Abstract
Oxidative stress could be involved in the pathophysiology of schizophrenia, a major psychiatric disorder. Glutathione (GSH), a redox regulator, is decreased in patients’ cerebrospinal fluid and prefrontal cortex. The gene of the key GSH-synthesizing enzyme, glutamate cysteine ligase modifier (GCLM) subunit, is strongly associated with schizophrenia in two case-control studies and in one family study. GCLM gene expression is decreased in patients’ fibroblasts. Thus, GSH metabolism dysfunction is proposed as one of the vulnerability factors for schizophrenia.
Schizophrenia (MIM 181500) is a major and frequent chronic psychiatric disorder with a strong genetic component.1,2 Converging evidence points to the involvement of oxidative stress3–7 and N-methyl d-aspartate (NMDA)–receptor hypofunction8,9 in the pathophysiology of the disease. As a main cellular nonprotein antioxidant and redox regulator,10 glutathione (GSH) plays a major role in protecting nervous tissue against reactive oxygen species11 and in modulating redox-sensitive sites, including NMDA receptors (NMDA-R).12,13 It was shown elsewhere that the GSH levels were decreased in patients’ cerebrospinal fluid (−27%), in medial prefrontal cortex in vivo (−52%),14,15 and in striatum postmortem tissue.16 GSH-deficient models reveal morphological, electrophysiological, and behavioral anomalies similar to those observed in patients.17–21 Here, we present strong evidence for an association between schizophrenia and the gene of the key GSH-synthesizing enzyme, glutamate cysteine ligase modifier (GCLM) subunit. The functional role of the GCLM gene variance in schizophrenia is supported by its low expression in patients’ fibroblasts and by the decreased stimulation of the enzyme activity when challenged by an oxidative stress.22 These findings are consistent with the concept that an abnormal GSH metabolism is a risk factor for schizophrenia.
To identify candidate gene(s) responsible for the low level of GSH observed in patients with schizophrenia, we studied steady-state levels of mRNA for 14 genes (data not shown) involved in GSH metabolism (fig. 1). Since GSH is ubiquitously present in cells, gene expression was studied in cultured skin fibroblasts. Two enzymes are responsible for GSH synthesis: glutamate cysteine ligase (GCL), also known as γ-glutamyl cysteine synthetase (Enzyme Commission number 6.3.2.2), and glutathione synthetase (GSS [Enzyme Commission number 6.3.2.3]).10 GCL, the first and rate-limiting enzyme,10 is composed of two subunits—GCL modifier (GCLM [light: 27.7 kDa])23 and GCL catalytic subunit (GCLC [heavy: 73 kDa])24—each encoded by separate genes.25 The specific mRNA steady-state levels were measured in fibroblasts obtained from 32 patients and 53 controls from a Swiss population (table 1). The subjects were recruited with fully informed written consent and guidelines for ethical treatment given by the University of Lausanne. All subjects were assessed using the Diagnostic Interview for Genetic Studies (DIGS) developed by the National Institute of Mental Health (NIMH).27 Additional measures of psychopathology of patients included the Positive and Negative Syndrome Scale (PANSS), which assessed the presence of symptoms within the same week as the blood collection and skin biopsy. Specific mRNA steady-state levels were measured in cultured skin fibroblasts grown for three passages, with the use of TaqMan chemistry and ABI Prism 7000 sequence detection system. cDNA corresponding to 10 ng of reverse-transcribed total RNA was amplified using TaqMan gene expression assays (Hs00155249 m1, Hs00157694 m1, and Hs00609286 m1) at the following amplification condition: 1 cycle for 2 min at 50°C, 1 cycle for 10 min at 95°C, and 50 cycles for 15 s at 95°C, followed by 1 min at 60°C. Human glyceraldehyde-3-phosphate dehydrogenase (Applied Biosystem 4333764F) was used as endogenous control.
Figure 1. .
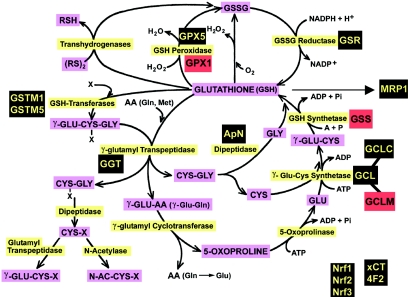
GSH metabolic pathway. The genes for which expression was studied in fibroblast cultures from skin biopsies are highlighted. Black indicates no difference between controls and patients, and red indicates lower expression in patients compared with controls. These genes code for the enzymes highlighted in yellow. Substrates and products are highlighted in purple. Genes not directly involved in GSH metabolism but included in the expression study: Nrf1, Nrf2, and Nrf3 = NF-E2–related transcription factors 1, 2, and 3, respectively; xCT and 4F2 = genes encoding proteins involved in cystine/glutamate exchange; and MRP1 = multidrug resistance protein 1. ADP = adenosine diphospate; GSSG = glutathione disulfide; RSH = reduced thiols; RS = disulfide; N-AC-CYS-X = N-acetyl-cysteine conjugate; Pi = inorganic phosphate.
Table 1. .
