York, UK, was a good choice for the venue of this workshop because the history of nucleolar research—like the city—falls into distinct eras. Founded by the Romans in 71 ad, York has been occupied at various points in time by the Saxons, Vikings and Normans. Our understanding of the nucleolus has also followed a pattern in which rapid advances have been interspersed with periods of relative dormancy. The nucleolus was first observed early in the nineteenth century, but it was not until the 1960s that its role in ribosome biogenesis was established. After that, it was assumed that the nucleolus did not do much else; however, to our surprise, many unexpected roles for the nucleolus have emerged in the past decade.

This EMBO Workshop on the Nucleolus: New Perspectives took place between 27 and 29 March 2006, in York, UK, and was organized by J. Wainwright, C. Rubbi, J. Milner and J. Hiscox.
What is in a nucleolus?
We now have working catalogues of the components of human and plant (Arabidopsis) nucleoli. A few years ago, collaboration between the laboratories of A. Lamond (Dundee, UK) and M. Mann (Odense, Denmark) led to the first major nucleolar proteomic study. Since then, at least 700 proteins have been identified in human nucleoli (Leung et al, 2006). Combining mass spectrometry with stable isotope labelling techniques, the Lamond group has also examined the redistribution kinetics of nucleolar proteins after drug treatment. For example, Lamond showed that actinomycin D causes some proteins to accumulate in the nucleolus, and others to redistribute to the nucleoplasm. Surprisingly—in response to treatment with the proteasome inhibitor MG132—ribosomal proteins accumulate in the nucleolus. Lamond interprets this to mean that the cell has a surplus of ribosomal proteins that are ready for an upregulation of ribosomal DNA transcription. In cells with lower rates of transcription, excess ribosomal proteins would be degraded through the proteasomal pathway.
New functions of the nucleolus have been unveiled by proteomic and RNomic analyses of Arabidopsis nucleoli by P. Shaw (Norwich, UK), in collaboration with J. Brown (Dundee, UK). Although the vast majority of the plant nucleolar proteins have human homologues, about 12% are unique to plant nucleoli. It is striking that several of the Arabidopsis nucleolar proteins are also present in the exon junction complex (Pendle et al, 2005), which couples mRNA processing to export and other downstream events such as nonsense-mediated decay. This suggests that nonsense-mediated decay occurs in the plant nucleolus, a possibility that is further supported by the presence of aberrantly spliced mRNA transcripts in these nucleoli. This lends credence to earlier suggestions that mRNAs spend part of their life cycles in the nucleolus, either for export or for quality control.
Nucleolar structure, assembly and disassembly
The textbook view of the nucleolus shows a fibrillar centre surrounded by a ring of dense fibrillar components, with granular components scattered throughout the body of the nucleolus. However, D. Lafontaine (Brussels, Belgium) reminded us that most eukaryotes have only two subcompartments and lack the fibrillar centre (Thiry & Lafontaine, 2005). This division between bipartite and tripartite nucleoli seems to have been a recent evolutionary event that occurred with the emergence of the amniota, which includes lizards, birds and mammals. The tripartite nucleoli of the amniota might have arisen from the much longer rDNA intergenic spacer regions than those found in lower eukaryotes.
A remarkable feature of the nucleolus is its mitotic disassembly and postmitotic reassembly. The latter uses complexes containing processing components associated with partially processed pre-rRNA; in telophase, these complexes are found in prenucleolar bodies (PNBs). During nucleolar reassembly, components from the PNBs are transferred to growing nucleoli. D. Hernandez-Verdun (Paris, France) showed that the timing of the transfer is dependent on the component (Angelier et al, 2005). Thus, proteins involved in early processing events, for example fibrillarin, are incorporated into nucleoli earlier than those implicated in later events, for example Nop52 or B23/NPM. This was reinforced by studies showing that Nop52 and B23/NPM do not interact during anaphase, but that an interaction signal could be detected when these proteins are recruited to PNBs during telophase (Fig 1). These complexes seem to be formed in the PNBs in preparation for the migration of the processing machinery to nucleoli. Nop52 might somehow facilitate cleavage of internal transcribed spacer 2 of pre-rRNA by the protein B23/NPM. This protein was shown by M. Olson (Jackson, MS, USA) to be phosphorylated by cyclin-dependent kinases in prophase and dephosphorylated in mid-anaphase. It is likely that dephosphorylation acts as a signal for the start of nucleolar assembly. At about the same time in the cell cycle, chromatin undergoes decondensation. E. Varela (Basel, Switzerland) reported that an interacting partner of the condensin complex, Lte1, is important in the chromatin decompaction step in preparation for interphase chromatin reorganization.
