Abstract
Drought and salinity are major abiotic stresses to crop production. Here, we show that overexpression of stress responsive gene SNAC1 (STRESS-RESPONSIVE NAC 1) significantly enhances drought resistance in transgenic rice (22–34% higher seed setting than control) in the field under severe drought stress conditions at the reproductive stage while showing no phenotypic changes or yield penalty. The transgenic rice also shows significantly improved drought resistance and salt tolerance at the vegetative stage. Compared with WT, the transgenic rice are more sensitive to abscisic acid and lose water more slowly by closing more stomatal pores, yet display no significant difference in the rate of photosynthesis. SNAC1 is induced predominantly in guard cells by drought and encodes a NAM, ATAF, and CUC (NAC) transcription factor with transactivation activity. DNA chip analysis revealed that a large number of stress-related genes were up-regulated in the SNAC1-overexpressing rice plants. Our data suggest that SNAC1 holds promising utility in improving drought and salinity tolerance in rice.
Keywords: Oryza sativa, abscisic acid, stomata, dehydration
Poor water management, increased competition for limited water resources, and the uncertain threats associated with global warming all highlight the looming water crisis that threatens agricultural productivity worldwide. In China alone, the estimated annual loss of national economy from water shortage alone reaches >$25 billion (1). In addition to altered water management practices, the ability to enhance the tolerance of crops to drought and salinity stress, particularly at the most sensitive reproductive stage of growth, can have a potentially huge impact on productivity in the years to come.
Plants can develop numerous physiological and biochemical strategies to cope with adverse conditions (2, 3). The major events of plant response to dehydration stresses are perception and transduction of the stress signals through signaling components, resulting in activation of a large number of stress-related genes and synthesis of diverse functional proteins that finally lead to various physiological and metabolic responses (4–6). Well characterized proteins involved in the protection of plant cells from dehydration stress damage include molecule chaperons, osmotic adjustment proteins (7), ion channels (8), transporters (9), and antioxidation or detoxification proteins (10). The expression of these functional proteins is largely regulated by specific transcription factors (4, 11).
More than 30 families of transcription factors have been predicted for Arabidopsis (12). Members of DREB or CBF, MYB, bZIP, and zinc-finger families have been well characterized with roles in the regulation of plant defense and stress responses (4–6, 13, 14). Most of these transcription factors regulate their target gene expression through binding to the cognate cis-elements in the promoters of the stress-related genes. Two well characterized dehydration stress-related cis-elements bound by transcription factors are the drought-responsible element (DRE) recognized by DREB or CBF transcription factors (15) and the abscisic acid (ABA)-responsive element (ABRE) recognized by bZIP domain transcription factors (16–18). Numerous reports suggest that overexpression of some stress-inducible transcription factors, such as DREB1A (19), CBF4 (20), SCOF (21), Tsi (22), and OSISAP1 (14), can increase the tolerance to drought, salinity, or low temperature in Arabidopsis or other plant species.
NAC (NAM, ATAF, and CUC) is a plant-specific gene family, and most NAC proteins contain a highly conserved N-terminal DNA-binding domain, a nuclear localization signal sequence, and a variable C-terminal domain. Ooka et al. (23) reported that 75 and 105 NAC genes were predicted in the Oryza sativa and Arabidopsis genomes, respectively. The cis-element of NAC transcription factor [NAC recognized sequence (NACRS)] was also identified in Arabidopsis (24). The first reported NAC genes were NAM from petunia (25) and CUC2 from Arabidopsis (26) that participate in shoot apical meristem development. Other development-related NAC genes have been suggested with roles in controlling cell expansion of specific flower organs [such as NAP (27)] or auxin-dependent formation of the lateral root system [such as NAC1 (28)]. Some of NAC genes, such as ATAF1 and ATAF2 genes from Arabidopsis (26) and the StNAC gene from potato (29), are induced by pathogen attack and wounding. More recently, a few NAC genes, such as AtNAC072 (RD26), AtNAC019, AtNAC055 from Arabidopsis (24, 30), and BnNAC from Brassica (31), were found to be involved in the response to various environmental stresses.
