Abstract
The cause of mental retardation in one-third to one-half of all affected individuals is unknown. Microscopically detectable chromosomal abnormalities are the most frequently recognized cause, but gain or loss of chromosomal segments that are too small to be seen by conventional cytogenetic analysis has been found to be another important cause. Array-based methods offer a practical means of performing a high-resolution survey of the entire genome for submicroscopic copy-number variants. We studied 100 children with idiopathic mental retardation and normal results of standard chromosomal analysis, by use of whole-genome sampling analysis with Affymetrix GeneChip Human Mapping 100K arrays. We found de novo deletions as small as 178 kb in eight cases, de novo duplications as small as 1.1 Mb in two cases, and unsuspected mosaic trisomy 9 in another case. This technology can detect at least twice as many potentially pathogenic de novo copy-number variants as conventional cytogenetic analysis can in people with mental retardation.
Mental retardation (MR) produces life-long disability, and its burden on affected families and society is enormous. Moderate-to-severe MR, which occurs in ∼1% of the population,1,2 is etiologically heterogeneous. Chromosomal abnormalities are the most common recognized cause, accounting for ∼10% of MR in most case series,3,4 but no etiology is recognized in at least one-third to one-half of all affected individuals. Accurate genetic counseling and prenatal diagnosis are not available for families of children with MR in whom no etiology is recognized. These children often endure a “diagnostic odyssey” of repeated testing for many different conditions, in an attempt to find the cause.
Chromosomal abnormalities are usually identified by cytogenetic analysis, a microscopic method of detecting gross gain, loss, or rearrangement of genetic material in dividing cells. There have been evolutionary improvements in karyotyping since its introduction as a routine clinical service >40 years ago,5–7 but cytogenetic analysis has been resistant to quantum improvements and to automation, because of the requirement for tissue culture and for highly skilled technologists to analyze the microscopic images. Standard cytogenetic analysis has the advantage of surveying the entire genome for gain or loss of genetic material in a single test, but it cannot detect imbalances of genetic segments <5–10 Mb.
Over the past several years, constitutional gain or loss of genomic segments containing only 1–5 Mb of DNA has been found to be another important cause of MR.8 These submicroscopic chromosomal alterations are usually diagnosed by locus-specific FISH,9 a test that provides much higher resolution than that of conventional cytogenetic analysis. However, locus-specific FISH is a labor-intensive microscopic technique that uses probes specifically designed for each locus (or for the relatively small number of loci) tested. FISH is, therefore, not suitable for genomewide searches for DNA copy-number changes. Better methods are needed to perform genomewide surveys for submicroscopic genomic copy-number changes in individuals with MR.
Array-based methods can provide high-resolution surveys of the entire genome for submicroscopic copy-number variants (CNVs). A few small studies using these methods have found apparently pathogenic CNVs among children with MR who had normal conventional cytogenetic analyses.10–17 These studies were done with arrays made with large-insert clones, usually BACs. The pathogenic submicroscopic deletions and duplications detected in these studies range in size from 0.5 to 15 Mb. However, smaller deletions and duplications can also cause MR.18–24 The ideal technique would, therefore, identify CNVs with an even greater genomewide resolution.
High-density whole-genome SNP arrays have been widely used for genotyping25 and can also be used to measure genomic copy number.26,27 Recent studies have shown that whole-genome sampling analysis (WGSA)28 with Affymetrix GeneChip Human Mapping 100K array sets can identify submicroscopic CNVs as well as uniparental disomy (UPD) without copy-number change.29–31 We studied 100 children with idiopathic MR and their parents, using WGSA with Mapping 100K arrays to look for potentially pathogenic submicroscopic genomic changes.
Methods
Patients and Families
We studied 100 children with idiopathic MR and both of their unaffected parents, eight unaffected siblings within these families (as negative controls), and eight trios in which the child had MR and a previously recognized chromosomal abnormality or UPD (as positive controls). Each of the children with idiopathic MR was assessed by a clinical geneticist who was unable to determine the cause of the child's MR despite thorough clinical evaluation and clinical testing that included routine karyotyping with at least 450-band resolution. The children were selected because they had moderate-to-severe MR or developmental delay with at least one of the following additional clinical features: one major malformation, microcephaly, abnormal growth, or multiple minor anomalies. Informed consent was obtained from each family, and assent was also obtained from the child, if possible. The study was approved by the University of British Columbia Clinical Research Ethics Board.
DNA Preparation
DNA was extracted from whole blood by use of a Gentra Puregene DNA Purification Kit by following the manufacturer's instructions. The DNA was precipitated in 70% alcohol, was resuspended in hydration solution, and was stored at 4ºC.
Hybridization to Mapping 100K Arrays
Genomic DNA sample quality was assessed by electrophoresis in a 0.7% agarose gel, followed by SYBR Green staining and visualization by use of a Typhoon 9400 variable mode imager. DNA concentration was measured with a Bio-Tek PowerWave X spectrophotometer. A sample of 500 ng of DNA was processed according to the instructions provided in the Affymetrix GeneChip Human Mapping 100K Assay Manual.31 In brief, 250 ng of high-quality genomic DNA was digested with XbaI or HindIII and was ligated to XbaI or HindIII adaptors. Adaptor-ligated restriction fragments were amplified by PCR, and the purified PCR products were quantified with a Bio-Tek PowerWave X spectrophotometer. Random fragmentation and labeling were performed as described elsewhere.31 Samples were hybridized to GeneChip Human Mapping 50K Xba240 or Hind240 arrays in an Affymetrix Hybridization Oven 640. Washes and staining of the arrays were performed with an Affymetrix Fluidics Station 450, and images were obtained using an Affymetrix GeneChip Scanner 3000.
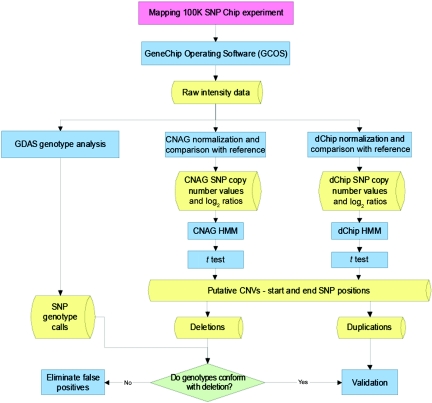
The protocol used to identify CNVs is summarized in figure 1. Initial analysis and quality assessment of the array data were performed using GeneChip DNA Analysis Software (GDAS) version 3.0.
Figure 1. .
Flow diagram showing algorithm used for detection of CNVs by use of WGSA with Mapping 100K arrays. HMM = hidden Markov model.
Copy-Number Analysis of Autosomes with CNAG
Detection of CNVs within trios consisting of an affected child and both unaffected parents was performed using Copy Number Analyser for GeneChip (CNAG)32 version 1.0. For each trio, three comparisons of SNP copy number were made: child versus father (as reference), child versus mother (as reference), and father versus mother (as reference). Regions of copy-number gain or loss in these comparisons were determined using the hidden Markov model output of CNAG. De novo and inherited deletions and duplications in the children were called using the rules described in table 1.
Table 1. .
CNV Detection with CNAG in Trios[Note]
| Line | Child vs. Father | Child vs. Mother | Father vs. Mother | CNV in Child | Type | Comment(s) |
| 1 | Loss | Loss | Loss | Deletion | De novo | The mother may have a duplication that was not inherited by the child. |
| 2 | Loss | Loss | No change | Deletion | De novo | Alternatively, both parents might have a duplication that was not inherited by the child. |
| 3 | Loss | Loss | Gain | Deletion | De novo | The father may have a duplication that was not inherited by the child. |
| 4 | Loss | No change | Gain | Deletion | Inherited from mother | Alternatively, the father may have a duplication that was not inherited by the child. |
| 5 | Loss | Gain | Gain | None | … | The father may have a duplication and the mother may have a deletion, but the child has a normal disomic copy number. Both of the child’s copies may be on one chromosome. |
| 6 | No change | Loss | Loss | Deletion | Inherited from father | Alternatively, the mother may have a duplication that was not inherited by the child. |
| 7 | No change | No change | No change | None | … | Alternatively, the child and both parents may all have the same deletion or duplication. |
| 8 | No change | Gain | Gain | Duplication | Inherited from father | Alternatively, the mother may have a deletion that was not inherited by the child. |
| 9 | Gain | Loss | Loss | None | … | The father may have a deletion and the mother may have a duplication, but the child has a normal disomic copy number. Both of the child’s copies may be on one chromosome. |
| 10 | Gain | No change | Loss | Duplication | Inherited from mother | Alternatively, the father may have a deletion that was not inherited by child. |
| 11 | Gain | Gain | Loss | Duplication | De novo | The father may have a deletion that was not inherited by the child. |
| 12 | Gain | Gain | No change | Duplication | De novo | Alternatively, both parents may have a deletion that was not inherited by the child. |
| 13 | Gain | Gain | Gain | Duplication | De novo | The mother may have a deletion that was not inherited by the child. |
Note.— “Loss,” “gain,” or “no change” of copy number refers to the first individual listed in comparison with the second individual listed. Our goal in this trio analysis was to identify de novo CNVs in the affected child. If the child showed the same copy-number change (gain or loss) in comparison with both parents, the child’s CNV was considered to have arisen de novo, regardless of whether the copy number in both parents was the same.
