Abstract
Nonsyndromic cleft lip with or without cleft palate (CL-P) is a common congenital anomaly with incidence ranging from 1 in 300 to 1 in 2,500 live births. We analyzed two Indian pedigrees (UR017 and UR019) with isolated, nonsyndromic CL-P, in which the anomaly segregates as an autosomal dominant trait. The phenotype was variable, ranging from unilateral to bilateral CL-P. A genomewide linkage scan that used ∼10,000 SNPs was performed. Nonparametric linkage (NPL) analysis identified 11 genomic regions (NPL>3.5; P<.005) that could potentially harbor CL-P susceptibility variations. Among those, the most significant evidence was for chromosome 13q33.1-34 at marker rs1830756 (NPL=5.57; P=.00024). This was also supported by parametric linkage; MOD score (LOD scores maximized over genetic model parameters) analysis favored an autosomal dominant model. The maximum LOD score was 4.45, and heterogeneity LOD was 4.45 (α=100%). Haplotype analysis with informative crossovers enabled the mapping of the CL-P locus to a region of ∼20.17 cM (7.42 Mb) between SNPs rs951095 and rs726455. Thus, we have identified a novel genomic region on 13q33.1-34 that harbors a high-risk variant for CL-P in these Indian families.
Nonsyndromic cleft lip with or without cleft palate (CL-P) is one of the most frequently occurring congenital malformations among live births. The prevalence varies widely, depending on the ethnicity and geographic location of the population, ranging from 1 in 300 to 1 in 2,500.1,2 In the United States, it affects 1 in 700–1,000 newborns each year and is the fourth most common birth defect. In India, cleft lip/palate occurs in nearly 1 in 500 live births; the majority of these defects are not surgically corrected.3 Although Asians have the highest rate of orofacial clefts (OFCs) at birth, the majority of the genetic studies have been conducted with whites. There are two types of CL-P: syndromic and nonsyndromic. Nonsyndromic CL-P represents almost half of facial malformations and could be familial. More than 400 recognized syndromes may include a facial cleft as one of the manifestations. Some of the common syndromes and/or anomalies associated with clefting include Apert,4 Meckel,5 Treacher Collins,6 and van der Woude syndromes.7 Dental anomalies such as supernumerary, hypoplastic, or congenitally missing teeth and malocclusion are common in patients affected with CL-P.
The genes responsible for one form of X-linked (CPX [MIM 303400]) and one form of autosomal dominant (CPI [MIM 119530]) CL-P have been mapped to chromosomes Xq13-q21.318–12 and 2q32,13 respectively. Pathogenic mutations were identified in the TBX22 gene on Xq21.1. Genomic regions with evidence of linkage for nonsyndromic OFC were identified at 6p24.3 (OFC1 [MIM 119530]),14,15 2p13 (OFC2 [MIM 602966]),16 19q13 (OFC3 [MIM 600757]),17 4q21-q31 (OFC4 [MIM 608371]),18,19 4p16.1 (OFC5 [MIM 608874]),20 and 1q32-q41 (OFC6 [MIM 608864]),21,22 but not all responsible genes are yet identified. Chromosomal aberrations involving chromosomes 13 and 18 were reported to cause an increased incidence of clefts.23–25 Genes associated with CL-P include MSX1, MSX2, PVRL1, IRF6, RARA, TGFA, TGFB3, TGFB2, MTHRF, GABRB3, FOXE1, GLI2, JAG2, LHX8, PHF8, SATB2, SKI, SPRY2, and TBX10.20,26–29 However, none of these seem to play a major role in nonsyndromic CL-P, and they appear to be responsible for only a fraction of CL-P cases.26
The ascertainment of occasional large multigenerational families segregating a “complex” trait such as CL-P is important, since it may reveal the existence of a single gene that, in these families, contributes significantly to the phenotype. There are only a few examples of such families with the CL-P phenotype used in linkage analysis who have revealed significant linkage to 4q,18 17p,30 and multiple loci.31 Here, we present genomewide linkage analysis of two multigenerational Indian families (UR017 and UR019) with CL-P, ascertained through a single proband with nonsyndromic CL-P. This analysis provided significant evidence of a susceptibility locus on a 7.42-Mb genomic region on chromosome 13q33.1-34.
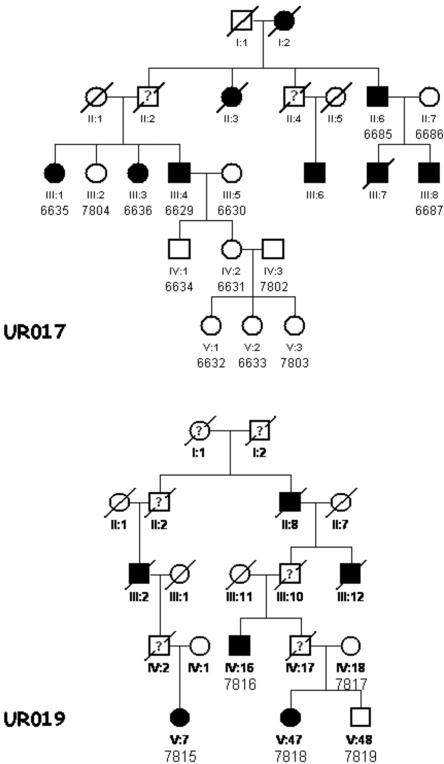
The families studied include family UR017, who were from Andhra Pradesh, the Southern state of India, and had an apparent autosomal dominant mode of inheritance (fig. 1). The original five-generation pedigree consists of 96 members with 12 affected individuals (6 females and 6 males). In the present study, we included five affected and nine unaffected individuals.
Figure 1. .