Demographic Characteristics of the Samples Used in Gene Expression and Association Studies
| Study and Population |
N | Agea (years) |
Sexb | Diagnostic Tool |
| Expression study: | ||||
| Switzerland: | ||||
| Patient | 32 | 35.5±10.5 | 3.0 | DSM-IV26/DIGS27 |
| Control | 53 | 37.2±13.4 | .9 | DIGS |
| Association study: | ||||
| Switzerland: | ||||
| Patient | 40 | 35.9±11.5 | 3.5 | DSM-IV/DIGS |
| Control | 31 | 37.7±13.31 | 1.6 | DIGS |
| Denmark: | ||||
| Patient | 349 | 38.8±12.1 | 1.5 | ICD-10c |
| Control | 348 | 40.2±10.5 | 1.5 | … |
Expressed as the mean and range.
Ratio of males to females.
International Classification of Diseases, 10th Revision.
Case-control comparisons showed significant differences in GCLM (t=1.989; P=.037) and GSS (t=1.997; P=.030) mRNA levels between patients and control subjects, without effect from sex or age. For the GCLC mRNA, a trend toward a decrease could be observed (t=2.000; P=.064) in patients.
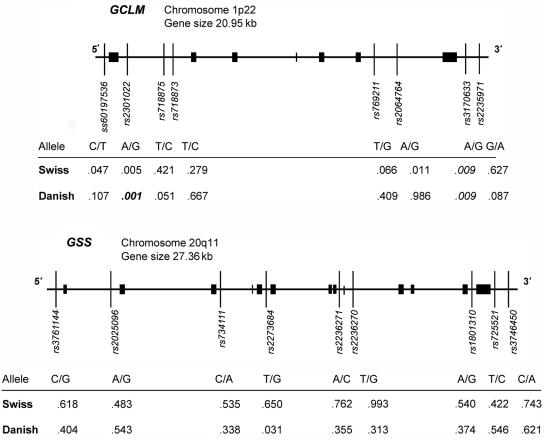
We tested, therefore, whether the reduced GCLM and GSS mRNA levels observed in patients with schizophrenia could be due to a primary defect. We studied eight SNPs in the GCLM gene and nine SNPs in the GSS gene for possible association with schizophrenia (fig. 2). Sixteen SNPs were chosen from a group of 60 SNPs selected from publicly available databases (SNP Consortium and dbSNP) on the basis of the estimated level of polymorphism in our population. One SNP (ss60197536) at the GCLM 5′ end, described by Nakamura et al.,28 is being submitted to dbSNP. The dbSNP-annotated SNP numbers and their positions in each gene are shown in figure 2. Genotyping was performed with DNA extracted from peripheral blood by the use of either Sequenom technology29 or sequencing. The list of specific primers is shown in table 2.
Figure 2. .
Map of GCLM and GSS genomic structures, with the positions of SNP markers and results of two case-control studies. Tables show P values for each SNP in case-control studies of subjects from two independent populations, from Switzerland and Denmark. Values in italics indicate a significant difference, and the value in bold italics indicates a significant difference after correction for multiple testing.
Table 2. .
Primers Used in Genotyping Studies
| PCR Amplification Primer(5′→3′ |
|||
| SNP | 1 | 2 | Extension Primer |
| rs2235971 | ACGTTGGATGCAGATCTGGTAACCACCATC | ACGTTGGATGAGTTCTCTGACGCATTTCCG | CACCATCTTTCCGGCTC |
| rs3170633 | ACGTTGGATGCTTTCTAGATTTTTCACCCAG | ACGTTGGATGAGGATGAACTGCTAGCCAAC | CAGTATTTTCAAAATTTGGGAAT |
| rs2064764 | ACGTTGGATGCCCTCTTCTAGCTTCACTTG | ACGTTGGATGAAACACTAGGAACCTTAATC | CTTTTACTAGTAGGAAAGGAA |
| rs769211 | ACGTTGGATGGATCATAAGCTTTTGTCTTAC | ACGTTGGATGCTGTATTTTTATCACTGTCC | CTTTTGTCTTACAAAAAGGTATTT |
| rs718873 | ACGTTGGATGTAACCTCTAGTTGGTTCTGC | ACGTTGGATGGGAGTTGAGTGTCATTCCAG | TTGGTTCTGCTCCTTCC |
| rs718875 | ACGTTGGATGCTTACCTTCCTGAATTGAGG | ACGTTGGATGAATTTCCCTCTGGAAGGATG | AATTCATCAGGAAAGCCTCA |
| rs2301022 | ACGTTGGATGTGATGCTCAGAGTCACACAC | ACGTTGGATGCCTACTGTTATGAAGCACCC | AAACATTGTTCAAAGGACTA |
| ss60197536 | ACGTTGGATGTGAGGTAGACACCGCCTCC | ACGTTGGATGAAAGAGACGTGTAGGAAGCC | GCCTGGTGAGGTCTCCC |
| rs3746450 | ACGTTGGATGCAGGACTTCTCTTTCTCCAG | ACGTTGGATGTTATCCTGGGTGACTACCTC | GTCCCCCTCCCTCTAGA |
| rs725521 | ACGTTGGATGTAGACCAGTCTCTACAGGTG | ACGTTGGATGTCTCATTCCTCCCTGTGATC | ATCCTTAGCCACCCACT |
| rs1801310 | ACGTTGGATGACGGTTGCAAAGGACTTCTC | ACGTTGGATGTTAAATGAGGCCAAGGACCC | TCATCTGATACCCTGGT |
