2004) Spatial positioning: a new dimension in genome function. Cell 119:153–156 10.1016/j.cell.2004.09.035 [DOI] [PubMed] [Google Scholar]
- 42.Parada L, Sotiria S, Misteli T (2004) Spatial genome organization. Exp Cell Res 296:64–70 10.1016/j.yexcr.2004.03.013 [DOI] [PubMed] [Google Scholar]
- 43.Parada L, McQueen P, Munson P, Misteli T (2002) Conservation of relative chromosome positioning in normal and cancer cells. Curr Biol 12:1692–1697 10.1016/S0960-9822(02)01166-1 [DOI] [PubMed] [Google Scholar]
- 44.Scherthan H, Weich S, Schweger H, Heyting C, Harle M, Cremer T (1996) Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol 134:1109–1125 10.1083/jcb.134.5.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng EY, Naluai-Cecchini T (2004) FISHing for acrocentric associations between chromosomes 14 and 21 in human oogenesis. Am J Obstet Gynecol 190:1781–1795 10.1016/j.ajog.2004.02.062 [DOI] [PubMed] [Google Scholar]
- 46.Cheng EY, Chen YJ, Disteche CM, Gartler SM (1999) Analysis of a paracentric inversion in human oocytes: nonhomologous pairing in pachytene. Hum Genet 105:191–196 10.1007/s004399900120 [DOI] [PubMed] [Google Scholar]
- 47.Stahl A, Luciani JM, Hartung M, Devictor M, Berge-Lefranc JL, Guichaoua M (1983) Structural basis for Robertsonian translocations in man: association of ribosomal genes in the nucleolar fibrillar center in meiotic spermatocytes and oocytes. Proc Natl Acad Sci USA 80:5946–5950 10.1073/pnas.80.19.5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bandyopadhyay R, Heller A, Knox-DuBois C, McCaskill C, Berend SA, Page SL, Shaffer LG (2002) Parental origin and timing of de novo Robertsonian translocation formation. Am J Hum Genet 71:1456–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Assenmacher N, Hopfner KP (2004) MRE11/RAD50/NBS1: complex activities. Chromosoma 113:157–166 [DOI] [PubMed] [Google Scholar]
- 50.Paull TT, Gellert M (1999) Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev 13:1276–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borde V, Lin W, Novikov E, Petrini JH, Lichten M, Nicolas A (2004) Association of Mre11 with double-strand break sites during yeast meiosis. Mol Cell 13:389–401 10.1016/S1097-2765(04)00034-6 [DOI] [PubMed] [Google Scholar]
- 52.Keeney S, Giroux C, Kleckner N (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88:375–384 10.1016/S0092-8674(00)81876-0 [DOI] [PubMed] [Google Scholar]
- 53.Paull TT, Gellert M (2000) A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc Natl Acad Sci USA 97:6409–6414 10.1073/pnas.110144297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maser RS, Mirzoeva OK, Wells J, Olivares H, Williams BR, Zinkel RA, Farnham PJ, Petrini JH (2001) Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol Cell Biol 21:6006–6016 10.1128/MCB.21.17.6006-6016.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mirozoeva OK, Petrini JH (2003) DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol Cancer Res 1:207–218 [PubMed] [Google Scholar]
- 56.Goedecke W, Eijpe M, Offenberg HH, van Aalderen M, Heyting C (1999) MRE11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat Genet 23:194–198 10.1038/13821 [DOI] [PubMed] [Google Scholar]
- 57.Lewis SM, Chen S, Strathern JN, Rattray AJ (2005) New approaches to the analysis of palindromic sequences from the human genome: evolution and polymorphism of an intronic site at the NF1 locus. Nucleic Acids Res 33:e186 10.1093/nar/gni189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gellert M (2002) V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem 71:101–132 10.1146/annurev.biochem.71.090501.150203 [DOI] [PubMed] [Google Scholar]