In this study, NAC gene SNAC1 (STRESS-RESPONSIVE NAC 1) was isolated and characterized in rice. This gene can be induced by drought specifically in guard cells. SNAC1-overexpressing transgenic plants showed significantly improved drought resistance under field conditions and strong tolerance to salt stress.
Results
The SNAC1 Gene Is Induced by Drought Predominantly in Guard Cells.
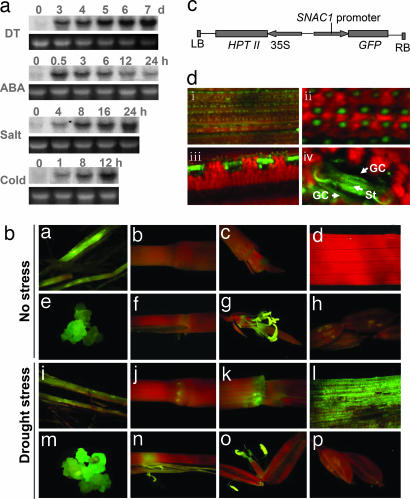
Based on the expression profiling of rice under drought stress using a cDNA microarray containing 9,216 unique cDNA sequences (unpublished data), an EST showing 5.6-fold increase of expression level in an upland rice cultivar IRAT109 (Oryza sativa L. ssp japonica) after drought stress was identified, and the EST showed homology to known NAC genes. A full-length cDNA (1,290 bp) of this gene, designated SNAC1, was isolated from the upland rice cultivar IRAT109 by RT-PCR. SNAC1 showed 98.6% sequence identity and the same location in the rice genome to the predicted gene ONAC044 (23). Northern blot analysis revealed that the expression of this gene could be induced by drought, salt, cold, and abscisic acid (Fig. 1a).
Fig. 1.
Stress-inducible expression of SNAC1. (a) RNA gel blot analysis of expression of the SNAC1 under drought (DT), salt (200 mM), cold (4°C), and ABA treatment (100 μM). (b) Expression pattern of GFP driven by the SNAC1 promoter in transgenic rice plants under normal conditions (ba–bh) or exposed to dehydration stress for 5 h (bi–bp). Shown are root (ba and bi); nodes (bb and bj); stems (bc and bk); leaves (bd and bl); calli (be and bm); ligule (bf and bn); stamen and pistil (bg and bo); and lemma (bh and bp). (c) Diagram of the PSNAC1:GFP construct. (d) Optical (di) and confocal (dii–div) microscopy analysis of GFP induction in guard cells by dehydration. GC, guard cell; ST, stomatal pore (closed). (Magnification: dii, ×10; diii, ×40; div, ×63.)
The temporal and spatial patterns of SNAC1 expression were investigated by transforming a japonica cultivar Nipponbare with a fusion gene of PSNAC1:GFP (Fig. 1c). A GFP signal was observed in callus, root, ligule, stamen, and pistil from transgenic plants under normal growth conditions (Fig. 1b). When transgenic plants were drought-stressed to the stage of leaf-rolling, strong induction of GFP was detected in leaves and minor induction was observed in roots and nodes, whereas no obvious change of GFP expression level was observed in callus, ligule, stamen, and pistil after drought stress (Fig. 1b). Further examination of the GFP signal in the stressed leaves revealed that the signal is localized predominantly in guard cells that constitute the stomata (Fig. 1d). This finding suggests that SNAC1 gene expression was specifically induced in guard cells, and this GFP signal was observed for guard cells on both the upper and lower sides of leaves.
Overexpression of SNAC1 Can Significantly Improve Drought Resistance.