Figure 1.
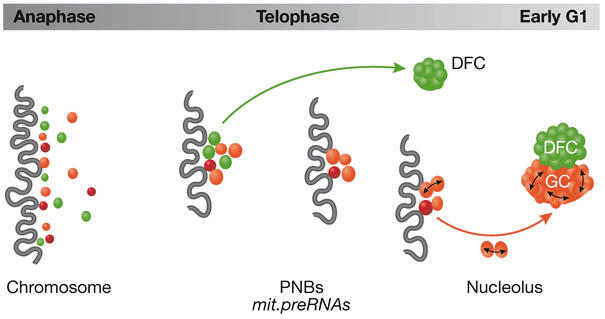
Dynamics of proteins during nucleolar reassembly. During anaphase, pre-rRNA processing proteins are located at the chromosome periphery (dark wavy line). In telophase these are incorporated into prenucleolar bodies (PNBs), which also contain mitotic pre-rRNAs (mit.preRNAs). Proteins involved in early processing events (green) are incorporated into the dense fibrillar component (DFC) before other proteins (red) that go to the granular component (GC). Bidirectional arrows indicate interactions detected in late telophase.
Ribosome biogenesis
Ribosome biogenesis is regulated by gene silencing and by modulating the transcriptional apparatus. In most eukaryotes, a significant fraction of the tandemly repeated rRNA genes is silenced. A key player in silencing is NoRC, an ATP-dependent nucleolar chromatin-remodelling complex that has been studied extensively by I. Grummt (Heidelberg, Germany). NoRC promotes the repression of rDNA transcription by recruiting a histone deacetylase and a DNA methylating enzyme to the promoter (Fig 2). NoRC associates with the promoter through the binding of its TIP5 subunit to the termination factor TTF-1, which is bound to a site adjacent to the rDNA promoter. Surprisingly, this interaction is RNA-dependent, but even more intriguing is the finding that the RNA is transcribed from the intergenic spacers (IGS) that separate the rRNA genes. IGS promoters were observed more than 20 years ago, but this is the first indication that RNA encoded by the IGS has a role in rDNA silencing.
Figure 2.
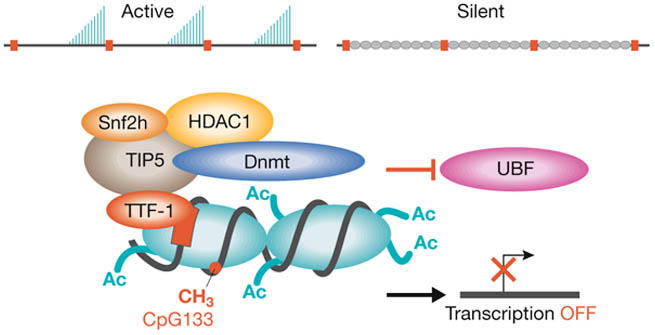
Assembly of the nucleolar remodelling complex NoRC at the rDNA promoter. NoRC, (subunits Snf2 and TIP5) associates with the rDNA promoter through the binding of TIP5 to termination factor TTF-1, which is bound to a site adjacent to the rDNA promoter. NoRC promotes the repression of rDNA transcription by recruiting a histone deacetylase (HDAC1) and a DNA methylating enzyme (Dnmt). Silencing results from histone H4 deacetylation, histone H3-Lys 9 dimethylation and DNA methylation. The transcription factor upstream binding factor (UBF) is prevented from associating with the promoter and transcription is blocked. Nucleosomes (turquoise ovals) contain histone acetyl groups (Ac) and DNA methylated (CH3) at CpG 133.
In plants, rDNA activation and silencing manifests itself in nucleolar dominance, in which there is preferential transcription of the rRNA genes from only one parent in interspecific hybrids. C. Pikaard (St Louis, MO, USA) showed that in Arabidopsis, the mechanism of silencing involves concerted changes in DNA methylation and histone modification; that is, H3 histones are methylated at Lys 9 and deacetylated, histone H4 is hypoacetylated and the promoter is methylated at cytosine. By using knockdown technology, Pikaard's group has identified the enzymes that cause the switch. Out of dozens of potential candidates, they found one DNA methyltransferase (DRM2) that aids in the silencing of genes, plus two methylcytosine-binding domain proteins (MBD6 and MBD10) and two histone deacetylases (HDT1 and HDA6) that are important for nucleolar dominance.