| rs2236270 | ACGTTGGATGCCAGTGAGAGCTGATTGTTG | ACGTTGGATGGAATCCTCAGGAATCCACAG | TCTGGAAACAGTGTAAATG |
| rs2236271 | ACGTTGGATGTTGCGTTTTCACCTTCACCC | ACGTTGGATGTTTCCACTGCTTAAAGCAGC | CCCTGCCATTAAAAATTTTTTCA |
| rs2273684 | ACGTTGGATGTCTGAGAATCAGCTGAGCAC | ACGTTGGATGCAGCCCAGCATATTCCAACC | CTCCCATCACATTCCTG |
| rs734111 | ACGTTGGATGCTCTGCAATCTTCCAGTTCC | ACGTTGGATGCAAACTCTTTCCAGGTAGGG | GCAGCTCCTGGCCCCCC |
| rs2025096 | ACGTTGGATGCGAGGTGATGACTGGTATAG | ACGTTGGATGTCTTTCTCCAATGAAGAGCC | TTGAACCCATGTCTCTG |
| rs3761144 | ACGTTGGATGCTTTTGCCTCTAATGCTTTCC | ACGTTGGATGAAGTCCCAGAAAAATCCCCC | TAATGCTTTCCCTGCTG |
A pilot association study was performed with a relatively small sample (40 patients and 31 controls from a Swiss population) (fig. 2). Most, although not all, of the subjects from the Swiss population were used in both gene expression and association studies. The details about subject groups are given in the tables: demographic data in table 1, genotype and allele frequencies for each SNP in table 3, and the allelic frequencies compared with those known for other populations in table 4. All SNPs were in Hardy-Weinberg equilibrium (HWE) in both subject groups (data not shown). No association was observed with the GSS gene. Two GCLM markers, rs2301022 in intron 1 and rs3170633 at the 3′ end, had P values that were low, but not significant after correction for multiple testing. However, these data suggested that a functional region in GCLM may be associated with schizophrenia. Consequently, we proceeded with a second case-control study of a larger sample from an independent population.
Table 3. .
Genotype and Allele Frequencies for Each SNP Studied in Two Populations[Note]
| Frequency |
||||||||||
| Swiss |
Danish |
|||||||||
| Genotype |
Allele |
Genotype |
Allele |
|||||||
| Gene and SNP | 1/1 | 1/2 | 2/2 | 1 | 2 | 1/1 | 1/2 | 2/2 | 1 | 2 |
| GCLM: | ||||||||||
| ss60197536 (C/T) | .73 | .23 | .04 | .84 | .16 | .71 | .27 | .03 | .84 | .16 |
| rs2301022 (A/G) | .13 | .40 | .47 | .33 | .67 | .13 | .42 | .45 | .34 | .66 |
| rs718875 (C/T) | .00 | .16 | .84 | .08 | .92 | .01 | .24 | .76 | .13 | .87 |
| rs718873 (C/T) | .00 | .33 | .67 | .16 | .84 | .03 | .24 | .73 | .15 | .85 |
| rs769211 (G/T) | .58 | .39 | .03 | .77 | .23 | .53 | .39 | .08 | .73 | .27 |
| rs2064764 (A/G) | .15 | .43 | .43 | .36 | .64 | .11 | .45 | .44 | .34 | .66 |
| rs3170633 (A/G) | .16 | .43 | .41 | .37 | .63 | .26 | .42 | .32 | .47 | .53 |
| rs2235971 (A/G) | .11 | .43 | .45 | .33 | .67 | .11 | .43 | .45 | .33 | .67 |
| GSS: | ||||||||||
| rs3761144 (C/G) | .40 | .46 | .14 | .63 | .37 | .37 | .50 | .13 | .62 | .38 |
| rs2025096 (A/G) | .09 | .38 | .53 | .28 | .72 | .03 | .30 | .67 | .18 | .82 |
| rs734111 (A/C) | .13 | .46 | .41 | .36 | .64 | .12 | .50 | .38 | .37 | .63 |
| rs2273684 (G /T) | .27 | .47 | .26 | .51 | .49 | .18 | .58 | .24 | .47 | .53 |
| rs2236270 (G /T) | .20 | .48 | .33 | .44 | .56 | .12 | .50 | .38 | .37 | .63 |
| rs1801310 (A/G) | .34 | .44 | .22 | .56 | .44 | .37 | .51 | .12 | .63 | .37 |
| rs2236271 (A/C) | .14 | .44 | .42 | .36 | .64 | .11 | .49 | .39 | .36 | .64 |
| rs725521 (C/T) | .41 | .45 | .14 | .64 | .36 | .40 | .49 | .12 | .64 | .36 |
| rs3746450 (A/C) | .28 | .50 | .22 | .53 | .47 | .39 | .49 | .12 | .63 | .37 |
Note.— The alternative alleles given in parentheses for each SNP correspond to 1 and 2, respectively, in the table.
Table 4. .