Copy-Number Analysis of Autosomes with dChip
An analysis complementary to that described above was performed using a reference set that included all 216 unaffected parents in this study. SNP copy number was assessed in each affected child and each unaffected parent in comparison with this large reference set, by use of DNA-Chip Analyzer (dChip) software (version release November 17, 2005).33 Regions of copy-number gain or loss were detected using the hidden Markov model output of dChip. De novo and inherited deletions and duplications in the children were called as described in table 2.
Table 2. .
CNV Detection with dChip in Trios by Use of a Reference Set[Note]
| Line | Child vs. Reference Set | Father vs. Reference Set | Mother vs. Reference Set | CNV in Child | Type | Comment(s) |
| 1 | Loss | Loss | Loss | Deletion | Inherited (from either parent) | The deletion is present in both parents, and the child could have inherited it from either parent. |
| 2 | Loss | Loss | No change | Deletion | Inherited from father | … |
| 3 | Loss | Loss | Gain | Deletion | Inherited from father | The mother has a duplication that was not inherited by the child. |
| 4 | Loss | No change | Loss | Deletion | Inherited from mother | … |
| 5 | Loss | No change | No change | Deletion | De novo | … |
| 6 | Loss | No change | Gain | Deletion | De novo | The mother has a duplication that was not inherited by the child. |
| 7 | Loss | Gain | Loss | Deletion | Inherited from mother | The father has a duplication that was not inherited by the child. |
| 8 | Loss | Gain | No change | Deletion | De novo | The father has a duplication that was not inherited by the child. |
| 9 | Loss | Gain | Gain | Deletion | De novo | Both parents have a duplication that was not inherited by the child. |
| 10 | No change | Loss | Loss | None | … | Both parents have a deletion that was not inherited by the child. |
| 11 | No change | Loss | No change | None | … | The father has a deletion that was not inherited by the child. |
| 12 | No change | Loss | Gain | None | … | The father has a deletion, the mother has a duplication, but the child has a normal disomic copy number. Both of the child’s copies may be on one chromosome. |
| 13 | No change | No change | Loss | None | … | The mother has a deletion that was not inherited by the child. |
| 14 | No change | No change | No change | None | … | … |
| 15 | No change | No change | Gain | None | … | The mother has a duplication that was not inherited by the child. |
| 16 | No change | Gain | Loss | None | … | The father has a duplication, the mother has a deletion, but the child has a normal disomic copy number. Both of the child’s copies may be on one chromosome. |
| 17 | No change | Gain | No change | None | … | The father has a duplication that was not inherited by the child. |
| 18 | No change | Gain | Gain | None | … | Both parents have a duplication that was not inherited by the child. |
| 19 | Gain | Loss | Loss | Duplication | De novo | Both parents have a deletion that was not inherited by the child. |
| 20 | Gain | Loss | No change | Duplication | De novo | The father has a deletion that was not inherited by the child. |
| 21 | Gain | Loss | Gain | Duplication | Inherited from mother | The father has a deletion that was not inherited by the child. |
| 22 | Gain | No change | Loss | Duplication | De novo | The mother has a deletion that was not inherited by the child. |
| 23 | Gain | No change | No change | Duplication | De novo | … |
| 24 | Gain | No change | Gain | Duplication | Inherited from mother | … |
| 25 | Gain | Gain | Loss | Duplication | Inherited from father | The mother has a deletion that was not inherited by the child. |
| 26 | Gain | Gain | No change | Duplication | Inherited from father | … |
| 27 | Gain | Gain | Gain | Duplication | Inherited (from either parent) | The duplication is present in both parents, and the child could have inherited it from either parent. |
Note.— The reference set comprised the 216 unaffected parents of all the studied children with idiopathic MR, known chromosomal abnormalities, or known UPD. “Loss,” “gain,” or “no change” of copy number refers to the individual listed in comparison with the large reference set. Our goal in this comparison was to identify de novo CNVs in the affected child. If the child showed a copy-number change (gain or loss) in comparison with the large reference set that was not present in either parent, the child’s CNV was considered to have arisen de novo.
Calculation of t Statistics for Identification of the Most Significant Aberrations
The software packages used to detect CNVs each employ a different algorithm to identify genomic regions of arbitrary size that have higher or lower copy number than adjacent regions. To compare CNV calls made by different packages and to identify the calls that were most likely to be biologically meaningful, we calculated t statistics and the corresponding P values for mean sample versus reference log2 copy-number ratios within each candidate deletion or duplication in comparison with the ratios outside the CNV on the same chromosome. The number of SNPs considered to be part of a CNV varies with the software and the parameters used to identify the aberrations, so we tried a range of window sizes (expressed as the number of contiguous SNPs) around the detected aberration and computed a t score for every window of the same size on the chromosome. We tested all reasonable window sizes for each candidate aberration and defined the optimal window size as the one that produced the best t score or equivalent P value for that CNV. Choice of window size for graphical display of t scores is arbitrary. We chose the optimal window size with small CNVs to maximize the t score and to make the aberration more apparent in the plot. Longer aberrations are generally associated with t scores that differ greatly from the rest of the chromosome, and, in these cases, we chose window sizes smaller than the optimal to produce smoother plots.
Combined Copy-Number and Genotype Analysis for Deletions
SNP genotype calls were generated from signal intensity data with GDAS version 3.0 by use of a confidence score threshold of 0.05 for genotype accuracy. Genotypes were assessed for each putative deletion identified by CNAG or dChip. If the number of heterozygous SNPs exceeded 10% of the total within a putatively deleted segment, the deletion call was considered to be a false-positive result. Deletions were accepted as hemizygous if at least 90% of the genotype calls within the segment were either “AA” or “BB.” Genotype calls within a hemizygous deletion in a child often exhibited Mendelian errors (e.g., an “AA” genotype result in the child and a “BB” genotype result in one parent), and such errors were used to determine the parental origin of the child's remaining inherited allele.
Copy-Number Analysis of X Chromosomes with dChip
Copy numbers for SNPs on the X chromosome were analyzed with dChip software33 as follows. Each male (child or father) was compared with a reference set comprising all 108 unaffected fathers included in the study. Each female (child or mother) was compared with a reference set comprising all 108 unaffected mothers included in the study. Regions of copy-number gain or loss were detected using the hidden Markov model output of dChip.
Screen for UPD
A search for UPD was performed using SNP genotypes generated with GDAS version 3.0, with a confidence score threshold of 0.05. SNP genotypes for each child were compared with those of both parents, and Mendelian errors (SNPs homozygous for one allele in the child and the opposite allele in one parent) were identified. Mendelian errors are very infrequent in the absence of deletions or UPD. When such Mendelian errors were found, other SNPs within the same chromosomal region or chromosome were evaluated for the presence of isodisomy or heterodisomy. Uniparental isodisomy was identified as a stretch of homozygous SNPs in the child with exclusively maternal or exclusively paternal origin. Diagnosis of uniparental isodisomy also required confirmation of a normal disomic copy number. Uniparental heterodisomy was detected when a continuous region within a window of 200–1,000 SNPs in the child showed genotypes identical to the same chromosomal region in the mother or father.
Validation of CNVs
Putative CNVs identified by WGSA were validated by FISH with cytogenetic pellets prepared according to standard clinical procedures. FISH was performed with BAC or fosmid probes selected using the University of California at Santa Cruz (UCSC) Genome Browser34 and the May 2004 assembly of the human genome sequence.
The genomic content of BAC inserts used for FISH confirmation of putative CNVs was verified by end sequencing, as necessary. BAC DNA was prepared as described elsewhere,35 was precipitated, and was resuspended in 35 μl of Ultrapure water (Gibco). DNA sequencing reactions were assembled in 384-well clear optical reaction plates (Applied Biosystems) by use of a Biomek FX workstation (Beckman-Coulter). Each reaction was performed in a total volume of 8 μl, consisting of 5 μl of purified BAC DNA, 0.7 μl of T7 or SP6Wan sequencing primer (5 pmol/μl [Invitrogen]), 0.3 μl of Ultrapure water (Gibco), and 2 μl of BigDye v.3.1 Ready Reaction Mix (Applied Biosystems). Thermal cycling was performed on a PTC-225 or DNA Engine Tetrad 2 thermal cycler (MJ Research) with parameters of 85 cycles at 96°C for 10 s, melting temperature (which was specific to each primer) for 5 s, and 60°C for 4 min, followed by incubation at 4°C. Reaction products were precipitated by adding 2 μl of 125-mM EDTA (pH 8.0) and 18 μl of 95% ethanol per well, followed by centrifugation at 2,750 g for 30 min in an Eppendorf 5810R centrifuge. The EDTA/ethanol was immediately decanted, and reaction products were washed with 30 μl of 70% ethanol. The 384-well cycle plates were allowed to dry inverted for 15 min. Samples were resuspended in 10 μl of Ultrapure water and were analyzed using a 3730XL DNA analyzer (Applied Biosystems). The reads were processed, and their quality was assessed, using Phred.36,37 The end sequences were aligned with the human genomic sequence (May 2004 assembly) by use of BLAT,38 via the UCSC Genome Browser.