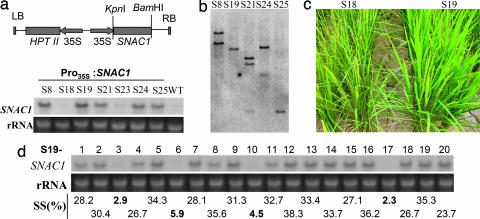
To test the effect of SNAC1 overexpression on drought resistance, the full-ength cDNA of SNAC1 under the control of the CaMV 35S promoter (Fig. 2a) was transformed into japonica cultivar Nipponbare. Of 33 independent T0 transgenic plants generated, 29 were positive transformants as detected by PCR of hygromycin resistance gene, and all of them exhibited a normal phenotype under normal growth conditions. Northern blot analysis of the transgene in seven independent positive transgenic plants showed that five (S8, S19, S21, S24, and S25) plants had high levels of transgene expression whereas the other two (S18 and S23) had no expression of transgene (Fig. 2a). S18 was thus used as a negative control for further analysis. Southern blot analysis suggested that all of the five expression-positive plants had 1–2 copies of T-DNA [portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells] (Fig. 2b). Positive transgenic plants were chosen for testing with three drought treatments at the anthesis stage: severe stress in the field (with soil water content ≈15%), moderate stress in the field (soil water content ≈28%), and moderate stress in poly(vinyl chloride) (PVC) pipes in which plants were individually stressed according to a defined protocol (see Materials and Methods).
Fig. 2.
Improved drought resistance of SNAC1-overexpressing transgenic rice at reproductive stage. (a) Overexpression contruct (Upper) and RNA gel blot analysis of SNAC1 in transgenic plants and the WT (Lower). (b) Southern blot analysis of transgenic plants using hygromycin resistance gene as a probe. (c) Appearance of one positive (S19) and one negative (S18) transgenic families in the field with severe drought stress. (d) Cosegregation of SNAC1-overexpressing (RNA gel blot analysis) with the improved drought tolerance in the T1 family of S19. SS(%), seed-setting rate
During the process of stress development, the transgenic plants showed much delayed leaf-rolling compared with the negative control S18 (Fig. 2c) and the WT (data not shown). All of the SNAC1-overexpressing plants produced significantly (t test, P < 0.01) higher spikelet fertility than the negative control under all three treatments (Table 1). Under severe drought stress in which the WT and the negative control produced almost no seeds, the five transgenic lines had 23.0–34.6% spikelet fertility. While the moderate drought stress was conducted in the drought-prone field, SNAC1-overexpressing plants exhibited 17.4–22.3% higher spikelet fertility than the WT or the negative transgenic line S18. Under the stress conditions created using the PVC pipes, the transgenic lines showed 17.2–24.0% higher seed setting than the control. Under well irrigated conditions, all transgenic and control plants had similar performance for spikelet fertility (Table 1). Moreover, no significant difference was detected between the transgenic plants and the control as evaluated by a number of agronomic traits such as plant height, number of panicles per plant, number of spikelets per panicle, and grain yield per plant, as well as root depth and root volume under unstressed conditions (Table 2, which is published as supporting information on the PNAS web site). This finding clearly indicates that overexpression of SNAC1 does not affect growth and productivity of the rice plant.
Table 1.
