Abstract
Using single particle electron cryomicroscopy that does not impose icosahedral averaging, we determined the structure of the entire infectious Salmonella phage Epsilon151, including both icosahedral and non-icosahedral components. At least three layers of condensed viral DNA were observed to pack in coaxial coils with local 25 Å hexagonal inter-strand spacing. At one of the five-fold vertices, a portal complex with twelve subunits replaces a capsid pentamer. A tail hub with six projecting trimeric tailspikes sits on the external face of the portal. Below the portal is a cylindrical protein core. An extended shaft of density fills the central channel of the protein core and the portal complex and appears to consist of about 90 nucleotides at the terminus of the packaged DNA poised for injection. Using an icosahedral symmetry imposed reconstruction, the fold of the capsid shell protein is seen to resemble the capsid protein fold of other tailed double-stranded DNA phages2–5 and human herpesvirus6. These common structural features suggest a common evolutionary origin among these viruses.
Double-stranded DNA (dsDNA) phages are vectors for gene transfer among enteric bacteria, including important human pathogens7. For all the well-studied tailed dsDNA phages, a preformed procapsid shell is assembled, and the DNA is pumped into the shell through a portal complex located at a single vertex8. The phage tails are also assembled at this vertex. The portal complex together with packaging enzymes have been shown to function as components of a very powerful molecular motor9, but it has not been possible to visualize the complex within the intact virion.
The inability to visualize the packed DNA and the portal vertex in the virion reflects the difficulties in determining these structural features which lack icosahedral symmetry and are lost in any icosahedral averaging used in X-ray crystallography or electron cryomicroscopy (cryoEM). Using a cryoEM single particle reconstruction technique without symmetry imposition, we have been able to determine the structure of these critical features of Salmonella phage Epsilon15, some of which are unexpected.
Epsilon15 is a short tailed dsDNA bacteriophage that infects Salmonella anatum. Its genome (NCBI accession number: NC_004775) contains 39,671 base pairs with 49 open reading frames (Supplementary Fig. 1) among which six, coding for structural proteins, were resolved by SDS-PAGE and identified by tryptic-digest/mass spectrometry (Supplementary Fig. 2).
CryoEM of frozen hydrated Epsilon15 shows intact particles consisting of isometric heads and a protruding density (Fig. 1a). ~15,000 particle images were used for image reconstruction without symmetry imposition. The 20 Å resolution density map of the whole phage is shown in Fig. 1b (Supplementary Movie 1). The angular-shaped capsid has a T=7l shell, with a larger diameter (700 Å) along the icosahedral 5-fold axes and a smaller diameter (650 Å) along the 3-fold axes. The virion capsid is made up of 11 pentons and 60 hexons.