Frequencies of the Rare Allele in Swiss and Danish Populations Compared with Reported Frequencies in Other White Populations
| Frequency |
|||
| Gene and SNP | Swiss | Danish | Other White Populationsa |
| GCLM: | |||
| ss60197536 (C/T) | .16 | .16 | ND |
| rs2301022 (A/G) | .33 | .34 | .37 (.31–.44) |
| rs718875 (C/T) | .08 | .13 | .09 |
| rs718873 (C/T) | .16 | .15 | .23 |
| rs769211 (G/T) | .23 | .27 | .17 |
| rs2064764 (A/G) | .36 | .34 | ND |
| rs3170633 (A/G) | .37 | .47 | .18 (.11–.25) |
| rs2235971 (A/G) | .33 | .33 | .35 |
| GSS: | |||
| rs3761144 (C/G) | .37 | .38 | .37 (.35–.39) |
| rs2025096 (A/G) | .28 | .18 | ND |
| rs734111 (A/C) | .36 | .37 | .34 (.21–.49) |
| rs2273684 (G/T) | .49 | .47 | .48 (.46–.51) |
| rs2236270 (G/T) | .44 | .37 | .39 (.3–.44) |
| rs1801310 (A/G) | .44 | .37 | .28 (.14–.42) |
| rs2236271 (A/C) | .36 | .36 | .35 (.17–.58) |
| rs725521 (C/T) | .36 | .36 | .33 (.21–.46) |
| rs3746450 (A/C) | .47 | .37 | .42 |
Mean and range shown when data for more than one population were available in dbSNP. ND = No data available.
The second study included 349 patients with schizophrenia from the Danish Psychiatric Biobank and 348 unrelated anonymous blood donors serving as unaffected control subjects (table 1). Both control and affected groups were tested for HWE. Whereas the control group was in equilibrium for all markers, the SNP rs2301022 showed a deviation from HWE (P=.026) in patients. This SNP also showed an association in our Swiss population. Genotype frequency analysis confirmed a strong association of SNP rs2301022 (χ2=13.2, 2 df; P=.023) after correction for multiple testing (fig. 2). Thus, the deviation from HWE in patients but not in controls must be viewed as additional evidence of association. Here again, no association with GSS SNPs was found; thus, we excluded this gene from further studies.
Our results showed that rs2301022 is strongly associated with schizophrenia. This marker is localized between markers rs718875 and ss60197536, which showed weaker association with this disease in one of two populations that we studied (fig. 2). This fact suggests that there is a functional variant associated with the disease in the proximity of these three markers. SNP ss60197536 is 343 bp upstream of the transcription initiation site, and rs718875 is localized, like rs2301022, in intron 1. We analyzed different genotype pattern classes for these three markers (ss60197536, rs2301022, and rs718875) in a region of ∼3,000 bp that includes GCLM exon 1. We identified 14 different genotype patterns (table 5), among which 2 were specifically present in affected individuals. Nine patterns of these three SNPs showed different frequencies between patients and unaffected controls, with χ2=30.39 and P=.004. Two particular combinations, TT/GG/TC and CC/GG/TT, had odds ratios (ORs) of 4.89 and 4.17, respectively.
Table 5. .
Diplotype Analysis of the SNP Patterns Associated with Schizophrenia[Note]
| Genotype Pattern |
Frequency |
||||
| rs718875 | rs2301022 | ss60197536 | Patients | Controls | OR |
| CC | GG | TT | .035 | .009 | 4.17 |
| TC | GG | TT | .006 | .014 | .4 |
| TT | GG | CC | .263 | .282 | .91 |
| TT | GG | TC | .041 | .009 | 4.89 |
| TT | AG | TC | .05 | .035 | 1.46 |
| TT | AG | CC | .339 | .3 | 1.2 |
| TC | GG | TC | .137 | .138 | .99 |
| TT | AA | CC | .044 | .121 | .33 |
| TC | AG | TC | .067 | .078 | .85 |
| Five rare patterns | .018 | .014 | 1.22 | ||
Note.— χ2=30.40, 9 df; P=.000375. Cells highlighted in bold show significantly different values.
At present, we cannot completely exclude the possibility of a second functionally associated region in the vicinity of SNP rs3170633. The association test for this SNP showed low P values in both populations (P=.009) (fig. 2). However, these values are not significant after correction for multiple testing.
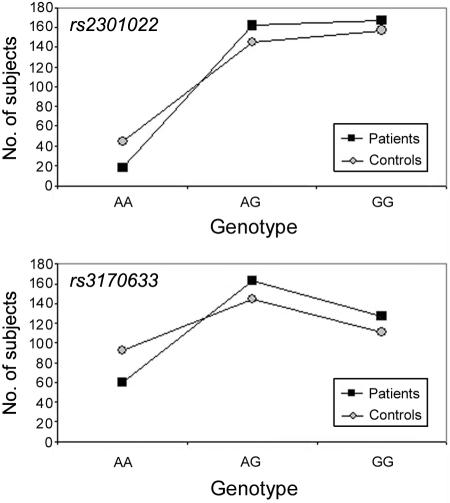
Genotype frequency distribution in the Danish population suggests a dominant mode of transmission for the two SNPs of interest (rs2301022 and rs3170633), with the G allele dominant in each of them (fig. 3). For rs2301022, genotypes AA, AG, and GG show respective ORs of 0.37, 1.22, and 1.12. Thus, we pooled AG and GG genotypes, since both are associated with disease, to approximately the same degree. The corresponding OR for disease then was 2.72 (P=.0005). For rs3170633, which showed weaker association, OR=1.77 (P=.002).
Figure 3. .