Although transcription of rDNA might be the first event in ribosome biogenesis, the association of assembly and processing factors with the transcribed RNA is not far behind. S. Baserga (New Haven, CT, USA) showed that U3 small nucleolar ribonucleoprotein (snoRNP) and more than 40 other proteins comprise the small subunit processome, which is the 'terminal knob' near the 5′ end of the nascent transcripts in yeast nucleoli (reviewed by Granneman & Baserga, 2004). There are 18 putative RNA helicases involved in ribosome biogenesis and five of them are present in the small subunit processome. One of them, Dbp8, is a DExD/H box protein that is required for synthesis of the 18S rRNA. Baserga's group also identified another nucleolar protein, Esf2, as a binding partner of Dbp8 and showed that it stimulates its ATPase activity. Although the precise functions of the helicases are not known, their presence reinforces the idea that numerous specialized proteins are required to correctly fold these very long RNAs.
Viral interactions in the nucleus
The second half of the meeting was devoted to non-traditional aspects of nucleolar research, starting with how viruses use nucleolar proteins and functions for their own ends. It has been known for some years that human immunodeficiency virus (HIV) transports its mRNA through the nucleolus, but J. Rossi (Duarte, CA, USA) outlined how this property can be used against HIV. He showed that U16 snoRNAs carrying anti-HIV RNAs to the nucleolus interfere with the transport of HIV RNA. In the first example presented by Rossi, an anti-HIV-1-targeted hammerhead ribozyme fused to the U16 snoRNA effectively ablated the cytoplasmic accumulation of some of the HIV RNA transcripts. This approach was used to generate T-cell lines that could not be infected with HIV-1. The second approach was to place a transactivation response (TAR) decoy sequence into a second hybrid U16 snoRNA. The TAR sequence is recognized by the HIV protein Tat, which has a key role in viral mRNA transcription and is localized to the nucleolus. Flooding the nucleolus with false TAR sequences severely compromised the ability of the virus to replicate. Again, T-cell lines expressing this decoy could not be infected by HIV-1. Clinical trials are planned in which haematopoietic stem cells will be transduced by a viral vector expressing a snoRNA TAR decoy and small interfering RNAs to knock down the cellular co-receptor for HIV, CCR5. The resulting cells will be reintroduced into the patient, in whom the transduced stem cells would repopulate the body with immune cells resistant to infection by HIV (Li et al, 2005). The team plans to use HIV-1-based lentiviral gene therapy vectors to introduce the anti-HIV RNAs into the stem cells.
A. Whitehouse (Leeds, UK) presented evidence that herpesvirus saimiri also uses the nucleolus for mRNA transport (Williams et al, 2005). This prototype gamma-2 herpesvirus belongs to a group that includes Karposi's sarcoma herpesvirus and is closely related to Epstein–Barr virus. Most of its mRNA lacks introns and so export of viral mRNA from the nucleus to the cytoplasm is inefficient. This problem is avoided by the viral protein, ORF57, which binds herpesvirus saimiri transcripts and efficiently transports them to the cytoplasm through the nucleolus—ablation of a nucleolar localization signal within ORF57 effectively abolishes the function of this protein. ORF57 seems to mediate its effects through interactions with Aly/TAP and the Tho complexes, which are recently established members of the normal mRNA export pathway.
Nucleolar targeting signals in the N proteins from the coronavirus family were explored by J. Hiscox (Leeds, UK). Coronaviruses include the severe acute respiratory syndrome (SARS) virus as well as economically important avian coronaviruses. The primary role of the N protein is to form a complex with the viral RNA genome. Although these viruses replicate in the cytoplasm, the N protein accumulates in the nucleoli of infected cells (You et al, 2005). In addition to isolating the motifs responsible for this phenomenon, Hiscox presented evidence that this protein shuttles between the cytoplasm and the nucleolus. Moreover, the N protein interacts with nucleolin and can mediate a cell-cycle arrest, favouring viral replication. Thus, viruses that traditionally have been thought not to need the nucleus do have something to gain from nucleolar interactions.