Figure 1. Structure of Epsilon15 phage.
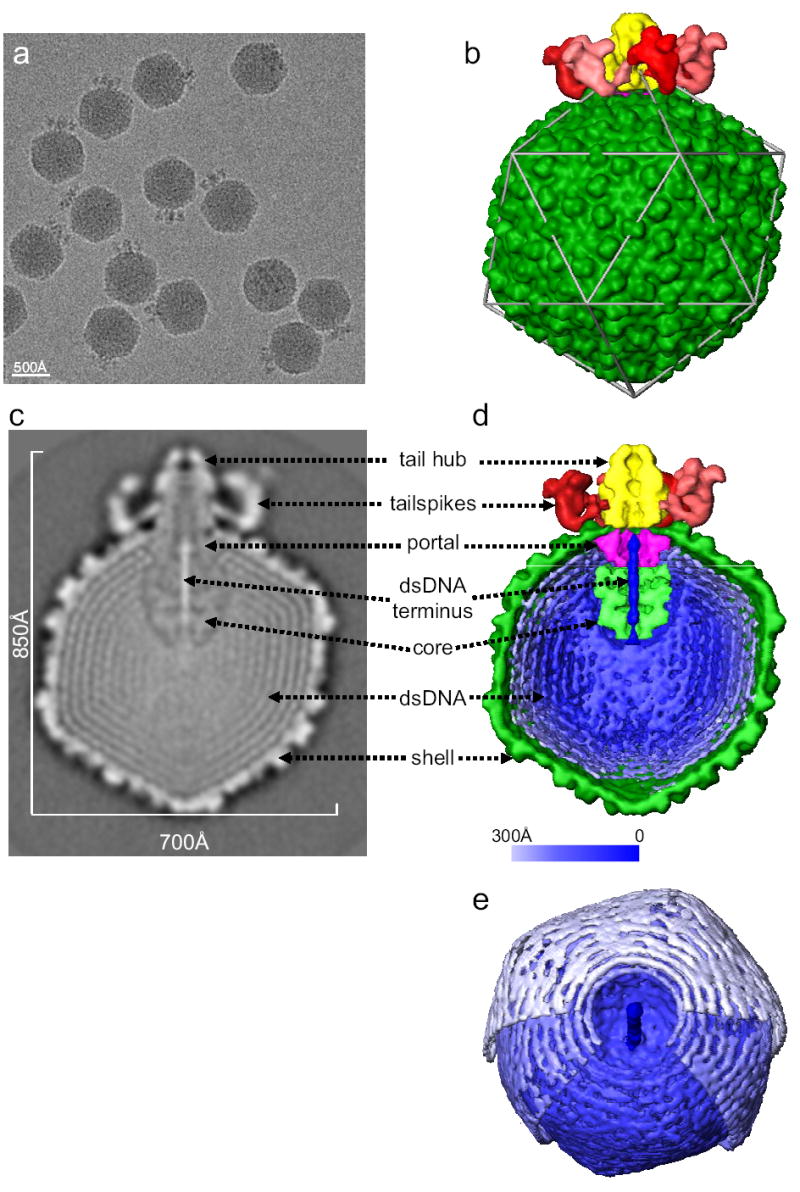
(a) 200 kV CCD image of Epsilon15 particles embedded in vitreous ice. (b) Surface rendering of the 20 Å resolution 3-D map of entire Epsilon15 phage reconstructed without symmetry imposition. The capsid (dark green) exhibits good icosahedral symmetry as indicated by the icosahedral lattice (gray). The structural components of Epsilon15 phage are annotated in the central section density (c) and the cut-away surface view (d) of the 3-D density map. Each of the structural components is colored differently and the same color scheme is used in Figures 1–3. (e) A slightly tilted view of the coaxially packed dsDNA genome. Only portions of the first and second layers of dsDNA were shown so that the three outermost layers can be viewed. Color gradient (light blue to darker blue) is used in (d) and (e) to indicate the likely packaging order of the dsDNA starting from outer layer towards inner layers and ending as the central straight segment of terminus.
One of the 5-fold vertices is occupied by six tailspikes (red) surrounding an external tail hub (yellow). A cross-section of the reconstruction (Fig. 1c) and cut-away view (Fig. 1d) reveal, inside the capsid shell (dark green), the densities of the portal complex (purple), a protein core (light green), and the packaged viral DNA (blue). Figure 1e shows three well-resolved concentric shells representing the dsDNA packed coaxially around the 5-fold axis.
The ~40kb Epsilon15 dsDNA genome has a length of 14 μm in an extended form and needs to assume a compact arrangement when it is packaged into the capsid. This compact arrangement must also be topologically organized to facilitate efficient dsDNA release during infection. The Epsilon15 genome structure is sufficiently resolved to see the individual dsDNA strands with a ~25 Å separation for at least the 3 outermost layers (Figs. 1d,e), which are packed coaxially around the vertical axis in Figure 1c,d coincident with the capsid 5-fold/portal/tail axis. Neighboring dsDNA strands are packed hexagonally, which appears to be the most space efficient and energetically favorable packing pattern for the cylindrically shaped dsDNA strands. The coaxial dsDNA rings are better resolved in the outermost layers closest to the capsid shell than those in the inner layers (Figs. 1c,d). This suggests that the dsDNA packaging starts from the inner surface of the capsid shell (light blue in Figure 1d,e), spools towards the inner radius region and ends as a terminal shaft in the portal channel (dark blue in Figure 1d,e). This type of packing agrees qualitatively with the coaxial spooling model based on scattering10, simulations11 and previous cryoEM reconstruction12. Our observed spool axis is consistent with the T7 spooling model13, but orthogonal to an earlier model of T410, and does not appear to vary in different layers11.