Genotype distribution of the two most common SNPs at the GCLM gene in the controls and patients sampled from a Danish population. The graph shows a dominance of the G allele for both markers.
Thus, case-control studies of two independent populations provided strong evidence of an association of the GCLM gene and schizophrenia. These data were supported with an additional linkage study of the families from the NIMH cohort, shown in table 6. Genotyping of 275 individuals from 72 families for seven SNPs in the GCLM gene showed supportive (although by no means significant) evidence of linkage between schizophrenia and two GCLM markers. The highest LOD score, 1.382 (corresponding to P=.012), was obtained under assumed dominant inheritance for rs2064764, which is located within 2 kb (∼0.002 cM) of rs3170633.
Table 6. .
Family Linkage Analysis of the GCLM Gene[Note]
| Dominant Model |
Recessive Model |
|||||
| SNP | LODa | χ2 | P | LODa | χ2 | P |
| rs2235971 | 0 | 0 | 1 | .038 | .176 | .675 |
| rs3170633 | .568 | 2.614 | .106 | .215 | .991 | .320 |
| rs2064764 | 1.382 | 6.363 | .012 | .470 | 2.163 | .141 |
| rs769211 | .271 | 1.248 | .264 | .081 | .371 | .542 |
| rs718873 | 1.106 | 5.093 | .024 | 1.206 | 5.553 | .019 |
| rs718875 | .516 | 2.375 | .123 | .109 | .501 | .479 |
| rs2301022 | .034 | .157 | .692 | .170 | .785 | .376 |
Note.— Cells highlighted in bold show significantly different values.
LOD score is log10 (OR).
To test whether there is association in the presence of linkage, we ran a family-based association statistic test (FBAT v. 1.7.2) in NIMH families (table 7). The results showed that there is significant evidence (Z=3.247; P=.0012) to accept the alternative hypothesis of association in the presence of linkage between SNP marker rs2301022 and the schizophrenia phenotype: allele G at the marker appeared as significantly overtransmitted to the affected offspring. Moreover, the P value remains significant, experiment-wise, after correction for multiple testing (table 7).
Table 7. .
Family-Based Association Test for SNP Markers Linked to GCLM[Note]
| Marker | Allele | Allele Frequency | Z | P |
| rs2301022 | G | .567 | 3.247 | .0012 |
| rs718875 | T | .892 | .202 | .8399 |
| rs718873 | T | .862 | .258 | .7963 |
| rs769211 | T | .209 | .784 | .4332 |
| rs2064764 | A | .428 | .935 | .3496 |
| rs3170633 | A | .441 | .577 | .5637 |
| rs2235971 | A | .266 | .832 | .4054 |
Note.— Cells highlighted in bold show significantly different values.
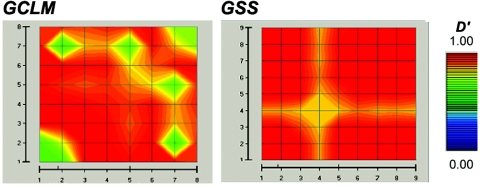
To define possible functional variants associated with the disease, we estimated pairwise linkage disequilibrium (LD) for all 17 SNPs of GCLM and GSS genes in 348 unaffected subjects from Denmark. The resulting LD map showed very strong association among all markers within each of the two genes (fig. 4). Thus, any association with this region might suggest the presence of a functional variant associated with schizophrenia.
Figure 4. .
GOLD plot of pairwise LD for 17 SNPs that belong to genes GCLM and GSS. The 17 SNPs used in association and linkage studies and graphically presented in figure 2 are labeled on both axes. The color differences represent regions of the local map that are in disequilibrium and the magnitude of that disequilibrium. Regions of high and low LD are presented in red and blue, respectively. GOLD plot shows very strong LD within each gene.
On the basis of the strong association of two SNPs (rs2301022 and rs718875) in the 5′ region of the GCLM gene and schizophrenia, we examined the relationship of these variants and the GCLM expression level in the Swiss population. The mRNA steady-state level in cultured fibroblasts showed a significant correlation with these two SNPs (P=.040). This result confirms the functional effect of the GCLM variants on the schizophrenia phenotype. However, it is still unknown in which way these variants affect the GCLM gene expression, since their localization is not directly related to any of the currently known regulatory sequences. It is worth noting that the GCLM gene is localized on chromosome 1p21, the region shown by previous linkage studies to be one of the several regions critical for schizophrenia.30,31
Several genes involved in GSH metabolism have already been considered as potential candidates for schizophrenia. Association of the glutathione-S-transferase M 1 gene was shown in a subgroup of patients with schizophrenia in Japanese32 and Korean33 populations. Case-control studies of glutathione-S-transferase P1 and glutathione peroxidase 1 showed no association.34,35
Although GCLM is not essential for survival, its interaction with the GCLC subunit increases, by four- to fivefold, the catalytic efficiency of the holoenzyme.36 The GCLM knockout (KO) mice exhibit an increased sensitivity to oxidative stress37; mouse fetal fibroblasts are 10-fold more sensitive to an oxidative stress than are the wild type. Similarly, we observed lower GCL activity in patients’ fibroblasts exposed to an oxidative stress, compared with that in control fibroblasts.22 Thus, a GCLM defect could lead to GCL-activity dysregulation, consistent with the involvement of oxidative stress–induced impairment of neuronal processes and mitochondrial function3–7 reported for schizophrenia.