BAC or fosmid DNA was isolated by small-scale (miniprep) preparation and was labeled with Spectrum Red or Green (Vysis) by use of a Vysis nick translation reagent kit. The labeled product was mixed with 3 μg of human Cot-1 DNA (Invitrogen) and was isolated by means of a standard DNA precipitation method. Chromosomes and nuclei were visualized by counterstaining with 4′,6-diamidino-2-phenylindole. For deletions, at least 10 metaphase cells were analyzed, and interphase nuclei were examined but not counted. For duplications, at least 10 metaphase cells and at least 50 interphase nuclei were analyzed. All FISH probes were tested on metaphase spreads from unaffected individuals to assure proper hybridization. Of 118 FISH probes successfully tested, 2 hybridized to the wrong location, and 14 produced cross-hybridization signals.
Results
We performed WGSA with Mapping 100K arrays on 100 children with idiopathic MR and on both parents of each affected child. We also tested eight unaffected siblings in these families (as negative controls) and eight trios in which the fetus or child had MR as the result of a previously recognized chromosomal abnormality (as positive controls). In this last group, the Mapping 100K arrays demonstrated the expected findings in all cases, which included the following known segmental aneusomies or UPDs: del(7)(q11.2q11.2), del(11)(p12p14.1), del(10)(q24.32q25.1), dup(10)(p12.2pter), dup(15)(pter), UPD 7, UPD 15, UPD 16, and UPD 22.
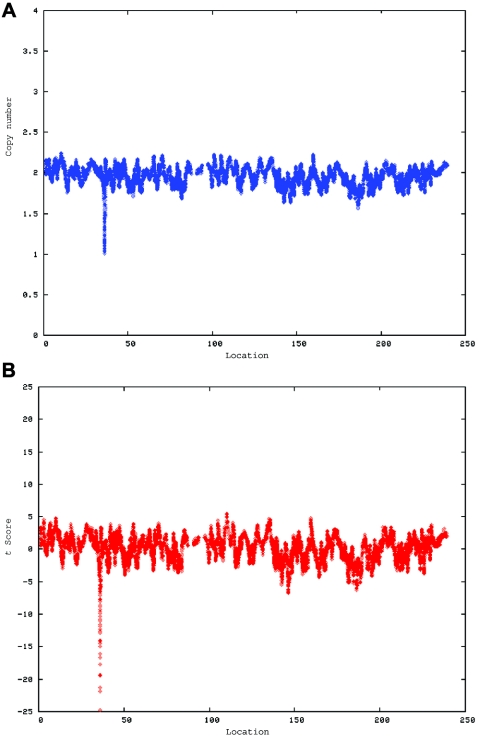
Data were analyzed to determine copy number along the length of all chromosomes, and the statistical significance of variations from the expected copy number was estimated in windows of various sizes by t scores calculated in comparison with the estimated copy number of the remainder of the chromosome (fig. 2). The findings for each child were then compared with those for his or her parents. Apparent CNVs seen in the child and in at least one parent were considered likely to be benign polymorphisms. Apparent CNVs found in the child but not in either parent were evaluated by FISH, to confirm the presence of the CNV and its de novo occurrence. Paternity was confirmed in all trios studied by use of the SNP genotyping calls produced in the array experiments.
Figure 2. .
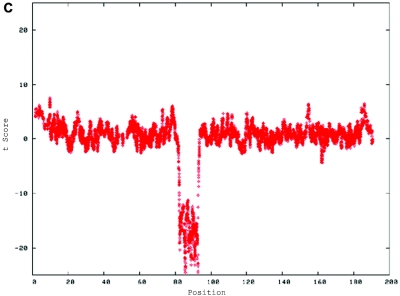
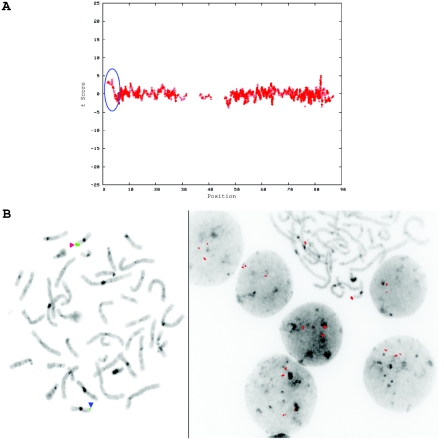
A CNV on chromosome 2 in the affected child of family 7551. A, dChip-smoothed copy-number (CN) estimate with a window of 38 SNPs, showing a 1-copy deletion. B, Diagram showing t scores calculated for a sliding window of 38 SNPs that compare the copy number estimated by dChip for SNPs within the window with the copy number estimated for all other SNPs on the same chromosome. The high (negative) t scores associated with the deletion indicate that this deviation in copy number is unlikely to have occurred by chance. In both panels, the X-axis shows the nucleotide position in Mb.
In the course of this study, we identified a total of 3,125 putative CNVs by WGSA in the 100 studied children with idiopathic MR. Apparent CNVs were found in all individuals, with a range of 19–43 per child with idiopathic MR. The median number of apparent CNVs per child was 30 (5th percentile = 22; 95th percentile = 38). Of these variants, 2,669 occurred only once in this data set, and the majority of false-positive CNV calls is likely to be in this subgroup. The putative CNVs with the highest statistical significance—that is, those that were least likely to have occurred by chance alone—were tested by FISH. Of the 3,125 apparent CNVs found in children with MR, 12 were confirmed both in the child and in one of his or her parents. An example is shown in figure 3. Some other apparent CNVs correspond to known polymorphisms (Database of Genomic Variants), but most are unique and have not been independently confirmed. All CNVs confidently detected in the eight unaffected siblings of children with idiopathic MR were inherited from one of the parents.
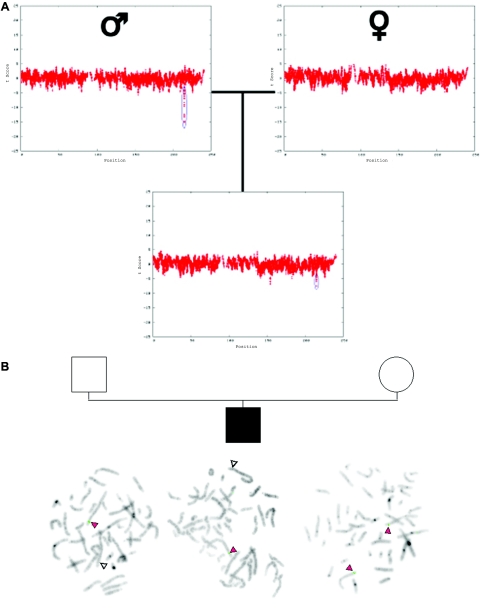
Figure 3. .
Inherited CNV demonstrated by WGSA with Mapping 100K arrays. A, Plot of t scores for chromosome 2 in father, mother, and child in family 0674, by use of a 21-SNP window. The SNPs are ordered along the X-axis from pter to qter, with the location shown in Mb. The child with MR inherited the CNV (blue oval) from his unaffected father. B, FISH confirmation of the inherited CNV on 2q34. In the father and child, one chromosome 2 (white arrowheads) lacks a signal for probe RP11-93G7, whereas the signal is present on the other chromosome 2 (red arrowheads). In the mother, the signal for probe RP11-93G7 is seen on both chromosomes 2 (red arrowheads).
CNVs that were confirmed by FISH in a child with MR and that were found by FISH to be not present in either parent were considered to have arisen de novo. We found such de novo alterations in 11 of the 100 children with idiopathic MR (table 3). Table 4 summarizes the phenotypic abnormalities observed in each of these children.
Table 3. .
De Novo CNVs Detected by WGSA with Mapping 100K Arrays and Confirmed by FISH
| Size of CNV |
SNP Genotype Summary |
||||||||
| Family | CNV Type | Chromosome and Band |
Starting SNP (Locationa in bp) |
Ending SNP (Locationa in bp) |
bp | SNPs | Heterozygous SNPs in |
Informative SNPs Inherited from |
OMIM Genes with Abnormal Phenotypes within CNV |
| 1895 | Deletion | 13q12.11-q12.13 | rs3929856 (18867056) | rs10507342 (24517730) | 5,650,674 | 262 | Child, 1; father, 61; mother, 72 | Father, 19; mother, 0 | SGCG, GJA3, GJB6, GJB2, and SACS |
| 3476 | Deletion | 4q21.21-q22.1 | rs1493182 (82146749) | rs1586340 (93214433) | 11,067,684 | 627 | Child, 1; father, 137; mother, 135 | Father, 0; mother, 60 | DSPP, PKD2, and SNCA |
| 4794 | Duplication | 16p13.3 | rs1544799 (925718) | rs2239318 (3864938) | 2,939,220 | 29 | … | … | GNPTAG, TSC2, CLCN7, IGFALS, PKD1, ABCA3, MEFV, and CREBBP |
| 4818 | Deletion | 12q14.2-q15 | rs10506536 (63342649) | rs10492198 (66780095) | 3,437,446 | 199 | Child, 0; father, 15; mother, 17 | Father, 4; mother, 0 | GNS |
| 5003 | Deletion | 2p16.3 | rs10490220 (50857428) | rs2193411 (51178791) | 321,363 | 25 | Child, 0; father, 10; mother, 2 | Father, 0; mother, 2 | None |
| 5566 | Deletion | 14q11.2 | rs10483251 (20741117) | rs9322978 (20918741) | 177,624 | 5 | Child, 0; father, 0; mother, 0 | Father, 0; mother, 0 | RPGRIP1 |
| 5994b | Mosaic trisomy | 9 | … | … | … | 4,782 | … | … | Many (whole chromosome) |
| 6168 | Duplication | 17q21.33 | rs10514971 (45093545) | rs8081154 (46196038) | 1,102,493 | 15 | … | … | SGCA, DLX3, and COL1A1 |
| 6545 | Deletion | 7p22.1-p22.2 | rs10499339 (3304850) | rs1368052 (6940933) | 3,636,083 | 61 | Child, 1; father, 13; mother, 6 | Father, 0; mother, 3 | PMS2 |
| 7807 | Deletion | 22q12.1 | rs10483135 (26138764) | rs6005907 (27552525) | 1,413,761 | 41 | Child, 0; father, 10; mother, 4 | Father, 0; mother, 0 | CHEK2 and XBP1 |
| 8326 | Deletion | 14q11.2 | rs10484227 (19584863) | rs10483256 (21207935) | 1,623,072 | 88 | Child, 0; father, 36; mother, 27 | Father, 0; mother, 3 | NP and RPGRIP1 |
Location is given using coordinates from National Center for Biotechology Information assembly 35 of the human genome sequence (June 2004).