On the surface of the capsid, six tailspikes, composed of gp20, connect to a central tail hub and extend out from one of capsid 5-fold vertices (Figs. 1b and 2a). This symmetry mismatch with a six-fold tail complex located at a five-fold vertex is a characteristic feature of tailed dsDNA phages. Each tailspike consists of two structural domains separated by a bend: a capsid proximal stem bound to the central tail hub and a distal flower-like domain composed of three petals around a central knob (Fig. 2a, Supplementary Fig. 4b). The distal end of the flower-like domain is clearly trimeric, suggesting a homo-trimer of gp20. These tailspikes bind and cleave the O-antigen component of the host’s cell surface lipopolysaccharide1. The petal-like densities are candidates for this enzymatic function. The bend may represent a functional hinge of the tailspike that transmits a recognition signal to activate the tail hub for viral genome injection.
Figure 2. The tail structure.
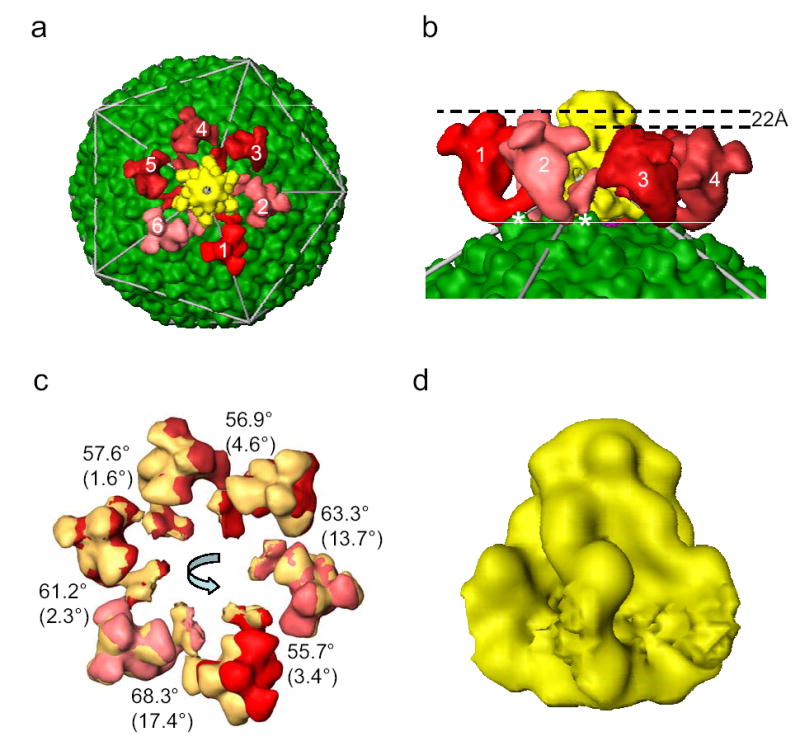
(a) Top view of the tail in the Epsilon15 reconstruction. Each of the 6 tailspikes is uniquely colored with a random shade of red in order to illustrate the inexact 6-fold arrangement around the central tail hub (yellow) with good 6-fold symmetry. (b) Zoom-in side view of the tailspikes. The contact sites for two (labeled as 1 and 2) out of the six tailspikes are on the capsid surface protrusion densities at local 2-fold positions (labeled with *). (c) Each of the tailspikes is aligned to its neighbor (counter-clockwise) with indicated amount of rotation around an axis that is tilted away from the 6-fold axis in different degrees as indicated in parenthesis. The tailspikes in the original positions are displayed in the same shades of red colors as in (a) while the rotated tailspikes in the new positions are displayed in yellow. (d) Side view of the tail hub. The segmentation of the tail hub at the interacting regions with the tailspikes is arbitrary because of their tight binding and limited resolution.