Partial pedigrees of families UR017 and UR019 with CL-P. Affected individuals are shown with blackened symbols, and unaffected individuals are shown with unblackened symbols. Data are not available for deceased individuals shown with a question mark. Samples included in the analysis are numbered under their symbols in the pedigree.
Family UR019 is a six-generation family, also from Andhra Pradesh, with CL-P (fig. 1). The original pedigree is much larger than what is shown in figure 1. The family includes seven affected members (five males and two females). The mode of inheritance is apparently autosomal dominant. We analyzed three affected and two unaffected individuals from this family.
Each individual was evaluated by an experienced dysmorphologist and a clinical geneticist. Clinical and x-ray photographs were taken of selected individuals. All affected individuals had severe CL-P, and no syndromic anomalies were observed in either pedigree. Some of the affected individuals had dystrophic or congenitally missing teeth (due to severe clefting) and speech problems. The phenotype of a few deceased individuals is unknown, and they were considered “affected status unknown” in the linkage analysis. Blood samples were obtained from all available cooperative family members, with their informed consent. Apparently, these two families are unrelated, since we could not establish any link between them. They live in a small village with <15,000 inhabitants.
Blood DNA was purified, and the whole-genome genotyping scan was performed using the GeneChip Mapping 10K 2.0 SNP Array, which contains 11,555 SNPs. These are equally distributed in the genome, with a mean intermarker distance of 210 kb and an average heterozygosity of 0.38. The assay was performed using 250 ng of genomic DNA for each sample. For each sample, >99% of the SNPs were determined unequivocally. Scan images were processed with Affymetrix Micro Array Suite Software. Data were analyzed with GDAS v2 software. PedCheck was used for the detection of Mendelian errors.32
In the parametric linkage analysis, the trait model (mode of inheritance, disease-allele frequency, and penetrance of genotypes) must be specified, which is a disadvantage when the true disease-model parameters are unknown.33 Since the parameters of the disease model were uncertain, in the initial genome scan, we assessed the evidence of linkage with a nonparametric, penetrance-independent, affected-only, and allele-sharing model. On finding significant evidence of linkage by exceeding the predetermined threshold (P<.005) with an allele-sharing method, we fitted a range of parametric models to the data. The linkage package MERLIN, which can efficiently handle thousands of genotypes,34 was used for nonparametric linkage (NPL) analysis. We calculated multipoint LOD scores maximized over genetic model parameters (MOD score analysis). For this analysis, we used the program Genehunter-Modscore,35 which calculates MOD scores by varying the disease-allele frequency and penetrance. The genomic positions of SNPs are derived from the National Center for Biotechnology Information (NCBI) (Build 35.1).
A genomewide NPL scan revealed NPL>3.5 and P<.0005 for CL-P loci at 11 chromosomal regions: 1p32, 3q26, 4q28, 6p12, 7p21, 9p23, 10q25, 11p11, 13q33, 14q32, and 18q21 (table 1). Among them, the best evidence was found for chromosome 13q33.1-34. A maximum multipoint NPL was yielded for SNP marker rs1830756 (106,878,224 bp) on chromosome 13q33.1-34 (NPL=5.57 and P=.00024). These results are also supported by parametric linkage analysis (MOD score analysis). We used the “modcalc single” option, under which Genehunter-Modscore performs a separate maximization for each genetic position assumed for the putative disease locus. This procedure yields the MOD score in conjunction with the best-fitting penetrance and disease-allele frequency at each genetic position. Under the best-fitted autosomal dominant model (100% penetrance and disease-allele frequency 0.00001), the LOD score was 4.45 and heterogeneity LOD (HLOD) was 4.45 (α=1.0). The family-specific results, the combined parametric and nonparametric multipoint linkage results at the peak SNP (rs1830756), and adjacent SNPs are shown in table 2.
Table 1. .
Initial Genome-Scan Results, Indicating Suggestive Evidence at Various Chromosomes Obtained with Parametric and Nonparametric Linkage (P<.005)
| Position |
Nonparametric |
Parametric |
|||||
| SNP Marker | Cytogenetic | Physicala (bp) |
NPL | P | LOD | HLOD | α |
| rs1417367 | 1p32.3 | 52,010,490 | 3.74 | .0046 | 1.48 | 1.48 | 1.00 |
| rs1402229 | 3q26.33 | 181,552,274 | 4.44 | .0013 | 1.72 | 1.50 | .52 |
| rs1878989 | 4q28.1 | 125,204,845 | 3.93 | .0039 | 1.61 | 1.61 | 1.00 |
| rs1372568 | 6p12.3 | 46,413,413 | 4.22 | .0027 | 1.59 | 1.59 | 1.00 |
| rs717698 | 7p21.3 | 8,167,488 | 3.55 | .0066 | .31 | .71 | .47 |
| rs1343535 | 9p23 | 10,004,143 | 3.84 | .0042 | .28 | .00 | .00 |
| rs765651 | 10q25.1 | 109,599,290 | 4.08 | .0031 | .12 | .78 | .48 |
| rs2204184 | 11p11.12 | 50,418,021 | 4.08 | .0031 | .78 | 1.76 | .50 |
| rs1830756 | 13q33.3 | 106,878,224 | 5.57 | .0002 | 4.44 | 4.44 | 1.00 |
| rs719252 | 14q32.32 | 102,682,945 | 4.44 | .0013 | 2.89 | 2.89 | 1.00 |
| rs959655 | 18q21.1 | 48,132,862 | 3.64 | .0066 | .98 | 1.75 | .50 |
Derived from the NCBI (Build 35.1).
Table 2. .