GCLM KO mice showed an increased feedback inhibition of GCL activity, apparently resulting in brain GSH levels ∼40% of normal.36,37 This is strikingly similar to the levels 52% below normal reported for patients with schizophrenia.15 Thus, GCLM KO mice can be used as a model for further studies of GSH deficit in schizophrenia.
A GCL dysregulation could lead to cellular alterations in the surroundings of dopaminergic terminals, affecting the synaptic contacts on dendritic spines of prefrontal cortical neurons that are particularly rich in dopamine innervations. Indeed, the metabolism of dopamine generates reactive oxygen species (e.g., hydrogen peroxide and quinones), which, in GSH-deficit conditions, are not adequately neutralized and, thus, induce cellular damage.11 Interestingly, in rat models, GSH deficit and excess dopamine during development mimic structural and functional anomalies observed in patients: they exhibited a decrease in spine density of pyramidal neurons (F. Gheorghita, unpublished data) and, selectively, in GABA-parvalbumin immunoreactivity of prefrontal cortex,21 similar to patients.38,39 The same rat model presented an impairment in object recognition18,19 and in integration of olfactory information,40 reproducing some cognitive deficits of schizophrenia.
Furthermore, NMDA-R hypofunction is implicated in schizophrenia, since the NMDA-R antagonist phencyclidine induces a psychotic syndrome.41 In the case of GSH deficit, NMDA-R activity could be depressed through interaction at their redox sites.12,13 Similarly, in rat hippocampal slices, GSH depletion impaired NMDA-dependent synaptic plasticity.20
In conclusion, these studies provide converging evidence of a link between schizophrenia and GCLM genetic variations, which affect the function of the encoded protein in its ability to promote GSH synthesis when challenged by an oxidative stress. They support the new concept that a dysregulation of GSH metabolism is one of the vulnerability factors contributing to the development of the disease.
Acknowledgments
We are grateful to Marinette Blanc, Sylviane Raymond, and Beatrice Benz, for technical help; to Dr. Messod Benathan, for advice concerning skin biopsy and fibroblast cell cultures; to Dr. Franziska Gamma, for participation in patient recruitment; and to Dr. Graham Knott, for critical reading of the manuscript. We are thankful to Prof. Olivier Halfon, for financial support of F.G., and to Prof. Pierre Magistretti, for his constant support. This work was supported by Swiss National Research Foundation grant 31-55924.98, by the Novartis Research Foundation, and by NIMH grant MH44292.
Web Resources
The URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- International Classification of Diseases (ICD-10), http://www3.who.int/icd/currentversion/fr-icd.htm
- Online Mendelian Inheritance in Man (OMIM) http://www.ncbi.nlm.nih.gov/Omim/ (for schizophrenia)
- SNP Consortium, http://snp.cshl.org/
References
- 1.Tsuang MT (2000) Schizophrenia: genes and environment. Biol Psychiatry 47:210–220 10.1016/S0006-3223(99)00289-9 [DOI] [PubMed] [Google Scholar]
- 2.Harrison PJ, Owen MJ (2003) Genes for schizophrenia? Recent findings and their pathophysiological implications. Lancet 361:417–419 10.1016/S0140-6736(03)12379-3 [DOI] [PubMed] [Google Scholar]
- 3.Mahadik SP, Mukherjee S (1996) Free radical pathology and antioxidant defense in schizophrenia: a review. Schizophr Res 19:1–17 10.1016/0920-9964(95)00049-6 [DOI] [PubMed] [Google Scholar]
- 4.Herken H, Uz E, Ozyurt H, Sogut S, Virit O, Akyol O (2001) Evidence that the activities of erythrocyte free radical scavenging enzymes and the products of lipid peroxidation are increased in different forms of schizophrenia. Mol Psychiatry 6:66–73 10.1038/sj.mp.4000789 [DOI] [PubMed] [Google Scholar]
- 5.Marchbanks RM, Ryan M, Day I, Owen M, McGuffin P, Whatley S (2003) A mitochondrial DNA sequence variant associated with schizophrenia and oxidative stress. Schizophr Res 65:33–38 10.1016/S0920-9964(03)00011-2 [DOI] [PubMed] [Google Scholar]
- 6.Prabakaran S, Swatton J, Ryan M, Huffaker SJ, Huang JJ, Griffin JL, Wayland M, Freeman T, Dudbridge F, Lilley KS, Karp NA, Hester S, Tkachev D, Mimmack ML, Yolken RH, Webster MJ, Torrey EF, Bahn S (2004) Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry 9:684–697 [DOI] [PubMed] [Google Scholar]