The tail deviates from exact six-fold azimuthal symmetry (Fig. 2a, 2c and Supplementary Fig. 4a). Each of the tailspikes has a slightly different orientation (Supplementary Fig. 4a) but similar conformation except tailspike 3 (Supplementary Fig. 4b). These differences among individual tailspikes may arise from the different interaction sites on the capsid shell surface and the symmetry mismatches between the strict 5-fold shell and the quasi 6-fold tail. Two neighboring tailspikes (labeled* in Fig. 2b) contact the protrusions at the local 2-fold positions of the shell, while the other 4 spikes have tenuous interactions with the shell at different locations at smaller radii. The two elevated contact points force these two tailspikes to a higher radius by 22Å than the other four tailspikes, and result in a slight overall tilt of the tail (Fig. 2b).
At the center of the six tailspikes is the tail hub of size ~170 Å (height) ×140 Å (width) (Fig. 2d). The tail hub has good 6-fold symmetry (Fig. 2a). It appears to anchor the tailspikes as well as cap the central opening of the portal to prevent premature DNA leakage. Upon host cell surface binding by the distal petal region of the tailspikes, the induced conformational changes might trigger the opening of the capped central channel (Figs. 1c,d) allowing for DNA injection into the host cell in a manner analogous to a gated ion channel.
Just below the tail hub lies the portal. While the oligomeric state of portals within intact phages has long been thought to be 12, it has not been measured until now. Our Epsilon15 density map provides unambiguous structural evidence that there are 12 densities at the portal in situ (Figs. 1c,d, Figs. 3a,b,c and Supplementary Figs. 5a,b), which is consistent with previous studies of portal complexes biochemically extracted from phage particles14, 15.
Figure 3. Structure of portal complex and internal core.
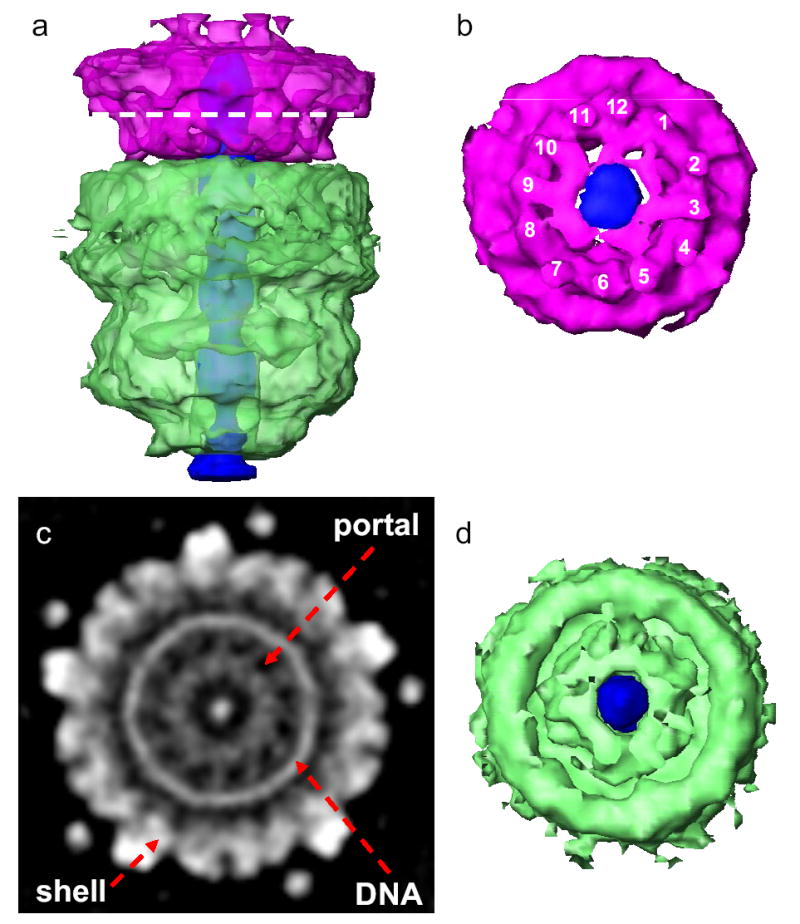
(a) Side view of the portal complex (pink) and internal core (light green) and the putative straight dsDNA terminus (dark blue). The portal complex and internal core are colored semi-transparently to show their central channels are filled with the putative dsDNA terminus. (b) Bottom view of the portal complex with the nearly 12-fold structural features labeled around the filled central channel. (c) The apparent 12-fold arrangement of densities for the portal complex in a section image of the 3-D map. The location of the section is indicated by the dashed line in (a). (d) Top view of the internal core (light green) showing central channel filled with the putative dsDNA terminus (dark blue).