Family-Specific Results with Combined Parametric and Nonparametric Linkage Scores for the Peak Region at Chromosome 13q33.1-34[Note]
| Family UR019 |
Family UR017 |
Total (UR019 and UR017) |
||||||||||
| Position |
Parametric |
Nonparametric |
Parametric |
Nonparametric |
Parametric |
Nonparametricb |
||||||
| SNP Marker | Cytogenetic | Physicala (bp) |
LOD | Z Mean | P | LOD | Z Mean | P | LOD | HLOD | Z Mean | P |
| rs1923862 | 13q33.1 | 103,629,222 | −3.63 | .74 | .52344 | 2.11 | 3.08 | .03125 | −1.52 | 1.51 | 2.70 | .03912 |
| rs728555 | 13q33.2 | 103,755,724 | 1.55 | 1.42 | .01563 | 2.11 | 3.08 | .03125 | 3.65 | 3.66 | 3.19 | .02393 |
| rs1590919 | 13q33.2 | 103,813,224 | 1.62 | 1.54 | .01563 | 2.11 | 3.08 | .03125 | 3.72 | 3.72 | 3.27 | .02393 |
| rs1590918 | 13q33.2 | 103,813,401 | 1.62 | 1.54 | .01563 | 2.11 | 3.08 | .03125 | 3.72 | 3.72 | 3.27 | .02393 |
| rs726039 | 13q33.2 | 103,947,454 | 1.71 | 1.72 | .01563 | 2.11 | 3.08 | .03125 | 3.81 | 3.81 | 3.40 | .02393 |
| rs930268 | 13q33.2 | 104,050,671 | 1.81 | 1.99 | .01563 | 2.10 | 3.08 | .03125 | 3.92 | 3.92 | 3.58 | .02393 |
| rs951095 | 13q33.2 | 104,276,645 | 1.95 | 2.43 | .00781 | 2.10 | 3.08 | .03125 | 4.05 | 4.05 | 3.89 | .02130 |
| rs952018 | 13q33.2 | 104,988,136 | 2.20 | 3.69 | .00781 | 2.10 | 3.07 | .03125 | 4.30 | 4.30 | 4.78 | .00098 |
| rs1933331 | 13q33.2 | 105,099,046 | 2.22 | 3.82 | .00781 | 2.10 | 3.07 | .03125 | 4.32 | 4.32 | 4.87 | .00079 |
| rs2149144 | 13q33.2 | 105,246,932 | 2.25 | 4.01 | .00781 | 2.10 | 3.07 | .03125 | 4.35 | 4.35 | 5.01 | .00058 |
| rs1927571 | 13q33.2 | 105,263,158 | 2.26 | 4.03 | .00781 | 2.10 | 3.07 | .03125 | 4.35 | 4.35 | 5.02 | .00058 |
| rs718831 | 13q33.3 | 105,841,769 | 2.33 | 4.63 | .00781 | 2.10 | 3.08 | .03125 | 4.43 | 4.43 | 5.45 | .00024 |
| rs1986664 | 13q33.3 | 106,099,421 | 2.34 | 4.73 | .00781 | 2.10 | 3.08 | .03125 | 4.44 | 4.44 | 5.53 | .00024 |
| rs2391376 | 13q33.3 | 106,199,369 | 2.34 | 4.76 | .00781 | 2.10 | 3.08 | .03125 | 4.44 | 4.44 | 5.55 | .00024 |
| rs288742 | 13q33.3 | 106,299,805 | 2.34 | 4.77 | .00781 | 2.10 | 3.09 | .03125 | 4.45 | 4.45 | 5.55 | .00024 |
| rs1927756 | 13q33.3 | 106,515,980 | 2.34 | 4.78 | .00781 | 2.11 | 3.09 | .03125 | 4.45 | 4.45 | 5.56 | .00024 |
| rs1323672 | 13q33.3 | 106,870,516 | 2.34 | 4.79 | .00781 | 2.10 | 3.08 | .03125 | 4.44 | 4.44 | 5.57 | .00024 |
| rs1830756 | 13q33.3 | 106,878,224 | 2.34 | 4.79 | .00781 | 2.10 | 3.08 | .03125 | 4.44 | 4.44 | 5.57 | .00024 |
| rs1935132 | 13q33.3 | 107,125,954 | 2.34 | 4.78 | .00781 | 2.09 | 3.09 | .03125 | 4.44 | 4.44 | 5.56 | .00024 |
| rs1935134 | 13q33.3 | 107,126,104 | 2.34 | 4.78 | .00781 | 2.09 | 3.09 | .03125 | 4.44 | 4.44 | 5.56 | .00024 |
| rs1935135 | 13q33.3 | 107,126,178 | 2.34 | 4.78 | .00781 | 2.09 | 3.09 | .03125 | 4.44 | 4.44 | 5.56 | .00024 |
| rs816958 | 13q33.3 | 107,320,896 | 2.34 | 4.75 | .00781 | 2.10 | 3.09 | .03125 | 4.44 | 4.44 | 5.54 | .00024 |
| rs725122 | 13q33.3 | 107,856,400 | 2.34 | 4.74 | .00781 | 2.11 | 3.09 | .03125 | 4.44 | 4.44 | 5.53 | .00024 |
| rs2391644 | 13q33.3 | 107,949,230 | 2.33 | 4.71 | .00781 | 2.11 | 3.09 | .03125 | 4.44 | 4.44 | 5.51 | .00024 |
| rs726449 | 13q33.3 | 107,949,490 | 2.33 | 4.71 | .00781 | 2.11 | 3.09 | .03125 | 4.44 | 4.44 | 5.51 | .00024 |
| rs984300 | 13q33.3 | 108,253,836 | 2.32 | 4.57 | .00781 | 2.11 | 3.09 | .03125 | 4.42 | 4.42 | 5.41 | .00024 |
| rs719737 | 13q34 | 109,148,350 | 2.27 | 4.20 | .00781 | 2.10 | 3.07 | .03125 | 4.37 | 4.37 | 5.14 | .00031 |
| rs496916 | 13q34 | 109,649,015 | 2.22 | 3.80 | .00781 | 2.10 | 3.07 | .03125 | 4.32 | 4.32 | 4.86 | .00079 |
| rs1961495 | 13q34 | 109,679,374 | 2.22 | 3.78 | .00781 | 2.10 | 3.07 | .03125 | 4.31 | 4.31 | 4.85 | .00098 |
| rs953386 | 13q34 | 109,741,693 | 2.21 | 3.76 | .00781 | 2.10 | 3.07 | .03125 | 4.31 | 4.31 | 4.83 | .00098 |
| rs1411040 | 13q34 | 109,741,922 | 2.21 | 3.76 | .00781 | 2.10 | 3.07 | .03125 | 4.31 | 4.31 | 4.83 | .00098 |
| rs2391882 | 13q34 | 110,377,552 | 2.16 | 3.36 | .00781 | 2.10 | 3.08 | .03125 | 4.25 | 4.25 | 4.55 | .00171 |
| rs1894756 | 13q34 | 110,816,774 | 2.00 | 2.52 | .00781 | 2.11 | 3.08 | .03125 | 4.10 | 4.10 | 3.96 | .00894 |