Spikelet fertility (%) of SNAC1-overexpressing transgenic rice plants under different drought stress conditions and mRWC for establishing leaf turgor pressure
| Line | Well irrigated condition | Severe stress in sheltered field | Moderate stress in open field | Drought stress in PVC pipes | mRWC |
|---|---|---|---|---|---|
| WT | 87.2 ± 3.4 | 0.8 ± 0.7 | 54.3 ± 5.6 | 48.2 ± 5.3 | 50.9 ± 1.4 |
| S18 | 89.1 ± 3.5 | 0.9 ± 0.6 | 57.7 ± 5.3 | 46.3 ± 6.7 | ND |
| S8 | 85.4 ± 4.2 | 24.1 ± 3.4** | 74.2 ± 6.4** | 65.8 ± 7.5** | 46.3 ± 1.6* |
| S19 | 88.6 ± 2.8 | 34.6 ± 6.1** | 78.3 ± 5.1** | 68.3 ± 6.8** | 42.2 ± 2.1** |
| S21 | 84.7 ± 3.7 | 23.3 ± 2.8** | 75.1 ± 4.3** | 71.3 ± 8.4** | 44.4 ± 2.0* |
| S24 | 89.2 ± 3.4 | 24.0 ± 3.5** | 73.4 ± 5.8** | 64.5 ± 6.1** | 45.5 ± 1.5* |
| S25 | 86.5 ± 3.5 | 23.0 ± 3.1** | 74.1 ± 4.6** | 65.1 ± 4.4** | 43.6 ± 2.6* |
S18 was used as a negative transgenic control (no expression of transgene). Values are the means ± SD (16 plants per line for spikelet fertility and 6 plants per line for mRWC). The * and ** indicate significantly higher values of transgenic plants than WT or negative transgenic control at the probability levels of P = 0.05 and P = 0.01, respectively (t test). ND, no data.
We also performed a cosegregation analysis between drought resistance and the transgene expression using the T1 family of the transformant S19, which was identified as having a single copy of transgene (Fig. 2b). Of the 20 plants analyzed, 16 positive plants had spikelet fertility ≈23.7–38.3%, whereas the spikelet fertility of the 4 negative plants ranging 2.3–5.9%, demonstrating perfect cosegregation between the transgene and fertility under drought stressed conditions (Fig. 2d).
Increased Stomatal Closure and ABA Sensitivity May Provide Partial Explanation for the Observed Drought Resistance.
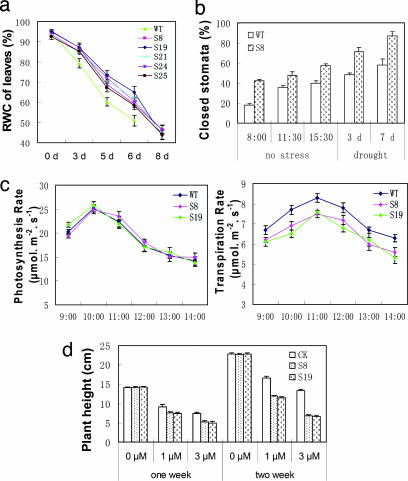
During the development of drought stress, the leaves of SNAC1-overexpressing plants lost water more slowly (Fig. 3a). Because SNAC1 showed a strongly localized expression in guard cells by drought stress, we investigated the response of stomata under drought stress. Significantly (P < 0.01) more stomatal pores were closed in transgenic rice than in the WT under both normal and drought-stressed conditions (Fig. 3b). Interestingly, photosynthesis rate was not significantly affected in the transgenic plants, but transpiration rate was lower in the transgenic plants than the WT (Fig. 3c). Furthermore, transgenic seedlings were significantly (P < 0.05) more sensitive to ABA treatment (Fig. 3d). These results suggested that the enhanced drought resistance of the transgenic plants was at least partly due to the increased stomatal closure and/or ABA sensitivity to prevent water loss. In addition, the rolled transgenic leaf segments, when rehydrated, could establish turgor pressure with significantly (P < 0.05) lower minimum relative water content (mRWC) than the WT (Table 1), suggesting an increased dehydration tolerance of the transgenic rice.
Fig. 3.
Increased stomatal closure and ABA sensitivity of transgenic rice. (a) Change of RWC in leaves during drought development. The last point of each curve indicates the mRWC with which the rolled leaves can reexpand when water is supplied. Values are the mean ± SD (n = 5). (b) Percentages of closed stomatal pores observed under SEM in the leaves of transgenic and WT plants under normal (three time points within a day) and drought stress (3 days or 7 days after water deprivation) conditions. Values are the mean ± SD (n = 4), with ≈100 stomatal pores on the adaxial side (similar result from the abaxial side was not shown) randomly counted for each sample (three samples for each time point). (c) Photosynthesis rate (Left) and transpiration rate (Right). Values are the mean ± SD (n = 8 flag leaves). (d) ABA sensitivity of SNAC1-overexpressing transgenic plants. Seeds germinated on MS medium containing 100 mg/liter hygromycin were transferred to the medium with 0, 1, or 3 μM ABA, and plant height was measured at 7 and 14 days after transplanting. Values are the mean ± SD (n = 8) for each line. CK, vector control.