- 7.Altar CA, Jurata LW, Charles V, Lemire A, Liu P, Bukhman Y, Young TA, Bullard J, Yokoe H, Webster MJ, Knable MB, Brockman JA (2005) Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry 58:85–96 10.1016/j.biopsych.2005.03.031 [DOI] [PubMed] [Google Scholar]
- 8.Olney JW, Newcomer JW, Farber NB (1999) NMDA receptor hypofunction model of schizophrenia. J Psychiatr Res 33:523–533 10.1016/S0022-3956(99)00029-1 [DOI] [PubMed] [Google Scholar]
- 9.Coyle JT, Tsai G (2004) NMDA receptor function, neuroplasticity, and the pathophysiology of schizophrenia. Int Rev Neurobiol 59:491–515 [DOI] [PubMed] [Google Scholar]
- 10.Meister A, Anderson ME (1983) Glutathione. Annu Rev Biochem 52:711–760 10.1146/annurev.bi.52.070183.003431 [DOI] [PubMed] [Google Scholar]
- 11.Rabinovic AD, Hastings TG (1998) Role of endogenous glutathione in the oxidation of dopamine. J Neurochem 71:2071–2078 [DOI] [PubMed] [Google Scholar]
- 12.Kohr G, Eckardt S, Luddens H, Monyer H, Seeburg PH (1994) NMDA receptor channels: subunit-specific potentiation by reducing agents. Neuron 12:1031–1040 10.1016/0896-6273(94)90311-5 [DOI] [PubMed] [Google Scholar]
- 13.Choi YB, Lipton SA (2000) Redox modulation of the NMDA receptor. Cell Mol Life Sci 57:1535–1541 10.1007/PL00000638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Do KQ, Lauer CJ, Schreiber W, Zollinger M, Gutteck-Amsler U, Cuenod M, Holsboer F (1995) Gamma-glutamylglutamine and taurine concentrations are decreased in the cerebrospinal fluid of drug-naive patients with schizophrenic disorders. J Neurochem 65:2652–2662 [DOI] [PubMed] [Google Scholar]
- 15.Do KQ, Trebesinger A, Kirsten-Kruger M, Lauer C, Dydak U, Hell D, Holsboer F, Boesinger P, Cuenod M (2000) Schizophrenia: glutathione deficit in cerebro spinal fluid and prefrontal cortex in vivo. Eur J Neurosci 12:3721–3728 10.1046/j.1460-9568.2000.00229.x [DOI] [PubMed] [Google Scholar]
- 16.Yao JK, Leonard S, Reddy R (2006) Altered glutathione redox state in schizophrenia. Dis Markers 22:83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grima G, Benz B, Parpura V, Cuenod M, Do KQ (2003) Dopamine-induced oxidative stress in neurons with glutathione deficit: implication for schizophrenia. Schizophr Res 62:213–224 10.1016/S0920-9964(02)00405-X [DOI] [PubMed] [Google Scholar]
- 18.Castagne V, Rougemont M, Cuenod M, Do KQ (2004) Low brain glutathione and ascorbic acid associated with dopamine uptake inhibition during rat’s development induce long-term cognitive deficit: relevance to schizophrenia. Neurobiol Dis 15:93–105 10.1016/j.nbd.2003.09.005 [DOI] [PubMed] [Google Scholar]
- 19.Castagne VV, Cuenod M, Do KQ (2004) An animal model with relevance to schizophrenia: sex-dependent cognitive deficits in osteogenic disorder-Shionogi rats induced by glutathione synthesis and dopamine uptake inhibition during development. Neuroscience 123:821–834 10.1016/j.neuroscience.2003.11.012 [DOI] [PubMed] [Google Scholar]
- 20.Steullet P, Neijt H, Cuenod M, Do KQ (2005) Synaptic plasticity impairment and hypofunction of NMDA receptors induced by glutathione deficit: relevance to schizophrenia. Neuroscience 137:807–819 10.1016/j.neuroscience.2005.10.014 [DOI] [PubMed] [Google Scholar]
- 21.Cabungcal J, Nicolas D, Kraftsik R, Cuenod M, Do KQ, Hornung JP (2006) Glutathione deficit during development induces anomalies in the rat anterior cingulate GABAergic neurons: relevance to schizophrenia. Neurobiol Dis 22:624–637 10.1016/j.nbd.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 22.Gysin R, Tosic M, Chappuis C, Deppen P, Ruiz V, Bovet P, Cuenod M, Do KQ (2005) Dysregulation of glutamate cysteine ligase in schizophrenia. Paper presented at the 36th Annual Meeting of the Society for Neuroscience, Washington, DC, November 12–16 [Google Scholar]
- 23.Huang CS, Anderson ME, Meister A (1993) Amino acid sequence and function of the light subunit of rat kidney gamma-glutamylcysteine synthetase. J Biol Chem 268:20578–20583 [PubMed] [Google Scholar]
- 24.Huang CS, Chang LS, Anderson ME, Meister A (1993) Catalytic and regulatory properties of the heavy subunit of rat kidney gamma-glutamylcysteine synthetase. J Biol Chem 268:19675–19680 [PubMed] [Google Scholar]
- 25.Tsuchiya K, Mulcahy RT, Reid LL, Disteche CM, Kavanagh TJ (1995) Mapping of the glutamate-cysteine ligase catalytic subunit gene (GLCLC) to human chromosome 6p12 and mouse chromosome 9D-E and of the regulatory subunit gene (GLCLR) to human chromosome 1p21-p22 and mouse chromosome 3H1-3. Genomics 30:630–632 10.1006/geno.1995.1293 [DOI] [PubMed] [Google Scholar]