Starting and ending SNPs are not shown because CNV involves the whole chromosome.
Table 4. .
Summary of Clinical Features in Children with de Novo CNVs Detected by WGSA with Mapping 100K Arrays and Confirmed by FISH
| Patient | Abnormality | Clinical Features | Evidence of Pathogenicity |
| 1895 | 5.7 Mb, del(13)(q12.11q12.13) | 6-year-old girl; microcephaly; mild growth retardation; several café-au-lait macules; mild anemia, thrombocytopenia, and neutropenia; moderate developmental delay | De novo CNV; large region of genetic imbalance |
| 3476 | 11.1 Mb, del(4)(q21.21q22.1) | 18-year-old girl; extremely short stature; deep-set eyes with narrow palpebral fissures; low-set, straight eyebrows; narrow nasal root and bridge; prominent columnella with receding alae nasae, short philtrum, and thin, downturned lips; small hands and feet; severe hypotonia; marked pes planus; mild scoliosis; severe cognitive impairment | De novo CNV; very large region of genetic imbalance |
| 4794 | 2.9 Mb, dup(16)(p13.3) | 12-year-old boy; blepharophimosis and ptosis; low-set, mildly dysplastic auricles; mild pectus excavatum; C5-C6 vertebral fusion; pes cavus and clawed toes; normal growth; full-scale IQ of 46 | De novo CNV; gene-rich region; striking similarity to other patients described with cytogenetically apparent 16p13 duplication |
| 4818 | 3.4 Mb, del(12)(q14.2q15) | 12-year-old boy; prenatal-onset growth retardation; partial anodontia; mild limitation of extension at elbows; mildly short and narrow fingers; tremor; mild developmental delay; osteopoikilosis on radiographic examination | De novo CNV; large region of genetic imbalance |
| 5003 | .32 Mb, del(2)(p16.3p16.3) | 7-year-old boy; full-scale IQ of 74; learning problems in both parents; attention deficit disorder; Asperger syndrome; frontal bossing, high anterior hairline, frontal hair whorl, and low posterior hairline; mild dorsal scoliosis with 13 ribs on left, bifid right second rib, hemivertebrae and fusions of T2, T3, T4, and fusion of L4 and L5; asthma | De novo CNV; no known polymorphic CNVs in region |
| 5566 | .18 Mb, del(14)(q11.2q11.2) | 2-year-old girl; large for gestational age at birth, head growth at 97th percentile subsequently, and normal height and weight; generalized hypotonia and joint hypermobility; short palpebral fissures; mildly dysplastic auricles; long toes with 2-3 cutaneous syndactyly; developmental delay, especially gross motor | De novo CNV; no known polymorphic CNVs in region; deletion lies within that of child 8326 and DECIPHER patient CAM126 |
| 5994 | Mosaic trisomy 9 (∼20%) | 17-mo-old boy; plagiocephaly, torticollis, and broad and prominent forehead with bifrontal narrowing; low-set ears, small mouth, and short philtrum; puffy hands and feet; undescended testes; hypotonia; tracheomalacia; enlarged cerebral ventricles; bicuspid aortic valve; moderate-to-severe developmental delay | De novo CNV; cytogenetic confirmation; phenotype characteristic of trisomy 9 mosaicism |
| 6168 | 1.1 Mb, dup(17)(q21.33) | 9-year-old girl; microcephaly; moderate-to-severe conductive hearing loss; bilateral preauricular skin tags, small ears with abnormal pinnae, and small ear canals; mild retrognathia; mild MR | De novo CNV; large region of genetic imbalance |
| 6545 | 3.6 Mb, del(7)(p22.2p22.1) | 14-mo-old girl; prenatal-onset growth retardation and microcephaly; patent ductus arteriosus and perimembranous ventricular septal defect; failure to thrive; severe developmental delay; brachycephaly, epicanthic folds, midface hypoplasia, and lateral flare of eyebrows; 2-3 syndactyly of toes | De novo CNV; large region of genetic imbalance; DECIPHER case UPP969 has 0.2-Mb deletion within deleted region of this patient |
| 7807 | 1.4 Mb, del(22)(q12.1q12.1) | 5-year-old boy; microcephaly; frontal upsweep, hypertrichosis of forehead, and prominent brows; bifid uvula; undescended testes; developmental delay, especially of speech | De novo CNV; large region of genetic imbalance; 1.8-Mb deletion in DECIPHER patient CHG758 overlaps the CNV in this patient |
| 8326 | 1.6 Mb, del(14)(q11.2q11.2) | 2-year-old boy; small ventricular septal defect that closed spontaneously; large patent ductus arteriosus that required surgical closure; plagiocephaly; pseudostrabismus with very broad nasal root and short nose; preauricular pit; undescended testes and hypoplastic scrotum; moderate-to-severe developmental delay, especially of speech | De novo CNV; DECIPHER patient CAM126 has similar deletion |
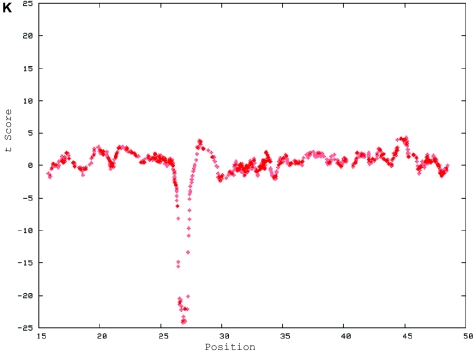
Eight of the 11 de novo cases had submicroscopic deletions that were confirmed by locus-specific FISH (figs. 4 and 5). These CNVs ranged in size from 178 kb (in family 5566) to >11 Mb (in family 3476) and involved 5–627 contiguous SNPs.
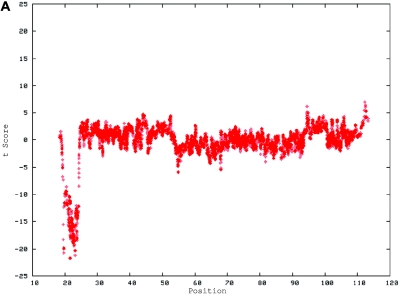
Figure 4. .
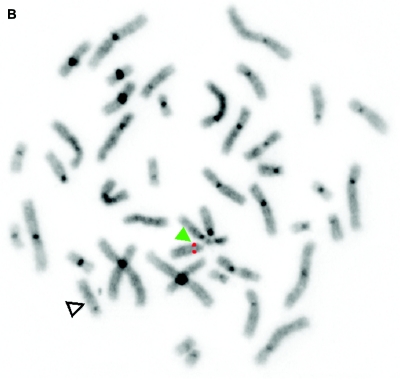
De novo submicroscopic genomic deletion, del(2)(p16.3p16.3), identified by WGSA with Mapping 100K arrays in family 5003. A, The t scores for the CNV in comparison with the rest of the chromosome, by use of a window size of 20 SNPs. The SNPs are ordered along the X-axis from pter to qter, with the location shown in Mb. B, FISH confirmation of the CNV in the affected child by use of probe RP11-1151G3. The white arrowhead indicates the deletion-containing chromosome 2 without the signal. The green arrowhead indicates the normal homologue, which shows the signal.
Figure 5. .