SNAC1-Overexpressing Transgenic Plants Significantly Improve Drought Resistance and Salt Tolerance at Vegetative Stage.
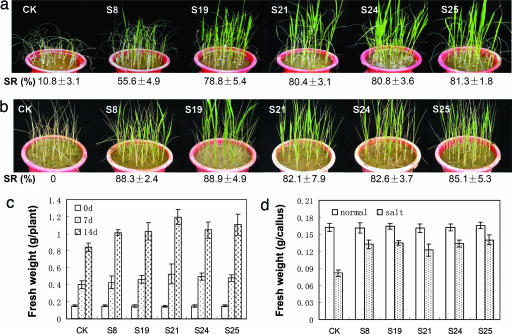
Positive transgenic plants of the five transgenic families were tested for drought resistance at the four-leaf stage. At 12 days after water-withholding, almost all leaves of the WT plants completely rolled, whereas only a small portion of the leaves of the transgenic plants slightly rolled. One week after rewatering, >50% of transgenic plants recovered, but only ≈10% of the control plants recovered (Fig. 4a and Table 3, which is published as supporting information on the PNAS web site), suggesting strong drought resistance of the transgenic rice at this stage.
Fig. 4.
Improved drought resistance and salt tolerance of SNAC1-overexpressing transgenic rice at vegetative stage. (a and b) Recovery of the SNAC1-overexpressing seedlings after drought stress (a; 12 days of water-withholding at four-leaf stage followed by 1 week of watering) or salt stress (b; 200 mM NaCl for 12 days). Survival rate is indicated below, and the values are based on three repeats (Table 3). CK, WT; SR, survival rate. (c) Fresh weight of hydroponic cultured transgenic seedlings measured during the recovery period of 0, 7, and 14 days after 5 days stress with 100 mM NaCl in the nutrient solution. Values are the means ± SD (n = 10). (d) Fresh weight of calli (starting with 0.1 g of callus with same size) grown in MS medium with 100 mM NaCl for 15 days. Values are the means ± SD (n = 10).
Salt tolerance of the transgenic lines was also tested. After treatment with 200 mM NaCl for 12 days, >80% of transgenic seedlings survived, whereas almost all of the control seedlings died (Fig. 4b and Table 3). After mild salt stress treatment (100 mM NaCl for 5 days, a condition that is not lethal to WT rice plants), fresh weight gain of transgenic plants was significantly (P < 0.01) higher than the WT, but no significant difference of fresh weight gain was observed under the normal conditions, indicating that the transgenic plants showed good performance under salt stress (Fig. 4c). Similarly, the calli generated from SNAC1-overexpressing rice seeds showed significantly (P < 0.01) higher fresh weight gain than the WT calli cultured on the medium containing 100 mM NaCl for 15 d (Fig. 4d). These results indicate that overexpression of SNAC1 significantly enhanced salt tolerance of the transgenic lines both at individual and cellular levels.
SNAC1 Encodes a NAC Transcription Factor that Regulates Many Stress Related Genes.