- 59.Hamer G, Roepers-Gajadien HL, van Duyn-Goedhart A, Gademan IS, Kal HB, van Buul PP, Ashley T, de Rooij DG (2003) Function of DNA-protein kinase catalytic subunit during the early meiotic prophase without Ku70 and Ku86. Biol Reprod 68:717–721 10.1095/biolreprod.102.008920 [DOI] [PubMed] [Google Scholar]
- 60.Nishant KT, Kumar C, Rao MR (2006) HUMHOT: a database of human meiotic recombination hot spots. Nucleic Acids Res 34:D25–D28 10.1093/nar/gkj009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou ZH, Akgun E, Jasin M (2001) Repeat expansion by homologous recombination in the mouse germ line at palindromic sequences. Proc Natl Acad Sci USA 98:8326–8333 10.1073/pnas.151008498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tapia-Páez I, O’Brien K, Kost-Alimova M, Sahlen S, Kedra D, Bruder C, Andersson B, Roe BA, Hu P, Imreh S, Blennow E, Dumanski JP (2000) Fine mapping of the constitutional translocation t(11;22)(q23;q11). Hum Genet 106:506–516 10.1007/s004390000287 [DOI] [PubMed] [Google Scholar]
- 63.Funke B, Edelmann L, McCain N, Pandita RK, Ferreira J, Merscher S, Zohouri M, Cannizzaro L, Shanske A, Morrow BE (1999) Der(22) syndrome and velo-cardio-facial syndrome/DiGeorge syndrome share a 1.5-Mb region of overlap on chromosome 22q11. Am J Hum Genet 64:747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dresser ME, Ewing DJ, Conrad MN, Dominguez AM, Barstead R, Jiang H, Kodadek T (1997) Dmc1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics 147:533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Plug AW, Peters AH, Keegan KS, Hoekstra MF, de Boer P, Ashley T (1998) Changes in protein composition of meiotic nodules during mammalian meiosis. J Cell Sci 111:413–423 [DOI] [PubMed] [Google Scholar]
- 66.Li M, Budarf ML, Chien P, Barnoski BL, Emanuel BS, Driscoll DA (1995) Clustering of DiGeorge/velocardiofacial-associated translocations suggestive of a translocation “hot spot”. Am J Hum Genet Suppl 57:A119 10.1002/ajmg.1320570124 [DOI] [Google Scholar]
- 67.Kehrer-Sawatzki H, Haussler J, Krone W, Bode H, Jenne DE, Mehnert KU, Tummers U, Assum G (1997) The second case of a t(17;22) in a family with neurofibromatosis type 1: sequence analysis of the breakpoint regions. Hum Genet 99:237–247 10.1007/s004390050346 [DOI] [PubMed] [Google Scholar]
- 68.Rhodes CH, Call KM, Budarf ML, Barnoski BL, Bell CJ, Emanuel BS, Bigner SH, Park JP, Mohandas TK (1997) Molecular studies of an ependymoma-associated constitutional t(1;22)(p22;q11.2). Cytogenet Cell Genet 78:247–252 [DOI] [PubMed] [Google Scholar]
- 69.Debeer P, Mols R, Huysmans C, Devriendt K, Van de Ven WJ, Fryns JP (2002) Involvement of a palindromic chromosome 22-specific low-copy repeat in a constitutional t(X;22)(q27;q11). Clin Genet 62:410–414 10.1034/j.1399-0004.2002.620510.x [DOI] [PubMed] [Google Scholar]
- 70.Spiteri E, Babcock M, Kashork CD, Wakui K, Gogineni S, Lewis DA, Williams KM, Minoshima S, Sasaki T, Shimizu N, Potocki L, Pulijaal V, Shanske A, Shaffer LG, Morrow BE (2003) Frequent translocations occur between low copy repeats on chromosome 22q11.2 (LCR22s) and telomeric bands of partner chromosomes. Hum Mol Genet 12:1823–1837 10.1093/hmg/ddg203 [DOI] [PubMed] [Google Scholar]
- 71.Nimmakayalu MA, Gotter AL, Shaikh TH, Emanuel BS (2003) A novel sequence-based approach to localize translocation breakpoints identifies the molecular basis of a t(4;22). Hum Mol Genet 12:2817–2825 10.1093/hmg/ddg301 [DOI] [PubMed] [Google Scholar]
- 72.Barlow AL, Benson FE, West SC, Hultén MA (1997) Distribution of Rad51 recombinase in human and mouse spermatocytes. EMBO J 16:5207–5215 10.1093/emboj/16.17.5207 [DOI] [PMC free article] [PubMed] [Google Scholar]