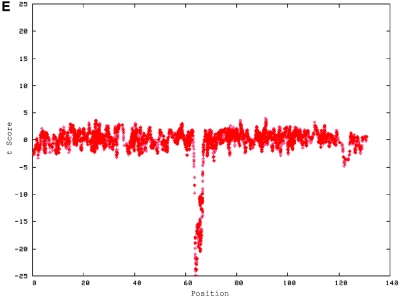
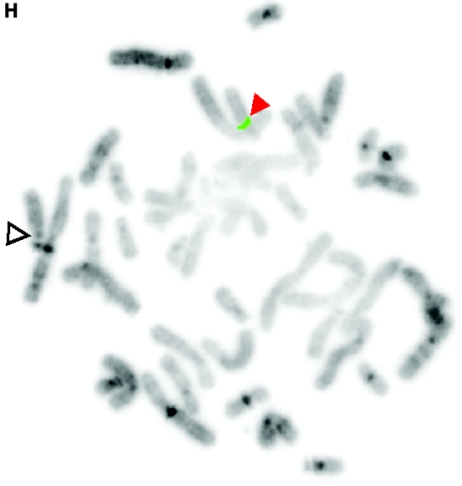
De novo submicroscopic genomic deletions identified by WGSA with Mapping 100K arrays in the affected child of families 1895, 3476, 4818, 5566, 6545, and 7807. Panels A, C, E, G, I, K, and M show the t scores for each CNV in comparison with the rest of the chromosome, by use of the window size given below. The SNPs are ordered along the X-axis from pter to qter, with the location shown in Mb. Panels B, D, F, H, J, L, and N show FISH confirmation of the CNV in the affected child, by use of the probe given below. The white arrowhead indicates the deleted chromosome without the signal. The red or green arrowhead indicates the normal homologue, which shows the signal. A, Family 1895, chromosome 13, window of 20 SNPs. B, Family 1895, probe RP11-26D3, del(13)(q12.11q12.13). C, Family 3476, chromosome 4, window of 30 SNPs. D, Family 3476, probe RP11-7G22, del(4)(q21.21q22.1). E, Family 4818, chromosome 12, window of 20 SNPs. F, Family 4818, probe RP11-91K23, del(12)(q14.2q15). G, Family 5566, chromosome 14, window of 5 SNPs. The de novo CNV is shown inside the blue oval. H, Family 5566, probe CTD-2136K6, del(14)(q11.2q11.2). I, Family 6545, chromosome 7, window of 20 SNPs. J, Family 6545, probe RP11-802L1, del(7)(p22.2p22.1). K, Family 7807, chromosome 22, window of 20 SNPs. L, Family 7807, probe RP11-79G21, del(22)(q12.1q12.1). M, Family 8326, chromosome 14, window of 20 SNPs. N, Family 8326, probe RP11-340M24, del(14)(q11.2q11.2).
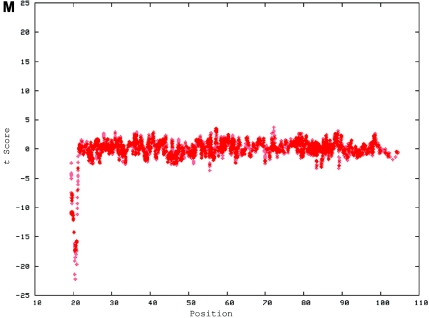
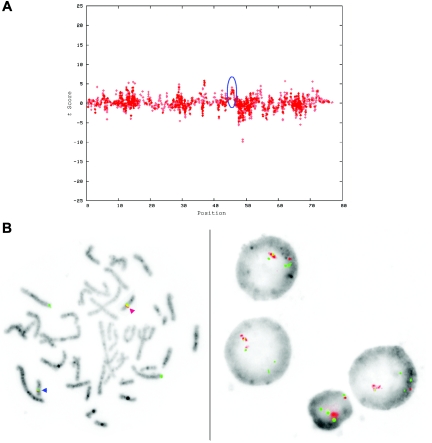
Two of the 11 de novo cases had submicroscopic duplications that were confirmed by locus-specific FISH (figs. 6 and 7). The sizes of these CNVs were 1.1 Mb (15 consecutive SNPs) in family 6168 and 2.9 Mb (29 consecutive SNPs) in family 4794.
Figure 6. .
De novo submicroscopic genomic duplication, dup(16)(p13.3p13.3), identified by WGSA with Mapping 100K arrays in family 4794. A, The t scores for the CNV in comparison with the rest of the chromosome, by use of a window size of 20 SNPs. The SNPs are ordered along the X-axis from pter to qter, with the location shown in Mb. The de novo CNV is shown inside the blue oval. B, FISH confirmation of the CNV in the affected child, by use of probe G248P82513E10 in metaphase cells (left) and probe RP11-397B22 in interphase cells (right). In the metaphase spread, the duplication is indicated by the red arrowhead, and the normal chromosome is indicated by the blue arrowhead. The signal is duplicated on one chromosome 16.
Figure 7. .
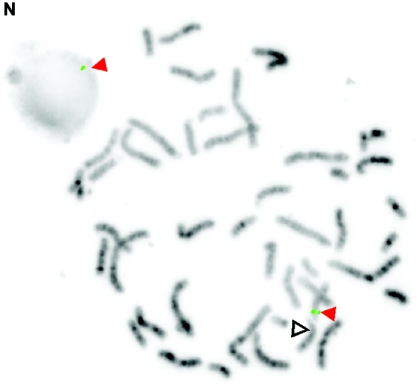
De novo submicroscopic genomic duplication, dup(17)(q21.33q21.33), identified by WGSA with Mapping 100K arrays in family 6168. A, The t scores for each CNV in comparison with the rest of the chromosome, by use of a window size of 8 SNPs. The SNPs are ordered along the X-axis from pter to qter, with the location shown in Mb. The de novo CNV is shown inside the blue oval. B, FISH confirmation of the CNV in the affected child, by use of probes RP11-962N5 (green) and RP11-121F10 (red) in metaphase (left) and interphase (right) cells. In the metaphase spread, the duplication is shown at the red arrowhead, and the normal chromosome is shown at the blue arrowhead. The red signal is duplicated on one chromosome 17; the green signal does not appear duplicated but cross-hybrizes to chromosome 6qter.
We also identified mosaic trisomy 9 in family 5994. This abnormality was not detected on initial cytogenetic analysis but was subsequently confirmed in 1 of 44 cells examined on an extended cytogenetic survey of peripheral-blood metaphases and in 60 of 300 peripheral-blood interphase cells studied by FISH with probes for chromosome 9.
Although we were able to demonstrate the UPD in all four control cases with known UPD, we did not find any copy-number–neutral alterations of familial genotype patterns indicative of UPD in the 100 studied children with idiopathic MR. We did not identify any de novo CNVs of the X chromosome or any inherited regions of X-chromosomal nullisomy in males with idiopathic MR.
Discussion
We have shown that WGSA with Affymetrix Mapping 100K arrays can identify apparently pathogenic, submicroscopic CNVs in individuals with idiopathic MR and normal results of conventional cytogenetic analysis. Our observed frequency of de novo submicroscopic CNVs is comparable to that seen in a similarly selected group of patients with idiopathic MR who were studied using comparative genomic hybridization with whole-genome BAC tiling-path arrays.12 However, using WGSA, we were able to detect smaller de novo CNVs than those that have been reported using the BAC tiling-path arrays. In addition, SNP arrays can identify copy-number–neutral UPD29–31 and can determine the parent of origin of a child's remaining allele in heterozygous deletions, neither of which can be done by BAC or nonpolymorphic oligonucleotide comparative genomic hybridization. Although we were able to detect the UPD in our control samples, we did not find UPD in any of the 100 studied children with idiopathic MR. The SNP genotypes provided by these mapping arrays were also useful for confirming paternity within the trios.
Because of the noise inherent in WGSA with Mapping 100K arrays, statistically significant increases or decreases in apparent copy number often occur by chance. Slater et al.31 estimated the false-positive copy-number call rate for WGSA with Mapping 100K arrays to be in the range 2.0%–3.5%. We employed a different approach for copy-number analysis and found that the false-positive call rate varied with the software, parameter settings, and comparison group. To increase the specificity of CNV calls, we calculated t scores for the variation from expected copy number in the region of interest in comparison with the copy number for the rest of a chromosome (fig. 2), and we accepted only calls that were very unlikely to have occurred by chance alone.
Another advantage of using SNP arrays that provide information on genotypes as well as copy number is that some false-positive deletion calls can be recognized by the presence of heterozygous genotypes within a putatively deleted region. More generally, the false-positive CNV call rate can be reduced by requiring a larger number of SNPs that are adjacent in the genome to demonstrate a similar apparent gain or loss of copy number, but doing so reduces the sensitivity and resolution of the analysis. The use of higher density arrays (e.g., 500K rather than 100K arrays) and critical assessment of the characteristics that permit CNVs to be identified by various software algorithms may help to reduce the false-positive call rate without substantially affecting the sensitivity of the assay.
Affymetrix Mapping 100K arrays have one SNP every 23.6 kb, on average, and an 8.5-kb median distance between adjacent SNPs, excluding centromeric and telomeric regions. We considered a false-positive rate of ⩽4 CNV calls per person to be manageable, given the usual error profiles observed in this study. To obtain this false-positive rate, we required that at least four adjacent SNPs exhibit a similar change in apparent copy number to call a CNV. A larger number of adjacent SNPs was needed to call a change in copy number when the hybridization results were unusually noisy.
The smallest confirmed inherited CNV we observed (a 37.6-kb deletion of chromosome 9 in family 9299) approaches the theoretical resolution of our analytical method. The smallest confirmed de novo CNV we found was 178 kb (in family 5566). The fact that we did not find smaller de novo CNVs is unlikely to reflect any biological limit on the minimal CNV size associated with MR. Smaller pathogenic deletions or duplications have been found in children with MR by use of other methods.18–23
SNPs included on the Mapping 100K arrays are not evenly spaced throughout the genome, and some genomic regions have denser representation than others. Thus, although we were able to confirm an inherited CNV as small as 37.6 kb, we may have failed to detect CNVs of several hundred kilobases if they happen to lie in regions that are poorly represented on the Mapping 100K arrays. Of particular concern is the fact that subtelomeric regions and regions rich in segmental duplications, which are frequent sites of benign and pathological copy-number variation,39–41 are poorly represented on these arrays. 500K SNP arrays have recently become available and should provide better resolution for WGSA, although subtelomeric regions and regions rich in segmental duplications are still underrepresented.42
As expected, we found many CNVs that probably represent normal polymorphisms.43 Our study was not designed to survey these normal variants in detail, and WGSA may not be the best technology to use for this purpose.42
We have used the occurrence of a CNV as a new mutation in a child with MR as a screen for pathogenicity of the variant, because this approach parallels that used clinically to assess cytogenetically apparent chromosomal abnormalities. We recognize that some CNVs that are inherited from an unaffected parent may nevertheless be pathogenic—for example, if an inherited deletion uncovers a region that is imprinted or includes a pathogenic recessive allele on the normal homologous chromosome.43,44 We do not know the frequency of de novo CNVs among unaffected individuals, although we did not observe any de novo CNVs in the eight unaffected siblings studied with these trios. We believe that all the de novo CNVs observed in this study of children with MR are likely to be pathogenic, because all exhibit features, in addition to their occurrence as new mutations, that have been associated with cytogenetically detectable deletions and duplications that cause MR.