| rs953360 | 13q34 | 110,972,580 | 1.95 | 2.32 | .00781 | 2.10 | 3.07 | .03125 | 4.06 | 4.06 | 3.81 | .02264 |
| rs1411628 | 13q34 | 110,997,322 | 1.95 | 2.30 | .01563 | 2.10 | 3.07 | .03125 | 4.05 | 4.05 | 3.80 | .02264 |
| rs1113337 | 13q34 | 111,132,750 | 1.91 | 2.13 | .01563 | 2.10 | 3.05 | .03125 | 4.01 | 4.01 | 3.67 | .02374 |
| rs1113336 | 13q34 | 111,132,873 | 1.90 | 2.13 | .01563 | 2.10 | 3.05 | .03125 | 4.01 | 4.01 | 3.67 | .02374 |
| rs1112333 | 13q34 | 111,133,498 | 1.90 | 2.13 | .01563 | 2.10 | 3.05 | .03125 | 4.01 | 4.01 | 3.67 | .02374 |
| rs726455 | 13q34 | 111,699,501 | −3.66 | .50 | .52344 | −∞ | 2.93 | .03125 | −∞ | .00 | 2.42 | .67631 |
Note.— The model was used under full-penetrance dominant inheritance with 0.000001 disease-allele frequency.
Derived from the NCBI (Build 35.1).
α=1 for all markers.
A second interesting region was identified at 14q32 (NPL=4.44; P=.0013). Parametric analysis under a dominant model yielded a LOD score of 2.89 at marker rs2024863, which is ∼19 cM away from the NPL peak. Similar MOD score analyses among the other suggested genomic positions gave nonsignificant results with a range of 0.12–1.72.
It has been demonstrated that applying linkage analyses that assume linkage equilibrium to dense markers may lead to bias,36,37 especially in the analysis of SNP linkage maps in data sets in which some parental genotypes are missing. Therefore, we assessed the impact of linkage disequilibrium (LD) on linkage at chromosome 13. We used MERLIN to accommodate marker-to-marker LD in both parametric and nonparametric analyses, by organizing closely located adjacent markers into clusters. Although many empirical studies have shown that the extent and distribution of LD are extremely variable throughout the genome, in most cases, significant LD does not influence markers separated by >0.1 cM in outbred populations.38–40 Accordingly, we used markers within 0.2 cM of each other in a cluster. Several clusters of two to six SNPs demonstrated LD. With the assumption of no LD within the cluster, MERLIN uses population haplotype frequencies while calculating linkage. At chromosome 13, the LOD score and NPL score are reduced to 3.46 and 3.03 (P=.002), respectively. However, this reduction in linkage scores might be due to both the effect of LD as well as the reduction of information content. Because of the clustering (hence, the reduction of markers), the information content was reduced from 89% to 74% at the peak region. Nonetheless, evidence of linkage, especially the parametric LOD score, at 13q33.1-34 is still very significant.
Haplotype analysis for the 13q-linked region was performed. A total of 45 informative SNP markers on 13q33.1-34 were used. Haplotype analysis (fig. 2) revealed informative recombination events in the affected individual V-7 (7815) of family UR019, with the candidate susceptibility locus confined to a region distal to rs951095 (map position 104,276,645 bp) and proximal to rs726455 (map position 111,699,501 bp), with a 20.17-cM genetic interval that corresponds to 7.42 Mb.
Figure 2. .
Partial pedigrees of UR017 and UR019, with genotypes and haplotypes of chromosome 13q. SNP markers are shown below selected individuals. Haplotypes associated with affected status are shown in red. Haplotype analysis indicated that the cosegregating segment of the CL-P locus is flanked proximally by rs951095 and distally by rs726455 on chromosome 13q33.1-34.
This interval could not be further narrowed because samples from additional individuals in these families were not available. The genomic interval between these two SNP markers contains 18 putative transcripts (Ensembl). Potential candidate genes in this region might include transcription factor DP1 (TFDP1 [MIM 189902]), inhibitor of growth 1 (ING1 [MIM 601566]), and a tumor suppressor, α-1 chain of collagen IV (COL4A1 [MIM 120130]). Mutation analysis for each transcript is required to first detect sequence variants and then to determine which of these are associated with the CL-P phenotype in families UR017 and UR019. In addition, all conserved noncoding elements need to be included in the mutation analysis.