- 26.American Psychiatric Association (1994) Diagnostic and statistical manual of mental disorders, 4th ed. American Psychiatric Association, Washington, DC [Google Scholar]
- 27.Nurnberger JI Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T (1994) Diagnostic interview for genetic studies: rationale, unique features, and training: NIMH Genetics Initiative. Arch Gen Psychiatry 51:849–859 [DOI] [PubMed] [Google Scholar]
- 28.Nakamura S, Kugiyama K, Sugiyama S, Miyamoto S, Koide S, Fukushima H, Honda O, Yoshimura M, Ogawa H (2002) Polymorphism in the 5′-flanking region of human glutamate-cysteine ligase modifier subunit gene is associated with myocardial infarction. Circulation 105:2968–2973 10.1161/01.CIR.0000019739.66514.1E [DOI] [PubMed] [Google Scholar]
- 29.Rodi CP, Darnhofer-Patel B, Stanssens P, Zabeau M, van den Boom D (2002) A strategy for the rapid discovery of disease markers using the MassARRAY system. Biotechniques 32:S62–S69 [PubMed] [Google Scholar]
- 30.Pulver AE, Mulle J, Nestadt G, Swartz KL, Blouin JL, Dombroski B, Liang KY, Housman DE, Kazazian HH, Antonarakis SE, Lasseter VK, Wolyniec PS, Thornquist MH, McGrath JA (2000) Genetic heterogeneity in schizophrenia: stratification of genome scan data using co-segregating related phenotypes. Mol Psychiatry 5:650–653 10.1038/sj.mp.4000814 [DOI] [PubMed] [Google Scholar]
- 31.Arinami T, Ohtsuki T, Ishiguro H, Ujike H, Tanaka Y, Morita Y, Mineta M, et al (2005) Genomewide high-density SNP linkage analysis of 236 Japanese families supports the existence of schizophrenia susceptibility loci on chromosomes 1p, 14q, and 20p. Am J Hum Genet 77:937–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harada S, Tachikawa H, Kawanishi Y (2001) Glutathione S-transferase M1 gene deletion may be associated with susceptibility to certain forms of schizophrenia. Biochem Biophys Res Commun 281:267–271 10.1006/bbrc.2001.4347 [DOI] [PubMed] [Google Scholar]
- 33.Pae CU, Yu HS, Kim JJ, Kim W, Lee CU, Lee SJ, Jun TY, Lee C, Paik IH, Serretti A (2004) Glutathione S-transferase M1 polymorphism may contribute to schizophrenia in the Korean population. Psychiatr Genet 14:147–150 10.1097/00041444-200409000-00005 [DOI] [PubMed] [Google Scholar]
- 34.Pae CU, Kim JJ, Lee SJ, Lee CU, Lee C, Paik IH, Park HR, Yang S, Serretti A (2003) Association study between glutathione S-transferase P1 polymorphism and schizophrenia in the Korean population. Prog Neuropsychopharmacol Biol Psychiatry 27:519–523 10.1016/S0278-5846(03)00043-5 [DOI] [PubMed] [Google Scholar]
- 35.Shinkai T, Luca VD, Zai G, Shaikh S, Matsumoto C, Arnold PD, Hwang R, King N, Trakalo J, Potapova N, Wong G, Hori H, Wong AH, Ohmori O, Nakamura J, Kennedy JL (2004) No association between the Pro197Leu polymorphism in the glutathione peroxidase (GPX1) gene and schizophrenia. Psychiatr Genet 14:177–180 10.1097/00041444-200409000-00012 [DOI] [PubMed] [Google Scholar]
- 36.Chen Y, Shertzer HG, Schneider SN, Nebert DW, Dalton TP (2005) Glutamate cysteine ligase catalysis: dependence on ATP and modifier subunit for regulation of tissue glutathione levels. J Biol Chem 280:33766–33774 10.1074/jbc.M504604200 [DOI] [PubMed] [Google Scholar]
- 37.Yang Y, Dieter MZ, Chen Y, Shertzer HG, Nebert DW, Dalton TP (2002) Initial characterization of the glutamate-cysteine ligase modifier subunit Gclm(−/−) knockout mouse: novel model system for a severely compromised oxidative stress response. J Biol Chem 277:49446–49452 10.1074/jbc.M209372200 [DOI] [PubMed] [Google Scholar]
- 38.Lewis DA, Hashimoto T, Volk DW (2005) Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci 6:312–324 10.1038/nrn1648 [DOI] [PubMed] [Google Scholar]
- 39.Kolluri N, Sun Z, Sampson AR, Lewis DA (2005) Lamina-specific reductions in dendritic spine density in the prefrontal cortex of subjects with schizophrenia. Am J Psychiatry 162:1200–1202 10.1176/appi.ajp.162.6.1200 [DOI] [PubMed] [Google Scholar]
- 40.Cabungcal J, Singer D, Hornung J-P, Cuenod M, Do KQ, Schenk F (2004) Special bias deficit in rats with low glutathione during development: a behaviour model with relevance to schizophrenia. Schizophr Res 67 Suppl 1:118 [Google Scholar]
- 41.Krystal JH, Karper LP, Seibyl JP, Freeman GK, Delaney R, Bremner JD, Heninger GR, Bowers MB Jr, Charney DS (1994) Subanesthetic effects of the noncompetitive NMDA antagonist, ketamine, in humans: psychotomimetic, perceptual, cognitive, and neuroendocrine responses. Arch Gen Psychiatry 51:199–214 [DOI] [PubMed] [Google Scholar]