D. Matthews (Bristol, UK), who works on the replication of human adenoviruses, also showed that nucleolar antigens can favour viral replication. Surprisingly, these viruses have the ability to sequester the nucleolar upstream binding factor (UBF) into viral DNA replication centres in the nucleus. Matthews showed that UBF interacts preferentially with the origins of viral DNA replication, and antibodies to UBF reduced the rate of viral DNA replication in vitro. UBF has a key role in rRNA synthesis initiation, but despite the sequestration of the majority of UBF into viral replication centres, rRNA synthesis was not markedly affected. The data suggest that, at least in this context, rRNA synthesis does not require the bulk of endogenous UBF.
M. Taliansky (Dundee, UK) examined the role of the nucleolar antigen fibrillarin in the replication of the plant virus groundnut rosette virus (GRV). GRV expresses a protein, ORF3, which shuttles between the nucleolus and cytoplasm of infected plants and interacts with fibrillarin. Nucleolar localization mutants showed that loss of transport to the nucleolus leads to a lack of formation of viral RNP particles and, consequently, the virus was unable to transport itself long distances through the phloem of infected plants. Thus, the availability of fibrillarin to interact with ORF3 enables the spread of this virus in plants.
The nucleolus and cellular disease
There is increasing evidence that the nucleolus is a stress sensor and a controller of p53 function (Rubbi & Milner, 2003). C. Rubbi (York, UK) proposed that p53 is stable in a cell unless the nucleolus promotes its degradation, which firmly places the nucleolus at the centre of control pathways influenced by p53 status (Fig 3). Degradation of p53 by the ubiquitin pathway seems to be intimately linked with the nucleolus. Using His-tagged ubiquitin and an enhanced green fluorescent protein-tagged p53, Rubbi showed that these two interact with each other in the nucleolus; moreover, micronucleated cell experiments indicated that ubiquitination of p53 only occurs in micronuclei that contain nucleoli. Together with heterokaryon assays, this establishes that p53 degradation or stabilization is dependent on the nucleolus. However, in a twist to this story, it was shown that export to the proteasome for degradation is the key stage in which the nucleolus has a role. Further insight into the relationship between p53 and the nucleolus was provided by J. Milner (York, UK) who reported that, under normal non-stress conditions, nucleolar co-localization of the transcription initiation factor TIF1A and UBF is dependent on both p53 and silent mating type information regulation 2 homologue 1 (SIRT1) status. The initiation factor TIF1A and UBF co-localize in RNA polymerase I (Pol I) initiation complexes, whereas p53 and SIRT1 are opposing modulators of the cell stress response. Perturbation of normal TIF1A localization dynamics, owing to the absence of functional p53, might contribute to oncogenic stress in cancer cells. Thus, the relationship between p53 and the nucleolus seems to be reciprocal: p53 influences the functional capacity of the nucleolus under normal growth conditions, whereas the nucleolus determines p53 function in response to stress.
Figure 3.
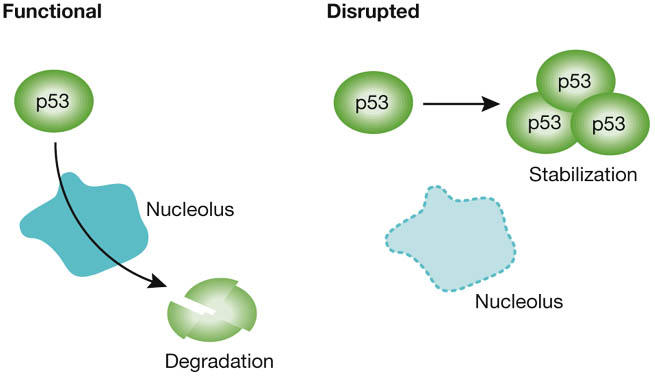
The nucleolus is a key regulator of p53 stability. The p53 protein is stable in a cell unless the nucleolus can promote its degradation through the ubiquitin pathway. Thus disruption of nucleolar functions can lead to stabilization of p53 with several knock-on effects through pathways controlled by p53 status.
Continuing the cancer theme, R. White (Glasgow, UK) proposed that a key checkpoint in cell transformation is RNA polymerase III (Pol III) output (Goodfellow et al, 2006). White presented several lines of evidence implicating the control of TFIIIB, which recruits Pol III onto promoters, as central to transformation. For example, the retinoblastoma protein (RB) binds and inactivates TFIIIB, which in turn restricts Pol III activity. This restriction on Pol III activity is overturned in many malignancies by hyperphosphorylation of RB or by the binding of oncoproteins to RB. Even more convincing was the engineering of cell lines that contained additional copies of the TFIIIB gene under the control of a doxycycline responsive promoter. An elegant series of experiments in mouse models concluded that such cells made more protein, were more proliferative, grew more colonies and were more tumorigenic.