Various parametric and nonparametric association and linkage studies of different populations provided evidence of several loci on various chromosomal regions contributing to CL-P.15–19,41–48 A number of additional candidate genes and regions also have been proposed through observations from chromosomal abnormalities in patients with CL-P.49,50 A few reports strongly supported the involvement of the IRF6 gene in some families with nonsyndromic CL-P21,22,51; however, in the present study, we did not observe any positive association. A previous linkage study conducted with 38 multiplex Indian CL-P families yielded weak evidence at multiple loci52; however, none of these previous linkage studies gave evidence involving chromosome 13 for the CL-P phenotype.
CL-P is very common in patients associated with trisomy involving all or part of chromosome 13.25 Increased incidence of chromosome 13–related anomalies are reported to be involved with CL-P.53–64 Najafzadeh et al.65 reported a newborn infant with CL-P and a 46,XX,13q+ karyotype derived from a paternal t(4;13)(q25;q32), with resulting del(13q) and dup(4q). A female with multiple congenital anomalies, including CL-P, and a karyotype of 46,XX,-13,+t(13q;13q) has been reported.66 A fetus with various developmental anomalies, including cleft lip, who had duplication of 13q32→qter due to unbalanced segregation of t(4;13)(p16;q32) in her father, was also reported.67
Multigenerational families with an autosomal dominant and an autosomal recessive inheritance of CL-P with reduced penetrance have been reported.18,31 An autosomal dominant form of inheritance is likely in the two pedigrees reported here. Families UR017 and UR019 live in the same village of <15,000 inhabitants. It is possible that there was a common ancestor for these families; however, for several generations, no marriages were reported between these two. Therefore, we could not establish any demonstrable relationship between them. Haplotype analysis from the 13q-linked region of the affected individuals did not find extensive common haplotypes shared between these two families, although there were some patches of identical alleles observed. Identification of the susceptibility variation at 13q33.1-34 for nonsyndromic CL-P in an Asian population will foster a better understanding of the molecular pathophysiology of this developmental anomaly.
Acknowledgments
We thank the patients for their cooperation in the study. We are grateful to the pediatric surgeons, plastic surgeons, and dentists, for referring patients. The study was supported in part by Green Cross Blood Bank, Ahmedabad, Gujarat, India. S.K.N. was supported by Oklahoma Medical Research Foundation institutional grant 9124, for linkage analysis.
Web Resources
The URLs for data presented herein are as follows:
- Affymetrix, http://www.affymetrix.com/products/arrays/specific/10k.affx
- Ensembl, http://www.ensembl.org/
- MERLIN, http://www.sph.umich.edu/csg/abecasis/Merlin/
- NCBI (Build 35.1), http://www.ncbi.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CPX, CPI, OFC1, OFC2, OFC3, OFC4, OFC5, OFC6, TFDP1, ING1, and COL4A1)
References
- 1.Murray JC (2002) Gene/environment causes of cleft lip and/or palate. Clin Genet 61:248–256 10.1034/j.1399-0004.2002.610402.x [DOI] [PubMed] [Google Scholar]
- 2.Stanier P, Moore GE (2004) Genetics of cleft lip and palate: syndromic genes contribute to the incidence of non-syndromic clefts. Hum Mol Genet 13:R73–R81 10.1093/hmg/ddh052 [DOI] [PubMed] [Google Scholar]
- 3.Ankola AV, Nagesh L, Hedge P, Karibasappa GN (2005) Primary dentition status and treatment needs of children with cleft lip and/or palate. J Indian Soc Pedod Prev Dent 23:80–82 [DOI] [PubMed] [Google Scholar]
- 4.Slaney SF, Oldridge M, Hurst JA, Moriss-Kay GM, Hall CM, Poole MD, Wilkie AO (1996) Differential effects of FGFR2 mutations on syndactyly and cleft palate in Apert syndrome. Am J Hum Genet 58:923–932 [PMC free article] [PubMed] [Google Scholar]
- 5.Paavola P, Salonen R, Weissenbach J, Peltonen L (1995) The locus for Meckel syndrome with multiple congenital anomalies maps to chromosome 17q21-q24. Nat Genet 11:213–215 10.1038/ng1095-213 [DOI] [PubMed] [Google Scholar]
- 6.Dixon MJ, Dixon J, Houseal T, Bhatt M, Ward DC, Klinger K, Landes GM (1993) Narrowing the position of the Treacher Collins syndrome locus to a small interval between three new microsatellite markers at 5q32-33.1. Am J Hum Genet 52:907–914 [PMC free article] [PubMed] [Google Scholar]
- 7.Houdayer C, Bonaiti-Pellie C, Erguy C, Soupre V, Dondon MG, Burglen L, Cougoureux E, Couderc R, Vazquez MP, Bahuau M (2001) Possible relationship between the van der Woude syndrome (vWS) locus and nonsyndromic cleft lip with or without cleft palate (NSCL/P). Am J Med Genet 104:86–92 [DOI] [PubMed] [Google Scholar]