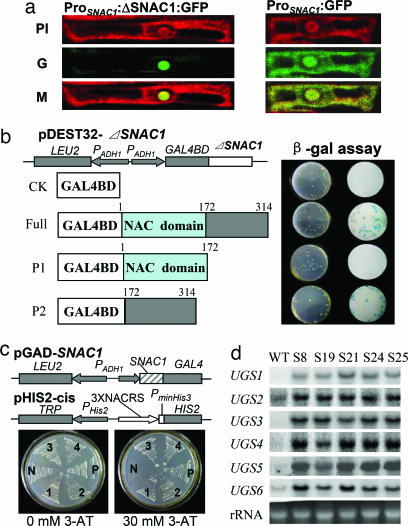
The predicted SNAC1 protein, 314 aa in length, contained a conserved DNA-binding domain found in the plant-specific NAC gene family (32), putative nuclear localization signal (NLS) sequences located at the regions of 77–89 aa and 114–130 aa, respectively, and a nonconserved C-terminal region (Fig. 6, which is published as supporting information on the PNAS web site). A construct PSNAC1:ΔSNAC1:GFP containing the first 90 N-terminal amino acids of SNAC1 fused to GFP under the control of the native SNAC1 promoter was transformed into the rice Nipponbare, and the GFP signal was observed only in nuclei of the cells (Fig. 5a), suggesting that the N-terminal NLS was sufficient to mediate the nuclear targeting of SNAC1 protein.
Fig. 5.
SNAC1 features a transcription factor. (a) Nuclear localization of SNAC1. Constructs of PSNAC1:ΔSNAC1:GFP (containing the first 90 aa of SNAC1) and PSNAC1:GFP were transformed into rice, and GFP was checked in root cell by confocal microscopy. PI, fluorescent image stained with propidium iodide; G, fluorescent image of GFP; M, merged image. (b) β-gal assay of the full or C-terminal SNAC1 in yeast MV203 to identify the transactivation activity (blue colonies in right column). (c) The pGAD-SNAC1 plasmid and the reporter construct pHIS-cis were cotransformed into yeast strain Y187. The transformants were examined by growth performance on SD/Leu−/Trp−/His− plates in the absence or presence of 3-AT. N, negative control (p53HIS2 plus pGAD-SNAC1); P, positive control (p53HIS2 plus pGAD-Rec2-53); labels 1–4, four different colonies containing pGAD-SNAC1 and pHIS-cis. (d) Northern blot analysis of six up-regulated genes in the SNAC1-overexpressing plants under normal growth conditions. The accession numbers of the up-regulated genes (UGS) are as follows: UGS1, AK065989; UGS2, AK103760; UGS3, AK073214; UGS4, AK102788; UGS5, AK103241; and UGS6 (OsERD1), AK068727.
Transactivation assay in the yeast strain MV203 using the full-length or C-terminal region (172–314 aa) sequences of SNAC1 fused to the DNA-binding domain of GAL4 produced the same transactivation activity (Fig. 5b). Serial deletion of SNAC1 in the C-terminal region from the end to the position of 271 aa did not affect the activation, whereas a further deletion to 241 aa abolished the transactivation activity (Fig. 7, which is published as supporting information on the PNAS web site). These results suggested that the predicted C-terminal activation domain had transactivation activity.
To test whether SNAC1 can bind to the putative NACRS (30) in the promoter of the rice ERD1 (early responsive to drought 1) homologue (OsERD1), vectors of pGAD-SNAC1 (containing the putative DNA-binding domain of SNAC1 fused to the GAL4 activation domain) and pHIS2-cis (containing triple tandem repeats of putative NACRS from the OsERD1 promoter) were cotransformed into the yeast strain Y187. The cotransformants could grow on the SD/Leu−/Trp−/His− medium with 30 mM 3-AT whereas the transformed control could not (Fig. 5c), indicating that the SNAC1 could bind to the similar NACRS in OsERD1 promoter.
The features of SNAC1 as a transcription factor prompted us to investigate the expression changes at the whole genome level using a rice DNA chip containing all putative genes in the rice genome (33). Compared with the WT, >80 cDNA-supported genes showed 2.1-fold or higher up-regulation in the transgenic plants under normal growth conditions (Table 4, which is published as supporting information on the PNAS web site). Northern blot analysis of six genes (including OsERD1) with different levels of up-regulation (3.1- to 14.4-fold) confirmed the up-regulation of the five genes (Fig. 5d). Further analysis showed that all of the six genes were induced by abiotic stresses including drought, salt, cold, and ABA treatment (Fig. 8, which is published as supporting information on the PNAS web site). Based on sequence analysis, ≈40 up-regulated genes encode proteins with predicted functions involved in the regulation of stress responses (such as transcription factors, protein kinases, and phosphatases), production of osmolytes (such as sorbitol transporter, exoglucanase, galactosidase, and glycosyltransferase), detoxification and redox homeostasis (such as OsERD1 and glutaredoxin), and protection of macromolecules [such as poly(A)-binding protein and N2, N2-dimethylguanosine tRNA methyltransferase], as well as proteins related to other stresses.