Circumscribed by a well-resolved dsDNA ring, the portal has the appearance of twelve well-resolved turbine-like densities (180Å in diameter) with nearly equivalent angular spacing (Fig. 3c, Supplementary Fig. 5a). This overall feature resembles that seen in other isolated portals with negligible sequence similarity16–19. The azimuthal density distribution of Epsilon15 portal deviates noticeably from perfect 12-fold symmetry (Fig. 3c and Supplementary Fig. 5a). The asymmetry of the portal ring may be caused by the non-equivalent interactions between the twelve portal subunits and the five surrounding shell protein subunits.
These observations require that the portal complex has the same spatial relationship with the tailspikes, tail hub, and the shell, from particle to particle, implying there are specific interactions among these components. Otherwise, the portal structure would not be resolved by averaging ~15,000 varying phage particles.
Figures 3a and 3d show an internal density (light green) organized immediately internal to the portal complex. This feature is ~200 Å in height and ~180 Å in width without obvious symmetry. We interpret this to be a protein core reminiscent of the phage T7 internal 8-fold symmetric structure observed in electron micrographs20. The lack of obvious symmetry for Epsilon15 core could be authentic or result from an azimuthal variation of the core among particles. The Epsilon15 capsid volume can accommodate up to 90kb dsDNA. Since the Epsilon15 genome is only ~40kb, there is ample space for a protein core of this size in the capsid chamber. The protein core may facilitate the topological ordering of the dsDNA genome during packaging and/or release as suggested for T7 core21.
A long straight segment of uniform density (~310 Å in length and ~30 Å in width) extends from near the capsid center along the 5-fold capsid-tail axis. Because these densities are significantly higher than those of the surrounding protein core and portal complex (Figs. 1c,d), we interpret this segment of density to be the last-in segment of packaged dsDNA (~90 base pairs). It appears to be poised for release and injection into host cell during infection.
This dsDNA terminus passes through the central opening of the portal and ends slightly beyond the portal to where it is capped by densities closing the tail hub’s central channel (Figs. 1c,d). The observation that Epsilon15’s portal channel is filled with a straight segment of putative dsDNA terminus is consistent with the observation that phage SPP1’s terminal segment of packaged dsDNA is tightly associated with its portal protein22.
Imposing the icosahedral symmetry, a 9.5 Å density map was generated from a separate dataset imaged to give a higher resolution reconstruction (Supplementary Movie 2, Supplementary Figs. 3b, 6a,b). This map is sufficiently resolved to delineate the molecular boundaries of each capsid protein subunit and the secondary structure elements (Figs. 4a,b). In the average subunit map, three helices could be identified (Fig. 4b). Helix H1 is ~40Å long and extends toward the 3-fold axes where three neighboring capsomeres interact intimately (Fig. 4a, Supplementary Fig. 8b). Parallel to helix H1 is a large β-sheet that contributes significantly to the local 3-fold interactions. The two shorter helices (H2 and H3) are located at the interfaces of adjacent subunits within the same hexon and are nearly parallel to each other.
Figure 4. Shell protein structure.