- 8.Moore GE, Ivens A, Chambers J, Farrall M, Williamson R, Page DC, Bjornsson A, Arnason A, Jensson O (1987) Linkage of an X-chromosome cleft palate gene. Nature 326:91–92 10.1038/326091a0 [DOI] [PubMed] [Google Scholar]
- 9.Moore GE, Williamson R, Jensson O, Chambers J, Takakubo F, Newton R, Balacs MA, Ivens A (1991) Localization of a mutant gene for cleft palate and ankyloglossia in an X-linked Icelandic family. J Craniofac Genet Dev Biol 11:372–376 [PubMed] [Google Scholar]
- 10.Gorski SM, Adams KJ, Birch PH, Friedman JM, Goodfellow PJ (1992) The gene responsible for X-linked cleft palate (CPX) in a British Columbia native kindred is localized between PGK1 and DXYS1. Am J Hum Genet 50:1129–1136 [PMC free article] [PubMed] [Google Scholar]
- 11.Stanier P, Forbes SA, Arnason A, Bjornsson A, Sveinbjornsdottir E, Williamson R, Moore G (1993) The localization of a gene causing X-linked cleft palate and ankyloglossia (CPX) in an Icelandic kindred is between DXS326 and DXYS1X. Genomics 17:549–555 10.1006/geno.1993.1370 [DOI] [PubMed] [Google Scholar]
- 12.Forbes SA, Richardson M, Brennan L, Arnason A, Bjornsson A, Campbell L, Moore G, Stanier P (1995) Refinement of the X-linked cleft palate and ankyloglossia (CPX) localisation by genetic mapping in an Icelandic kindred. Hum Genet 95:342–346 [DOI] [PubMed] [Google Scholar]
- 13.Brewer CM, Leek JP, Green AJ, Holloway S, Bonthron DT, Markham AF, FitzPatrick DR (1999) A locus for isolated cleft palate, located on human chromosome 2q32. Am J Hum Genet 65:387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carreno H, Paredes M, Tellez G, Palomino H, Blanco R (1999) Association of non-syndromic cleft lip and cleft palate with microsatellite markers located in 6p. Rev Med Chil 127:1189–1198 [PubMed] [Google Scholar]
- 15.Prescott NJ, Lees MM, Winter RM, Malcolm S (2000) Identification of susceptibility loci for nonsyndromic cleft lip with or without cleft palate in a two stage genome scan of affected sib-pairs. Hum Genet 106:345–350 10.1007/s004390051048 [DOI] [PubMed] [Google Scholar]
- 16.Pezzetti F, Scapoli L, Martinelli M, Carinci F, Bodo M, Carinci P, Tognon M (1998) A locus in 2p13-p14 (OFC2), in addition to that mapped in 6p23, is involved in nonsyndromic familial orofacial cleft malformation. Genomics 50:299–305 10.1006/geno.1998.5273 [DOI] [PubMed] [Google Scholar]
- 17.Stein J, Mulliken JB, Stal S, Gasser DL, Malcolm S, Winter R, Blanton SH, Amos C, Seemanova E, Hecht JT (1995) Nonsyndromic cleft lip with or without cleft palate: evidence of linkage to BCL3 in 17 multigenerational families. Am J Hum Genet 57:257–272 [PMC free article] [PubMed] [Google Scholar]
- 18.Beiraghi S, Foroud T, Diouhy S, Bixler D, Conneally PM, Delozier-Blanchet D, Hodes ME (1994) Possible localization of a major gene for cleft lip and palate to 4q. Clin Genet 46:255–256 [DOI] [PubMed] [Google Scholar]
- 19.Marazita ML, Field LL, Cooper ME, Tobias R, Maher BS, Peanchitlertkajorn S, Liu YE (2002) Genome scan for loci involved in cleft lip with or without cleft palate, in Chinese multiplex families. Am J Hum Genet 71:349–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jezewski PA, Vieira AR, Nishimura C, Ludwig B, Johnson M, O’Brien SE, Daack-Hirsch S, Schultz RE, Weber A, Nepomucena B, Romitti PA, Christensen K, Orioli IM, Castilla EE, Machida J, Natsume N, Murray JC (2003) Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate. J Med Genet 40:399–407 10.1136/jmg.40.6.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scapoli L, Palmieri A, Martinelli M, Pezzetti F, Carinci P, Tognon M, Carinci F (2005) Strong evidence of linkage disequilibrium between polymorphisms at the IRF6 locus and nonsyndromic cleft lip with or without cleft palate, in an Italian population. Am J Hum Genet 76:180–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, Caprau D, et al (2004) Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med 351:769–780 10.1056/NEJMoa032909 [DOI] [PubMed] [Google Scholar]
- 23.Perrotin F, de Poncheville LM, Marret H, Paillet C, Lansac J, Body G (2001) Chromosomal defects and associated malformations in fetal cleft lip with or without cleft palate. Eur J Obstet Gynecol Reprod Biol 99:19–24 10.1016/S0301-2115(01)00347-5 [DOI] [PubMed] [Google Scholar]
- 24.Adeyokunnu AA (1983) Autosomal trisomy 18 and 13 syndromes in Ibadan, Nigeria. Afr J Med Med Sci 12:81–89 [PubMed] [Google Scholar]
- 25.Berge SJ, Plath H, Van de Vondel PT, Appel T, Niederhagen B, Von Lindern JJ, Reich RH, Hansmann M (2001) Fetal cleft lip and palate: sonographic diagnosis, chromosomal abnormalities, associated anomalies and postnatal outcome in 70 fetuses. Ultrasound Obstet Gynecol 18:422–431 10.1046/j.0960-7692.2001.00575.x [DOI] [PubMed] [Google Scholar]