Discussion
Most reports that document enhancement of drought tolerance by means of overexpression of selected genes have been done with model systems, and there are only rare examples [e.g., ERA1 (34)] in which similar effects have been demonstrated through drought resistance testing in the field. With a long-term goal to improve drought resistance of rice, the stress-inducible SNAC1 gene was overexpressed in rice, and the transgenic plants were tested in the field for resistance to drought stress at the stage of anthesis. This stage was chosen for drought resistance testing because it is most sensitive to dehydration stress and very critical for the final yield trait of rice (35). Our data show that the seed-setting rate [one of the key components of yield trait and the best indicator of drought resistance at reproductive stage (36)] of transgenic rice was 22–34% higher than the control plants under severe drought-stressed conditions. With moderate drought stress applied at the early reproductive stage, the SNAC1-overexpressing transgenic rice also had 17–24% higher fertility than the WT. The SNAC1-overexpressing transgenic rice seedlings showed very significantly higher survival rate than WT under drought treatment, further supporting the usefulness of this gene in genetic improvement of drought resistance in rice.
Constitutive overexpression of some stress-responsive transcription factor genes, such as DREBs, controlled by the CaMV 35S promoter frequently caused unwanted phenotypes, such as reduced plant growth, that finally caused significant reduction of potential yield (37). However, transgenic plants (T0) or families (T1 and T2) of SNAC1 showed no obvious difference from the WT plants in all of the traits investigated. Such results provided another good feature for the usefulness of this gene in improving drought resistance.
Drought tolerance and drought avoidance are two major mechanisms for drought resistance in plants. Because SNAC1-overexpressing plants showed no difference from WT in root depth and root volume, the contribution of root traits should be very limited. This finding is corroborated by the results that transgenic plants had significantly higher seed settings than WT with the drought treatment in PVC pipes, in which transgenic and WT plants were individually stressed to the same degree of leaf-rolling; thus, the effects of drought avoidance by deep root is theoretically excluded. Compared with the WT or negative transgenic control, positive transgenic plants showed delayed leaf-rolling and lost water more slowly, which can be considered as another type of drought avoidance (38). The coincidence of delayed leaf-rolling and reduced rate of water loss with increased stomatal closure in transgenic leaves suggested a role for stomatal closure at the early stage of drought stress in preventing water loss from the plant. The strongly localized induction of SNAC1 in guard cells of WT plants suggests that increased stomatal closure is a likely target of regulation by SNAC1, although the details of the regulatory mechanism remain to be characterized. In this connection, it is interesting to note that quite a few rice homologs of genes related to stomatal movement [such as major intrinsic protein (39), calmodulin-binding protein (40), Rac-like GTP-binding protein (41), and WWE domain-containing protein (42)] are up-regulated in the SNAC1-overexpressing plants (Table 4). Recently, two guard-cell-specific R2R3-MYB transcription factors, AtMYB60 (negatively modulated by drought and reducing stomatal opening in null-mutant) (43) and AtMYB61 (positively regulating stomatal closure) (44), were characterized in Arabidopsis for their different roles in the regulation of stomatal closure. A rice R2R3-MYB gene (UGS5) containing core binding sequence of putative NACRS in the promoter region was also up-regulated in the SNAC1-overexpressing plants (Fig. 5). Further investigation of the relationship between SNAC1 and the MYB or other groups of genes is necessary for characterizing the regulatory mechanism involved in stomatal movement under drought stress. Despite having increased numbers of closed stomata in the transgenic rice, the photosynthesic rate between transgenic and WT plants was not significantly different, possibly because rice leaves function normally with more open stomata than may be optimal (S. Pen, personal communication). These results indicate that a more detailed characterization of the effect of SNAC1 overexpression on water use efficiency is warranted.