All 11 de novo CNVs we found in children with MR contain genes (or portions of genes) annotated by Ensembl and sequences that are expressed in human fetal brain (table 5). Almost all these de novo CNVs also contain genes from Online Mendelian Inheritance in Man (OMIM) that have been associated with phenotypic abnormalities (table 3), but none of the children has clinical features compatible with known mutations of these OMIM genes. The duplication in child 4794 includes a region that is deleted in some cases of Rubinstein-Taybi syndrome (MIM 180849), a well-characterized multiple congenital anomaly MR syndrome.45
Table 5. .
Ensembl Genes and Fetal Brain Gene Expression in Regions Defined by de Novo CNVs in Children with Idiopathic MR[Note]
| SAGE Tag Expression Bins |
|||||||||||||
| Family | De Novo CNV | Chromosome | Starting Breakpoint (bp) |
Ending Breakpoint (bp) |
No. of Ensembl Gene Predictions in Region | No. of Genes Mapped by SAGE Tags | 1–5 | 6–9 | 10–49 | 50–99 | 100+ | Total Tags | Total Tags in Genes |
| 1895 | Deletion | 13 | 18867056 | 24517730 | 63 | 16 | 70 | 20 | 6 | 1 | 0 | 97 | 45 |
| 3476 | Deletion | 4 | 82146749 | 93214433 | 112 | 22 | 113 | 29 | 15 | 8 | 4 | 169 | 57 |
| 4794 | Duplication | 16 | 925718 | 3864938 | 165 | 47 | 109 | 43 | 33 | 11 | 10 | 206 | 102 |
| 4818 | Deletion | 12 | 63342649 | 66780095 | 29 | 9 | 31 | 9 | 9 | 0 | 0 | 49 | 22 |
| 5003 | Deletion | 2 | 50857428 | 51178791 | 0 | 0 | 9 | 3 | 0 | 0 | 0 | 12 | 0 |
| 5566 | Deletion | 14 | 20741117 | 20918741 | 3 | 0 | 1 | 1 | 1 | 0 | 0 | 3 | 0 |
| 5994a | Mosaic trisomy | 9 | … | … | 1,362 | 433 | 1,085 | 344 | 284 | 50 | 23 | 1,786 | 1,113 |
| 6168 | Duplication | 17 | 45093545 | 46196038 | 37 | 12 | 47 | 15 | 15 | 5 | 1 | 83 | 32 |
| 6545 | Deletion | 7 | 3304850 | 6940933 | 73 | 15 | 56 | 20 | 20 | 2 | 2 | 100 | 43 |
| 7807 | Deletion | 22 | 26138764 | 27552525 | 17 | 3 | 31 | 7 | 3 | 0 | 0 | 41 | 12 |
| 8326 | Deletion | 14 | 19584863 | 21207935 | 72 | 12 | 34 | 11 | 11 | 2 | 1 | 59 | 26 |
Note.— Expression levels were estimated by the number of unique tag types per expression level bin (normalized to tags per million) in long SAGE Library LSAGE_Brain_fetal_normal_B_S.
Starting and ending breakpoints are not shown because CNV involves the whole chromosome.
The de novo deletions we observed that extend across >1 Mb of genomic sequence (in families 1895, 3476, 4818, 6545, 7807, and 8326) are almost certainly pathogenic, because of their large size and consequent gene content. These deletions are similar in size to the pathogenic CNVs that occur in recognized microdeletion syndromes41 and are larger than almost all the deletion polymorphisms reported in unaffected individuals.12,39,40,46–51
Two of the de novo deletions we confirmed by FISH are substantially smaller than the others (321 kb in family 5003 and 178 kb in family 5566), demonstrating the improved resolution of WGSA with Mapping 100K arrays in comparison with other array hybridization platforms used to identify CNVs in children with MR.10–17 Despite their small size, the deletions in families 5003 and 5566 are also probably pathogenic because, to the best of our knowledge, they have never been observed in unaffected individuals and because each of these CNVs arose as a new mutation in the affected child. Similar arguments regarding possible pathogenicity apply to the de novo duplications found in this study (figs. 6 and 7). Both of these duplications are >1 Mb in size, neither has been reported in unaffected individuals, and both apparently arose as a new mutation in the affected child.
The ability of WGSA to detect trisomy 9 mosaicism despite the failure of routine karyotyping to do so may reflect the fact that trisomy 9 cells are selected against under routine cytogenetic culture conditions or that the trisomy line happens to be much less frequent in the T-lymphocytes that were karyotyped than in other nucleated blood cells. Our observation that trisomy 9 was confirmed by FISH in 20% of interphase cells but by cytogenetic analysis in ∼2% of metaphase cells studied is consistent with both interpretations.
CNVs frequently occur as benign polymorphisms in healthy individuals,12,39,40,46–51 and it is not always clear whether a CNV found in a child with MR is actually pathogenic. There are several features that can be used to differentiate apparently benign polymorphic variants from CNVs that may cause MR. Our primary approach was based on family studies—the method that is used clinically to determine whether a novel chromosomal alteration identified cytogenetically is pathogenic. In this approach, a CNV that has been inherited from an unaffected parent is considered unlikely to be pathogenic, whereas a CNV that occurs de novo in a child with MR is probably causally related. Although there are exceptions—for example, in the case of recessive mutations, imprinted genes, or X-linked genes transmitted by a woman to her son—family studies are often very helpful in determining the likelihood that a CNV is pathogenic.
Another useful rule of thumb is that a CNV observed frequently in unaffected people is unlikely to be pathogenic in a child with MR. None of the de novo CNVs we found in affected children is listed in the Database of Genomic Variants as having been reported in an unaffected individual, and we did not find these changes in any of the unaffected parents or unaffected sibling controls in our study.
Some CNVs are known to be pathogenic, because they have been observed repeatedly in children who share a particular MR phenotype. The familiar forms of MR associated with microdeletions of 7q11.23 (Williams-Beuren syndrome [MIM 194050])52 and 17p11.2 (Smith-Magenis syndrome [MIM 182290])53 provide cogent examples of CNVs that were demonstrated to be pathogenic in this way. Thus, the phenotypic similarity of child 5994 to other children who have been described with mosaic trisomy 954,55 makes it virtually certain that his MR was caused by the trisomy 9 mosaicism that we discovered in him by WGSA.
Five of the other de novo CNVs we found have also been reported in other children with MR: dup(16)(p13.3p13.3) in child 4794, del(7)(p22.2p22.1) in child 6545, del(22)(q12.1q12.1) in child 7807, and del(14)(q11.2q11.2) in child 8326 and child 5566. The appearance of child 4794 is strikingly similar to that of other individuals with cytogenetically apparent duplications of 16p13 and especially to the patient recently described by Sommer et al.56 The de novo duplication we found in child 4794 also overlaps that of DECIPHER patient CHG237.
The 1.6-Mb deletion we demonstrated in child 8326 includes a 1.1-Mb deletion reported in DECIPHER patient CAM126, and both of these lesions include the 178-kb deletion that we found in child 5566. These three children share similar minor anomalies that may constitute a recurrent syndrome. The 3.6-Mb deletion in child 6545 includes a 0.2-Mb deletion reported in DECIPHER patient UPP969, and the 1.8-Mb deletion in DECIPHER patient CHG758 overlaps the 1.4-Mb deletion found in child 7807. A detailed comparison of the phenotypic features of these individuals is being undertaken.
In a few instances, a CNV found in a child with MR may include a gene that is known to be dosage sensitive and to produce the specific phenotype seen in the child. This is the case, for example, in children who have neurofibromatosis 1 as a result of deletion of the entire NF1 locus.57 When this occurs, the child’s MR may be attributed to the involved gene, but, even in these instances, there may be other genes included in the CNV that also contribute to the MR.
A most compelling demonstration that MR can be caused by a particular CNV is obtained if sequence mutations of a gene within the affected region produce the same phenotypic abnormalities as the CNV. This approach is illustrated by the finding of CHD7 sequence mutations in 58% of individuals with CHARGE syndrome (MIM 214800)58 after CHD7 was identified within the CNVs of two other children with this phenotype.59 Such investigations have not been performed on the novel CNVs we found in this study, but the small size of some of these CNVs makes the genes within them attractive candidates for mutational analysis in children with MR who do not have detectable CNVs.
The results of our study demonstrate that WGSA with high-density, whole-genome SNP arrays can identify apparently pathogenic CNVs as small as 178 kb in children with MR. This technology appears to be capable of finding the cause of MR in at least twice as many affected individuals as is conventional cytogenetic analysis.