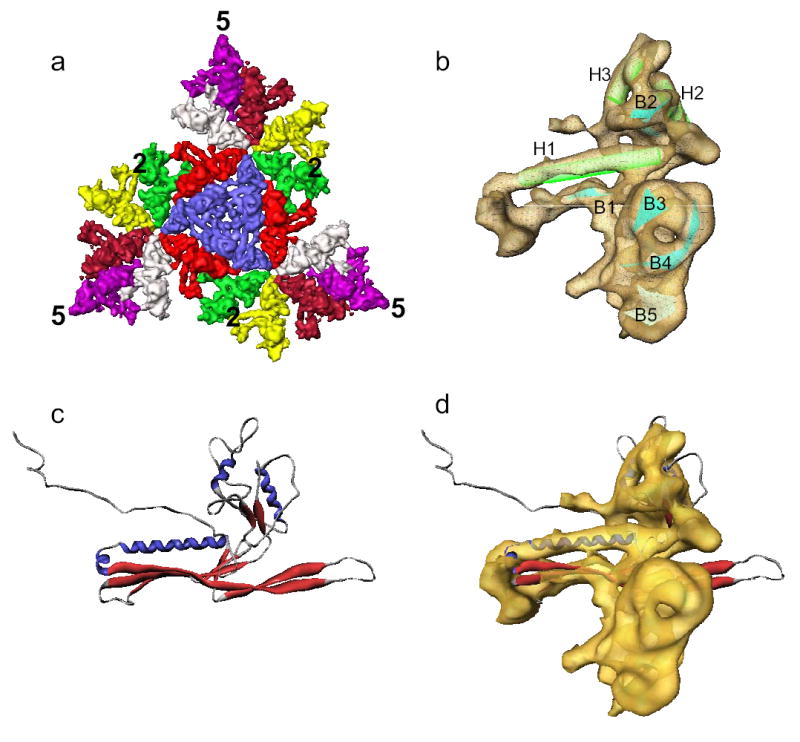
(a) Surface rendering of three asymmetric units forming one icosahedral face of the T=7l shell. Each of the 7 subunits is colored differently. (b) A single averaged shell protein subunit with 3 helices (H1-H3) and 5 sheets (B1-B5) annotated. (c) Ribbon representation of the atomic model of HK97 head protein gp5 (PDB accession ID: 1OHG). (d) Superposition of Epsilon15 shell protein average subunit and the HK97 head protein gp5.
The dispositions of the helices and sheets are similar to those observed in other phages and herpesvirus2–6 (Fig. 4c,d, Supplementary Fig. 7) despite little sequence identity. The similarity extends beyond the protein fold of individual subunits to the molecular interaction patterns between the subunits of a hexon or penton, and at the local 3-fold symmetry axes where three neighboring capsomeres meet (Supplementary Figs. 8a,b). The similarities of the protein fold and assembly of the capsid shell of these viruses suggest that these viruses share an ancient common ancestor and that the capsid protein fold and molecular interactions used to hold the icosahedral shell intact are strongly conserved during evolution.
Methods
See Supplementary methods for details in purification, cryoEM imaging23, 3-D reconstruction24–26 and structural analysis27, 28.
Supplementary Material
Acknowledgments
We acknowledge the support of grants from National Institutes of Health and the Robert Welch Foundation. We thank Matthew Dougherty for the production of the animations, Dr. Matthew Baker for the AIRS program for secondary structure element search, and Drs. Michael F. Schmid and Frazer Rixon for discussions.
Data deposition
The 3-D density maps have been deposited into the EBI-MSD EMD database with accession codes: EMD-1175 for the complete structure without symmetry imposition and EMD-1176 for the icosahedral shell structure.
References
- 1.McConnell M, Reznick A, Wright A. Studies on the initial interactions of bacteriophage epsilon15 with its host cell, Salmonella anatum. Virology. 1979;94:10–23. doi: 10.1016/0042-6822(79)90434-3. [DOI] [PubMed] [Google Scholar]
- 2.Jiang W, et al. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat Struct Biol. 2003;10:131–5. doi: 10.1038/nsb891. [DOI] [PubMed] [Google Scholar]
- 3.Wikoff WR, et al. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–33. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 4.Fokine A, et al. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci U S A. 2005;102:7163–8. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morais MC, et al. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol Cell. 2005;18:149–59. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79:14967–70. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neidhardt, F. C. & Curtiss, R. Escherichia coli and Salmonella: cellular and molecular biology (ASM Press, Washington, D.C., 1996).