R. Tsai (Houston, TX, USA), argued that nucleolar proteins have a role in cell proliferation and ageing, and presented data on nucleostemin. This nucleolar GTP-binding protein is enriched in cancer and stem cells and rapidly shuttles between the nucleolus and nucleoplasm. The biological significance of nucleostemin was examined in loss-of-function mice in which homozygous null mice embryos are aborted by the blastula stage. Additional heterozygous intercrosses helped establish the importance of this protein in early embryogenesis and in the delay of ageing in these mice. In support of this idea, Tsai presented evidence that nucleostemin interacts with the telomerase reverse transcriptase complex, TERT.
RNA editing has an essential role in some diseases, especially those related to the development of the nervous system. Editing enzymes that convert adenosine to inosine are found in a wide range of eukaryotes. This conversion leads to alterations in mRNA splicing and changes in protein function. M. Carmo-Fonseca (Lisbon, Portugal) showed that one of these enzymes, adenosine deaminase ADAR1, localizes to the nucleolus and is a substrate for conjugation with SUMO-1; this modification reduces its RNA-editing activity. ADAR2 also localizes to the nucleolus, but is not SUMOylated. Curiously, ADAR2 edits RNA in the nucleolus, but ADAR1 does not. For unknown reasons, the cell suppresses ADAR1 activity in the nucleolus and uses conjugation by SUMO-1 to accomplish this task.
Concluding remarks
This was the first meeting to focus on the non-traditional roles of the nucleolus, and so many have accumulated that only a few could be covered in the time allocated. What was once thought to be an isolated factory that simply cranks out ribosomes is now known to be intimately involved in many functions within the cell, including its own survival. The rapid pace of discovery of novel nucleolar functions makes it certain that more meetings of this nature will be held in the future.

J. Wainright [organizer], C. Rubbi [organizer], J. Milner [organizer], M. Olson [author], J. Hiscox [organizer] & D. Matthews [author]
Acknowledgments
The meeting was generously supported by the European Molecular Biology Organization, Carl Zeiss International and The Company of Biologists. Special thanks go to J. Milner, J. Hiscox, C. Rubbi and J. Wainwright for organizing the meeting. We also thank D. Hernandez-Verdun, I. Grummt and C. Rubbi for providing figures 1–3, respectively.
References
- Angelier N, Tramier M, Louvet E, Coppey-Moisan M, Savino TM, De Mey JR, Hernandez-Verdun D (2005) Tracking the interactions of rRNA processing proteins during nucleolar assembly in living cells. Mol Biol Cell 16: 2862–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow SJ, Innes F, Derblay LE, MacLellan WR, Scott PH, White RJ (2006) Regulation of RNA polymerase III transcription during hypertrophic growth. EMBO J 25: 1522–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Baserga SJ (2004) Ribosome biogenesis: of knobs and RNA processing. Exp Cell Res 296: 43–50 [DOI] [PubMed] [Google Scholar]
- Leung AK, Trinkle-Mulcahy L, Lam YW, Andersen JS, Mann M, Lamond AI (2006) NOPdb: nucleolar proteome database. Nucleic Acids Res 34: D218–D220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MJ, Kim J, Li S, Zaia J, Yee JK, Anderson J, Akkina R, Rossi JJ (2005) Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther 12: 900–909 [DOI] [PubMed] [Google Scholar]
- Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JW, Shaw PJ (2005) Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell 16: 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi CP, Milner J (2003) Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 22: 6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry M, Lafontaine DL (2005) Birth of a nucleolus: the evolution of nucleolar compartments. Trends Cell Biol 15: 194–199 [DOI] [PubMed] [Google Scholar]
- Williams BJ, Boyne JR, Goodwin DJ, Roaden L, Hautbergue GM, Wilson SA, Whitehouse A (2005) The prototype gamma-2 herpesvirus nucleocytoplasmic shuttling protein, ORF 57, transports viral RNA through the cellular mRNA export pathway. Biochem J 387: 295–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Dove BK, Enjuanes L, DeDiego ML, Alvarez E, Howell G, Heinen P, Zambon M, Hiscox JA (2005) Subcellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein. J Gen Virol 86: 3303–3310 [DOI] [PubMed] [Google Scholar]