Note Added in Proof.—De novo CNVs were identified by 50K Xba GeneChip WGSA in 2 of 10 children studied who had multiple congenital anomalies and normal results on conventional cytogenetic analysis.60
Acknowledgments
This study was supported by funding from Genome Canada, Genome British Columbia, and the Canada Foundation for Innovation, with additional support from Affymetrix. Research at the British Columbia Cancer Agency is also supported by the British Columbia Cancer Foundation. M.A.M., S.J.M.J., and R.A.H. are Michael Smith Foundation for Health Research Scholars. We thank Dr. Wendy Robinson and Dr. Michael Taylor for providing us samples from control trios with known chromosomal abnormalities. We appreciate the skilled technical assistance of Nassim Aliakbarli, David Chai, Darko Curman, Christie Garbutt, Noriko Vickerson, and Willie Xue. This study was done in partnership with the affected families, to whom we are extremely grateful.
Web Resources
The URLs for data presented herein are as follows:
- Affymetrix GeneChip Human Mapping 100K Assay Manual, http://www.affymetrix.com/support/downloads/manuals/100k_manual.pdf [DOI] [PMC free article] [PubMed]
- BLAT, http://genome.ucsc.edu/cgi-bin/hgBlat?command=start
- Database of Genomic Variants, http://projects.tcag.ca/variation/
- DECIPHER, http://www.sanger.ac.uk/PostGenomics/decipher/
- GeneChip DNA Analysis Software (GDAS) Version 3.0, http://www.affymetrix.com/Auth/support/downloads/manuals/gdas_manual.zip
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Rubinstein-Taybi, Williams-Beuren, Smith-Magenis, and CHARGE syndromes)
- SAGE Library LSAGE_Brain_fetal_normal_B_S1, http://cgap.nci.nih.gov/SAGE/SAGELibInfo?LID=656&ORG=Hs
References
- 1.Roeleveld N, Zielhuis G, Gabreels F (1997) The prevalence of mental retardation: a critical review of recent literature. Dev Med Child Neurol 39:125–132 [DOI] [PubMed] [Google Scholar]
- 2.Pope A, Tarlov A (1991) Disability in America: toward a national agenda for prevention. Institute of Medicine, Washington, DC [Google Scholar]
- 3.van Karnebeek CD, Jansweijer MC, Leenders AG, Offringa M, Hennekam RC (2005) Diagnostic investigations in individuals with mental retardation: a systematic literature review of their usefulness. Eur J Hum Genet 13:6–25 10.1038/sj.ejhg.5201279 [DOI] [PubMed] [Google Scholar]
- 4.Leonard H, Wen X (2002) The epidemiology of mental retardation: challenges and opportunities in the new millennium. Ment Retard Dev Disabil Res Rev 8:117–134 10.1002/mrdd.10031 [DOI] [PubMed] [Google Scholar]
- 5.Ford CE, Jones KW, Polani PE, de Almeida JC, Briggs JH (1959) A sex-chromosome anomaly in a case of gonadal dysgenesis (Turner's syndrome). Lancet 1:711–713 10.1016/S0140-6736(59)91893-8 [DOI] [PubMed] [Google Scholar]
- 6.Jacobs P, Strong J (1959) A case of human intersexuality having a possible XXY sex-determining mechanism. Nature 183:302–303 10.1038/183302a0 [DOI] [PubMed] [Google Scholar]
- 7.Lejeune J, Gautier M, Turpin M (1959) Étude des chromosomes somatiques de neuf enfants mongoliens. C R Acad Sci 248:1721–1722 [PubMed] [Google Scholar]
- 8.Devriendt K, Vermeesch JR (2004) Chromosomal phenotypes and submicroscopic abnormalities. Hum Genomics 1:126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J, Chen Z (2003) Advances in molecular cytogenetics for the evaluation of mental retardation. Am J Med Genet C Semin Med Genet 117:15–24 10.1002/ajmg.c.10016 [DOI] [PubMed] [Google Scholar]
- 10.Menten B, Maas N, Thienpont B, Buysse K, Vandesompele J, Melotte C, de Ravel T, Van Vooren S, Balikova I, Backx L, Janssens S, De Paepe A, De Moor B, Moreau Y, Marynen P, Fryns JP, Mortier G, Devriendt K, Speleman F, Vermeesch JR (2006) Emerging patterns of cryptic chromosomal imbalances in patients with idiopathic mental retardation and multiple congenital anomalies: a new series of 140 patients and review of the literature. J Med Genet 43:625–633 10.1136/jmg.2005.039453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyson C, Harvard C, Locker R, Friedman JM, Langlois S, Lewis ME, Van Allen M, Somerville M, Arbour L, Clarke L, McGilivray B, Yong SL, Siegel-Bartel J, Rajcan-Separovic E (2005) Submicroscopic deletions and duplications in individuals with intellectual disability detected by array-CGH. Am J Med Genet A 139:173–185 [DOI] [PubMed] [Google Scholar]
- 12.de Vries BB, Pfundt R, Leisink M, Koolen DA, Vissers LE, Janssen IM, Reijmersdal S, Nillesen WM, Huys EH, Leeuw N, Smeets D, Sistermans EA, Feuth T, van Ravenswaaij-Arts CM, van Kessel AG, Schoenmakers EF, Brunner HG, Veltman JA (2005) Diagnostic genome profiling in mental retardation. Am J Hum Genet 77:606–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenberg C, Knijnenburg J, Bakker E, Vianna-Morgante AM, Sloos W, Otto PA, Kriek M, Hansson K, Krepischi-Santos AC, Fiegler H, Carter NP, Bijlsma EK, van Haeringen A, Szuhai K, Tanke HJ (2006) Array-CGH detection of micro rearrangements in mentally retarded individuals: clinical significance of imbalances present both in affected children and normal parents. J Med Genet 43:180–186 10.1136/jmg.2005.032268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoumans J, Ruivenkamp C, Holmberg E, Kyllerman M, Anderlid BM, Nordenskjold M (2005) Detection of chromosomal imbalances in children with idiopathic mental retardation by array based comparative genomic hybridisation (array-CGH). J Med Genet 42:699–705 10.1136/jmg.2004.029637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaw-Smith C, Redon R, Rickman L, Rio M, Willatt L, Fiegler H, Firth H, Sanlaville D, Winter R, Colleaux L, Bobrow M, Carter NP (2004) Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J Med Genet 41:241–248 10.1136/jmg.2003.017731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vissers LE, de Vries BB, Osoegawa K, Janssen IM, Feuth T, Choy CO, Straatman H, van der Vliet W, Huys EH, van Rijk A, Smeets D, van Ravenswaaij-Arts CM, Knoers NV, van der Burgt I, de Jong PJ, Brunner HG, van Kessel AG, Schoenmakers EF, Veltman JA (2003) Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am J Hum Genet 73:1261–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyake N, Shimokawa O, Harada N, Sosonkina N, Okubo A, Kawara H, Okamoto N, Kurosawa K, Kawame H, Iwakoshi M, Kosho T, Fukushima Y, Makita Y, Yokoyama Y, Yamagata T, Kato M, Hiraki Y, Nomura M, Yoshiura K, Kishino T, Ohta T, Mizuguchi T, Niikawa N, Matsumoto N (2006) BAC array CGH reveals genomic aberrations in idiopathic mental retardation. Am J Med Genet A 140:205–211 [DOI] [PubMed] [Google Scholar]
- 18.Brooks EM, Branda RF, Nicklas JA, O'Neill JP (2001) Molecular description of three macro-deletions and an Alu-Alu recombination-mediated duplication in the HPRT gene in four patients with Lesch-Nyhan disease. Mutat Res 476:43–54 [DOI] [PubMed] [Google Scholar]
- 19.Fridman C, Hosomi N, Varela MC, Souza AH, Fukai K, Koiffmann CP (2003) Angelman syndrome associated with oculocutaneous albinism due to an intragenic deletion of the P gene. Am J Med Genet A 119:180–183 10.1002/ajmg.a.20105 [DOI] [PubMed] [Google Scholar]
- 20.Ren Y, Saijo M, Nakatsu Y, Nakai H, Yamaizumi M, Tanaka K (2003) Three novel mutations responsible for Cockayne syndrome group A. Genes Genet Syst 78:93–102 10.1266/ggs.78.93 [DOI] [PubMed] [Google Scholar]
- 21.Ariani F, Mari F, Pescucci C, Longo I, Bruttini M, Meloni I, Hayek G, Rocchi R, Zappella M, Renieri A (2004) Real-time quantitative PCR as a routine method for screening large rearrangements in Rett syndrome: report of one case of MECP2 deletion and one case of MECP2 duplication. Hum Mutat 24:172–177 10.1002/humu.20065 [DOI] [PubMed] [Google Scholar]
- 22.Kinning E, Tufarelli C, Winship WS, Aldred MA, Trembath RC (2005) Genomic duplication in Dyggve Melchior Clausen syndrome, a novel mutation mechanism in an autosomal recessive disorder. J Med Genet 42:e70 10.1136/jmg.2005.033829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zanni G, Saillour Y, Nagara M, Billuart P, Castelnau L, Moraine C, Faivre L, Bertini E, Durr A, Guichet A, Rodriguez D, des Portes V, Beldjord C, Chelly J (2005) Oligophrenin 1 mutations frequently cause X-linked mental retardation with cerebellar hypoplasia. Neurology 65:1364–1369 10.1212/01.wnl.0000182813.94713.ee [DOI] [PubMed] [Google Scholar]