- 8.Bazinet C, King J. The DNA translocating vertex of dsDNA bacteriophage. Annu Rev Microbiol. 1985;39:109–29. doi: 10.1146/annurev.mi.39.100185.000545. [DOI] [PubMed] [Google Scholar]
- 9.Smith DE, et al. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–52. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 10.Earnshaw WC, King J, Harrison SC, Eiserling FA. The structural organization of DNA packaged within the heads of T4 wild-type, isometric and giant bacteriophages. Cell. 1978;14:559–68. doi: 10.1016/0092-8674(78)90242-8. [DOI] [PubMed] [Google Scholar]
- 11.Arsuaga J, Tan RK, Vazquez M, Sumners de W, Harvey SC. Investigation of viral DNA packaging using molecular mechanics models. Biophys Chem. 2002;101–102:475–84. doi: 10.1016/s0301-4622(02)00197-7. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, et al. Visualization of the maturation transition in bacteriophage P22 by electron cryomicroscopy. J Mol Biol. 2000;297:615–26. doi: 10.1006/jmbi.2000.3601. [DOI] [PubMed] [Google Scholar]
- 13.Cerritelli ME, et al. Encapsidated conformation of bacteriophage T7 DNA. Cell. 1997;91:271–80. doi: 10.1016/s0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 14.Lurz R, et al. Structural organisation of the head-to-tail interface of a bacterial virus. J Mol Biol. 2001;310:1027–37. doi: 10.1006/jmbi.2001.4800. [DOI] [PubMed] [Google Scholar]
- 15.Carazo JM, Fujisawa H, Nakasu S, Carrascosa JL. Bacteriophage T3 gene 8 product oligomer structure. J Ultrastruct Mol Struct Res. 1986;94:105–13. doi: 10.1016/0889-1605(86)90056-x. [DOI] [PubMed] [Google Scholar]
- 16.Simpson AA, et al. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–50. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orlova EV, et al. Structure of a viral DNA gatekeeper at 10 Å resolution by cryo-electron microscopy. Embo J. 2003;22:1255–62. doi: 10.1093/emboj/cdg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agirrezabala X, et al. Structure of the connector of bacteriophage T7 at 8 Å resolution: structural homologies of a basic component of a DNA translocating machinery. J Mol Biol. 2005;347:895–902. doi: 10.1016/j.jmb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Tang L, Marion WR, Cingolani G, Prevelige PE, Johnson JE. Three-dimensional structure of the bacteriophage P22 tail machine. Embo J. 2005;24:2087–95. doi: 10.1038/sj.emboj.7600695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerritelli ME, et al. A second symmetry mismatch at the portal vertex of bacteriophage T7: 8-fold symmetry in the procapsid core. J Mol Biol. 2003;327:1–6. doi: 10.1016/s0022-2836(03)00117-7. [DOI] [PubMed] [Google Scholar]
- 21.Molineux IJ. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol Microbiol. 2001;40:1–8. doi: 10.1046/j.1365-2958.2001.02357.x. [DOI] [PubMed] [Google Scholar]
- 22.Tavares P, Lurz R, Stiege A, Ruckert B, Trautner TA. Sequential headful packaging and fate of the cleaved DNA ends in bacteriophage SPP1. J Mol Biol. 1996;264:954–67. doi: 10.1006/jmbi.1996.0689. [DOI] [PubMed] [Google Scholar]
- 23.Booth CR, et al. A 9 Å single particle reconstruction from CCD captured images on a 200 kV electron cryomicroscope. J Struct Biol. 2004;147:116–27. doi: 10.1016/j.jsb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Kivioja T, Ravantti J, Verkhovsky A, Ukkonen E, Bamford D. Local average intensity-based method for identifying spherical particles in electron micrographs. J Struct Biol. 2000;131:126–34. doi: 10.1006/jsbi.2000.4279. [DOI] [PubMed] [Google Scholar]
- 25.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 26.Jiang W, et al. Semi-automated icosahedral particle reconstruction at sub-nanometer resolution. J Struct Biol. 2001;136:214–25. doi: 10.1006/jsbi.2002.4439. [DOI] [PubMed] [Google Scholar]
- 27.Jiang W, Baker ML, Ludtke SJ, Chiu W. Bridging the information gap: computational tools for intermediate resolution structure interpretation. J Mol Biol. 2001;308:1033–44. doi: 10.1006/jmbi.2001.4633. [DOI] [PubMed] [Google Scholar]
- 28.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