- 26.Vieira AR, Avila JR, Daack-Hirsch S, Dragan E, Felix TM, Rahimov F, Harrington J, Schultz RR, Watanabe Y, Johnson M, Fang J, O’Brien SE, Orioli IM, Castilla EE, Fitzpatrick DR, Jiang R, Marazita ML, Murray JC (2005) Medical sequencing of candidate genes for nonsyndromic cleft lip and palate. PLoS Genet 1:e64 10.1371/journal.pgen.0010064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laumonnier F, Holbert S, Ronce N, Faravelli F, Lenzner S, Schwartz CE, Lespinasse J, Van Esch H, Lacombe D, Goizet C, Phan-Dinh Tuy F, van Bokhoven H, Fryns JP, Chelly J, Ropers HH, Moraine C, Hamel BC, Briault S (2005) Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J Med Genet 42:780–786 10.1136/jmg.2004.029439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sozen MA, Suzuki K, Tolarova MM, Bustos T, Fernandez Iglesias JE, Spritz RA (2001) Mutation of PVRL1 is associated with sporadic, non-syndromic cleft lip/palate in northern Venezuela. Nat Genet 29:141–142 10.1038/ng740 [DOI] [PubMed] [Google Scholar]
- 29.Suzuki Y, Jezewski PA, Machida J, Watanabe Y, Shi M, Cooper ME, Viet LT, Nguyen TD, Hai H, Natsume N, Shimozato K, Marazita ML, Murray JC (2004) In a Vietnamese population, MSX1 variants contribute to cleft lip and palate. Genet Med 6:117–125 [DOI] [PubMed] [Google Scholar]
- 30.Blanton SH, Bertin T, Patel S, Stal S, Mulliken JB, Hecht JT (2004) Nonsyndromic cleft lip and palate: four chromosomal regions of interest. Am J Med Genet A 125:28–37 10.1002/ajmg.a.20423 [DOI] [PubMed] [Google Scholar]
- 31.Wyszynski DF, Albacha-Hejazi H, Aldirani M, Hammod M, Shkair H, Karam A, Alashkar J, Holmes TN, Pugh EW, Doheny KF, McIntosh I, Beaty TH, Bailey-Wilson JE (2003) A genome-wide scan for loci predisposing to non-syndromic cleft lip with or without cleft palate in two large Syrian families. Am J Med Genet A 123:140–147 10.1002/ajmg.a.20283 [DOI] [PubMed] [Google Scholar]
- 32.O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clerget-Darpoux F, Bonaiti-Pellie C, Hochez J (1986) Effects of misspecifying genetic parameters in lod score analysis. Biometrics 42:393–399 10.2307/2531059 [DOI] [PubMed] [Google Scholar]
- 34.Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- 35.Strauch K, Fimmers R, Baur MP, Wienker TF (2003) How to model a complex trait. 2. Analysis with two disease loci. Hum Hered 56:200–211 10.1159/000076394 [DOI] [PubMed] [Google Scholar]
- 36.Schaid DJ, McDonnell SK, Wang L, Cunningham JM, Thibodeau SN (2002) Caution on pedigree haplotype inference with software that assumes linkage equilibrium. Am J Hum Genet 71:992–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Q, Shete S, Amos CI (2004) Ignoring linkage disequilibrium among tightly linked markers induces false-positive evidence of linkage for affected sib pair analysis. Am J Hum Genet 75:1106–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dawson E, Abecasis GR, Bumpstead S, Chen Y, Hunt S, Beare DM, Pabial J, et al (2002) A first-generation linkage disequilibrium map of human chromosome 22. Nature 418:544–548 10.1038/nature00864 [DOI] [PubMed] [Google Scholar]
- 39.Phillips MS, Lawrence R, Sachidanandam R, Morris AP, Balding DJ, Donaldson MA, Studebaker JF, et al (2003) Chromosome-wide distribution of haplotype blocks and the role of recombination hot spots. Nat Genet 33:382–387 10.1038/ng1100 [DOI] [PubMed] [Google Scholar]
- 40.Ke X, Hunt S, Tapper W, Lawrence R, Stavrides G, Ghori J, Whittaker P, Collins A, Morris AP, Bentley D, Cardon LR, Deloukas P (2004) The impact of SNP density on fine-scale patterns of linkage disequilibrium. Hum Mol Genet 13:577–588 10.1093/hmg/ddh060 [DOI] [PubMed] [Google Scholar]
- 41.Moreno LM, Arcos-Burgos M, Marazita ML, Krahn K, Maher BS, Cooper ME, Valencia-Ramirez CR, Lidral AC (2004) Genetic analysis of candidate loci in non-syndromic cleft lip families from Antioquia-Colombia and Ohio. Am J Med Genet A 125:135–144 10.1002/ajmg.a.20425 [DOI] [PubMed] [Google Scholar]
- 42.Schultz RE, Cooper ME, Daack-Hirsch S, Shi M, Nepomucena B, Graf KA, O’Brien EK, O’Brien SE, Marazita ML, Murray JC (2004) Targeted scan of fifteen regions for nonsyndromic cleft lip and palate in Filipino families. Am J Med Genet A 125:17–22 10.1002/ajmg.a.20424 [DOI] [PubMed] [Google Scholar]
- 43.Ardinger HH, Buetow KH, Bell GI, Bardach J, VanDemark DR, Murray JC (1989) Association of genetic variation of the transforming growth factor-alpha gene with cleft lip and palate. Am J Hum Genet 45:348–353 [PMC free article] [PubMed] [Google Scholar]
- 44.Eiberg H, Bixler D, Nielsen LS, Conneally PM, Mohr J (1987) Suggestion of linkage of a major locus for nonsyndromic orofacial cleft with F13A and tentative assignment to chromosome 6. Clin Genet 32:129–132 [DOI] [PubMed] [Google Scholar]
- 45.Blanton SH, Bertin T, Serna ME, Stal S, Mulliken JB, Hecht JT (2004) Association of chromosomal regions 3p21.2, 10p13, and 16p13.3 with nonsyndromic cleft lip and palate. Am J Med Genet A 125:23–27 10.1002/ajmg.a.20426 [DOI] [PubMed] [Google Scholar]
- 46.Marazita ML, Field LL, Cooper ME, Tobias R, Maher BS, Peanchitlertkajorn S, Liu YE (2002) Nonsyndromic cleft lip with or without cleft palate in China: assessment of candidate regions. Cleft Palate Craniofac J 39:149–156 [DOI] [PubMed] [Google Scholar]