- 24.Vissers LE, Veltman JA, van Kessel AG, Brunner HG (2005) Identification of disease genes by whole genome CGH arrays. Hum Mol Genet Spec No 2 14:R215–R223 10.1093/hmg/ddi268 [DOI] [PubMed] [Google Scholar]
- 25.Stoughton RB (2005) Applications of DNA microarrays in biology. Annu Rev Biochem 74:53–82 10.1146/annurev.biochem.74.082803.133212 [DOI] [PubMed] [Google Scholar]
- 26.Rauch A, Ruschendorf F, Huang J, Trautmann U, Becker C, Thiel C, Jones KW, Reis A, Nurnberg P (2004) Molecular karyotyping using an SNP array for genomewide genotyping. J Med Genet 41:916–922 10.1136/jmg.2004.022855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bignell GR, Huang J, Greshock J, Watt S, Butler A, West S, Grigorova M, Jones KW, Wei W, Stratton MR, Futreal PA, Weber B, Shapero MH, Wooster R (2004) High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res 14:287–295 10.1101/gr.2012304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kennedy GC, Matsuzaki H, Dong S, Liu WM, Huang J, Liu G, Su X, Cao M, Chen W, Zhang J, Liu W, Yang G, Di X, Ryder T, He Z, Surti U, Phillips MS, Boyce-Jacino MT, Fodor SP, Jones KW (2003) Large-scale genotyping of complex DNA. Nat Biotechnol 21:1233–1237 10.1038/nbt869 [DOI] [PubMed] [Google Scholar]
- 29.Bruce S, Leinonen R, Lindgren CM, Kivinen K, Dahlman-Wright K, Lipsanen-Nyman M, Hannula-Jouppi K, Kere J (2005) Global analysis of uniparental disomy using high density genotyping arrays. J Med Genet 42:847–851 10.1136/jmg.2005.032367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altug-Teber O, Dufke A, Poths S, Mau-Holzmann UA, Bastepe M, Colleaux L, Cormier-Daire V, Eggermann T, Gillessen-Kaesbach G, Bonin M, Riess O (2005) A rapid microarray based whole genome analysis for detection of uniparental disomy. Hum Mutat 26:153–159 10.1002/humu.20198 [DOI] [PubMed] [Google Scholar]
- 31.Slater HR, Bailey DK, Ren H, Cao M, Bell K, Nasioulas S, Henke R, Choo KH, Kennedy GC (2005) High-resolution identification of chromosomal abnormalities using oligonucleotide arrays containing 116,204 SNPs. Am J Hum Genet 77:709–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nannya Y, Sanada M, Nakazaki K, Hosoya N, Wang L, Hangaishi A, Kurokawa M, Chiba S, Bailey DK, Kennedy GC, Ogawa S (2005) A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res 65:6071–6079 10.1158/0008-5472.CAN-05-0465 [DOI] [PubMed] [Google Scholar]
- 33.Zhao X, Li C, Paez JG, Chin K, Janne PA, Chen TH, Girard L, Minna J, Christiani D, Leo C, Gray JW, Sellers WR, Meyerson M (2004) An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res 64:3060–3071 10.1158/0008-5472.CAN-03-3308 [DOI] [PubMed] [Google Scholar]
- 34.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002) The human genome browser at UCSC. Genome Res 12:996–1006 10.1101/gr.229102. Article published online before print in May 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schein J, Kucaba T, Sekhon M, Smailus D, Waterston R, Marra M (2004) High-throughput BAC fingerprinting. In: Zhao S, Stodolsky M (eds) Methods in molecular biology: bacterial artificial chromosomes. Humana Press, Totowa, NJ, pp 143–156 [DOI] [PubMed] [Google Scholar]
- 36.Ewing B, Green P (1998) Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res 8:186–194 [PubMed] [Google Scholar]
- 37.Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185 [DOI] [PubMed] [Google Scholar]
- 38.Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12:656–664 10.1101/gr.229202. Article published online before March 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C (2004) Detection of large-scale variation in the human genome. Nat Genet 36:949–951 10.1038/ng1416 [DOI] [PubMed] [Google Scholar]
- 40.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M (2004) Large-scale copy number polymorphism in the human genome. Science 305:525–528 10.1126/science.1098918 [DOI] [PubMed] [Google Scholar]
- 41.Stankiewicz P, Lupski JR (2002) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82 10.1016/S0168-9525(02)02592-1 [DOI] [PubMed] [Google Scholar]
- 42.Wirtenberger M, Hemminki K, Burwinkel B (2006) Identification of frequent chromosome copy-number polymorphisms by use of high-resolution single-nucleotide-polymorphism arrays. Am J Hum Genet 78:520–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feuk L, Carson AR, Scherer SW (2006) Structural variation in the human genome. Nat Rev Genet 7:85–97 10.1038/nrg1767 [DOI] [PubMed] [Google Scholar]
- 44.Feuk L, Marshall CR, Wintle RF, Scherer SW (2006) Structural variants: changing the landscape of chromosomes and design of disease studies. Hum Mol Genet Suppl 1 15:R57–R66 10.1093/hmg/ddl057 [DOI] [PubMed] [Google Scholar]
- 45.Petrij F, Dauwerse HG, Blough RI, Giles RH, van der Smagt JJ, Wallerstein R, Maaswinkel-Mooy PD, van Karnebeek CD, van Ommen GJ, van Haeringen A, Rubinstein JH, Saal HM, Hennekam RC, Peters DJ, Breuning MH (2000) Diagnostic analysis of the Rubinstein-Taybi syndrome: five cosmids should be used for microdeletion detection and low number of protein truncating mutations. J Med Genet 37:168–176 10.1136/jmg.37.3.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK (2006) A high-resolution survey of deletion polymorphism in the human genome. Nat Genet 38:75–81 [DOI] [PubMed] [Google Scholar]
- 47.Eichler EE (2006) Widening the spectrum of human genetic variation. Nat Genet 38:9–11 [DOI] [PubMed] [Google Scholar]
- 48.Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA (2006) Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat Genet 38:82–85 [DOI] [PubMed] [Google Scholar]
- 49.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, Dallaire S, Gabriel SB, Lee C, Daly MJ, Altshuler DM, International HapMap Consortium (2006) Common deletion polymorphisms in the human genome. Nat Genet 38:86–92 [DOI] [PubMed] [Google Scholar]
- 50.Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Segraves R, Oseroff VV, Albertson DG, Pinkel D, Eichler EE (2005) Segmental duplications and copy-number variation in the human genome. Am J Hum Genet 77:78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, Olson MV, Eichler EE (2005) Fine-scale structural variation of the human genome. Nat Genet 37:727–732 10.1038/ng1562 [DOI] [PubMed] [Google Scholar]
- 52.Tassabehji M (2003) Williams-Beuren syndrome: a challenge for genotype-phenotype correlations. Hum Mol Genet Spec No 2 12:R229–R237 10.1093/hmg/ddg299 [DOI] [PubMed] [Google Scholar]
- 53.Shelley BP, Robertson MM (2005) The neuropsychiatry and multisystem features of the Smith-Magenis syndrome: a review. J Neuropsychiatry Clin Neurosci 17:91–97 [DOI] [PubMed] [Google Scholar]
- 54.Wooldridge J, Zunich J (1995) Trisomy 9 syndrome: report of a case with Crohn disease and review of the literature. Am J Med Genet 56:258–264 10.1002/ajmg.1320560304 [DOI] [PubMed] [Google Scholar]
- 55.Cantu ES, Eicher DJ, Pai GS, Donahue CJ, Harley RA (1996) Mosaic vs. nonmosaic trisomy 9: report of a liveborn infant evaluated by fluorescence in situ hybridization and review of the literature. Am J Med Genet 62:330–335 [DOI] [PubMed] [Google Scholar]
- 56.Sommer A, Pastore M, Wenger G (2006) Trisomy 16p: a longitudinal profile and photo essay. Am J Med Genet A 140:174–179 [DOI] [PubMed] [Google Scholar]
- 57.Descheemaeker MJ, Roelandts K, De Raedt T, Brems H, Fryns JP, Legius E (2004) Intelligence in individuals with a neurofibromatosis type 1 microdeletion. Am J Med Genet A 131:325–326 10.1002/ajmg.a.30346 [DOI] [PubMed] [Google Scholar]
- 58.Lalani SR, Safiullah AM, Fernbach SD, Harutyunyan KG, Thaller C, Peterson LE, McPherson JD, Gibbs RA, White LD, Hefner M, Davenport SL, Graham JM, Bacino CA, Glass NL, Towbin JA, Craigen WJ, Neish SR, Lin AE, Belmont JW (2006) Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype-phenotype correlation. Am J Hum Genet 78:303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, Janssen IM, van der Vliet WA, Huys EH, de Jong PJ, Hamel BC, Schoenmakers EF, Brunner HG, Veltman JA, van Kessel AG (2004) Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet 36:955–957 10.1038/ng1407 [DOI] [PubMed] [Google Scholar]
- 60.Ming JE, Geiger E, James AC, Ciprero KL, Nimmakayalu M, Zhang Y, Huang A, Vaddi M, Rappaport E, Zackai EH, Shaikh TH (2006) Rapid detection of submicroscopic chromosomal rearrangements in children with multiple congenital anomalies using high density oligonucleotide arrays. Hum Mutat 27:467–473 10.1002/humu.20322 [DOI] [PubMed] [Google Scholar]