- 47.Wyszynski DF, Maestri N, McIntosh I, Smith EA, Lewanda AF, Garcia-Delgado C, Vinageras-Guarneros E, Wulfsberg E, Beaty TH (1997) Evidence for an association between markers on chromosome 19q and non-syndromic cleft lip with or without cleft palate in two groups of multiplex families. Hum Genet 99:22–26 10.1007/s004390050303 [DOI] [PubMed] [Google Scholar]
- 48.Martinelli M, Scapoli L, Pezzetti F, Carinci F, Carinci P, Baciliero U, Padula E, Tognon M (1998) Suggestive linkage between markers on chromosome 19q13.2 and nonsyndromic orofacial cleft malformation. Genomics 51:177–181 10.1006/geno.1998.5384 [DOI] [PubMed] [Google Scholar]
- 49.Yoshiura K, Machida J, Daack-Hirsch S, Patil SR, Ashworth LK, Hecht JT, Murray JC (1998) Characterization of a novel gene disrupted by a balanced chromosomal translocation t(2;19)(q11.2;q13.3) in a family with cleft lip and palate. Genomics 54:231–240 10.1006/geno.1998.5577 [DOI] [PubMed] [Google Scholar]
- 50.Wiktor A, Feldman GL, Kratkoczki P, Ditmars DM Jr, Van Dyke DL (1994) 10p duplication characterized by fluorescence in situ hybridization. Am J Med Genet 52:315–318 10.1002/ajmg.1320520312 [DOI] [PubMed] [Google Scholar]
- 51.Blanton SH, Cortez A, Stal S, Mulliken JB, Finnell RH, Hecht (2005) Variation in IRF6 contributes to nonsyndromic cleft lip and palate. Am J Med Genet A 137:259–262 [DOI] [PubMed] [Google Scholar]
- 52.Field LL, Ray AK, Cooper ME, Goldstein T, Shaw DF, Marazita ML (2004) Genome scan for loci involved in nonsyndromic cleft lip with or without cleft palate in families from West Bengal, India. Am J Med Genet A 130:265–271 10.1002/ajmg.a.30252 [DOI] [PubMed] [Google Scholar]
- 53.Escobar JI, Yunis JJ (1974) Trisomy for the proximal segment of the long arm of chromosome 13: a new entity? Am J Dis Child 128:221–222 [DOI] [PubMed] [Google Scholar]
- 54.Escobar JI, Sanchez O, Yunis JJ (1974) Trisomy for the distal segment of chromosome 13: a new syndrome. Am J Dis Child 128:217–220 [DOI] [PubMed] [Google Scholar]
- 55.Jotterand M, Juillard E (1976) A new case of trisomy for the distal part of 13q due to maternal translocation, t(9;13)(p21;q21). Hum Genet 33:213–222 10.1007/BF00286845 [DOI] [PubMed] [Google Scholar]
- 56.Kaye CI, Booth CW, Meeker D, Nadler HL (1977) Cleft palate and multiple anomalies in one of two siblings with partial 13 trisomy. Cleft Palate J 14:244–248 [PubMed] [Google Scholar]
- 57.Noel B, Quack B, Rethore MO (1976) Partial deletions and trisomies of chromosome 13; mapping of bands associated with particular malformations. Clin Genet 9:593–602 [DOI] [PubMed] [Google Scholar]
- 58.Schinzel A, Schmid W (1974) Different forms of incomplete trisomy 13: mosaicism and partial trisomy for the proxim. Humangenetik 22:287–298 10.1007/BF00295488 [DOI] [PubMed] [Google Scholar]
- 59.Schinzel A, Hayashi K, Schmid W (1976) Further delineation of the clinical picture of trisomy for the distal segment of chromosome 13: report of three cases. Hum Genet 32:1–12 10.1007/BF00569970 [DOI] [PubMed] [Google Scholar]
- 60.Habedank M (1979) Partial trisomy 13q21toqter de novo due to a recombinant chromosome rec(13)dup q. Hum Genet 52:91–99 [DOI] [PubMed] [Google Scholar]
- 61.Rao VV, Carpenter NJ, Gucsavas M, Coldwell J, Say B (1995) Partial trisomy 13q identified by sequential fluorescence in situ hybridization. Am J Med Genet 58:50–53 10.1002/ajmg.1320580111 [DOI] [PubMed] [Google Scholar]
- 62.Riccardi VM, Hittner HM, Francke U, Pippin S, Holmquist GP, Kretzer FL, Ferrell R (1979) Partial triplication and deletion of 13q: study of a family presenting with bilateral retinoblastomas. Clin Genet 15:332–345 [DOI] [PubMed] [Google Scholar]
- 63.Rivas F, Rivera H, Plascencia ML, Ibarra B, Cantu JM (1984) The phenotype in partial 13q trisomies, apropos of a familial (13;15)(q22;q26) translocation. Hum Genet 67:86–93 10.1007/BF00270563 [DOI] [PubMed] [Google Scholar]
- 64.Yanagisawa S, Yokoyama H, Agena N (1978) Partial distal trisomy 13q resulting from familial reciprocal 5/13 translocation. Hum Genet 45:345–350 10.1007/BF00278733 [DOI] [PubMed] [Google Scholar]
- 65.Najafzadeh TM, Littman VA, Dumars KW (1983) Familial t(4;13) with abnormal offspring in three generations. Am J Med Genet 16:15–22 10.1002/ajmg.1320160104 [DOI] [PubMed] [Google Scholar]
- 66.Ribeiro Noce T, de Pina-Neto JM, Happle R (2001) Phylloid pattern of pigmentary disturbance in a case of complex mosaicism. Am J Med Genet 98:145–147 [DOI] [PubMed] [Google Scholar]
- 67.Tapper JK, Zhang S, Harirah HM, Panova NI, Merryman LS, Hawkins JC, Lockhart LH, Gei AB, Velagaleti GV (2002) Prenatal diagnosis of a fetus with unbalanced translocation (4;13)(p16;q32) with overlapping features of Patau and Wolf-Hirschhorn syndromes. Fetal Diagn Ther 17:347–351 10.1159/000065383 [DOI] [PubMed] [Google Scholar]