The ability of transgenic leaves to reestablish turgor pressure at a lower mRWC upon reapplication of water indicates that the transgenic plants have also acquired enhanced dehydration tolerance, in which, according to current understanding, the following mechanisms might be involved: osmotic adjustment (OA) and cell membrane stability, protection of important macromolecules from degradation, and maintenance of redoxin homeostasis and detoxification. The functional categories of genes up-regulated in the SNAC1-overexpressing plants may provide supporting evidence for the actions of such mechanisms in drought tolerance of the transgenic plants. In fact, at least 18 genes (highlighted in Table 4) encoding proteins or enzymes related to OA (such as sorbitol transporter and exoglucanase) or cell membrane stability (such as phosphoethanolamine methyltransferase, and arabinoxylan arabinofuranohydrolase), mRNA and tRNA stability [such as poly(A)-binding protein and N2,N2-dimethylguanosine tRNA methyltransferase], and redoxin homeostasis and detoxification (such as glutaredoxin and reductase) were up-regulated in the transgenic plants.
Besides the improved drought resistance, SNAC1-overexpressing transgenic rice also showed enhanced salt tolerance. Among the genes up-regulated in the SNAC1-overexpressing transgenic plants, no gene was found to have homology to transporter or antiporter genes that were previously reported to function in salt tolerance (45), indicating that there was another potential mechanism of salt tolerance regulated by SNAC1.
In conclusion, this study elucidates that SNAC1 encodes a NAC transcription factor and is induced predominantly in guard cells under dehydration. The significantly enhanced drought resistance and salinity tolerance of the SNAC1-overexpressing rice plants suggest that this gene may show great promise for genetic improvement of stress tolerance in rice.
Materials and Methods
Constructs and Transformation.
The native promoter [an upstream fragment (1,373 bp) starting from the base next to the start codon of SNAC1] was amplified from genomic DNA of upland rice IRAT109 and placed to pCAMBIA1381xb-GFP to control GFP expression (Fig. 1b). The SNAC1 native promoter fragment with the first 270 bp of SNAC1-coding sequence (harboring a putative nuclear localization signal) was introduced to pCAMBIA1381xb-GFP for GFP fusion expression. The full-length cDNA of SNAC1 was amplified from the rice IRAT109 by RT-PCR and inserted into pCAMBIA1301 under the control of the CaMV 35S promoter for overexpression (Fig. 2a). All of the constructs were transformed into the japonica rice Nipponbare by the Agrobacterium-mediated transformation method (46).
Stress Treatments and Measurements.
Transgenic plants of T1 or T2 families were selected by germinating seeds on MS medium containing 50 mg/liter hygromycin for stress testing. Drought resistance at reproductive stage was evaluated under three drought-stressed conditions. The first one was in the refined paddy field (with sand/paddy soil of 1/3) facilitated with a movable rain-off shelter. The second one was conducted in the natural drought-prone field. For these two testings, twenty positive transgenic plants from each transgenic family were planted in two rows (one plot) along with the WT and negative transgenic controls after a randomized complete block designation with three replicates. The third drought testing was conducted in PVC tubes (1 m in height and 20 cm in diameter) filled with well mixed sandy soil (with sand/paddy soil of 1/3), one plant per PVC tube. The detail drought treatment and trait measurement for these testings followed our previous study (36). To determine the mRWC of leaf for reestablishing turgor pressure, RWC was progressively measured (47) until the rolled leaf cannot reexpand when being supplied with water. Photosynthesis rate was measured by using the LI-6400 Photosynthesis System (LI-COR).
Northern Blot and Southern Blot Analysis.
Total RNA was isolated from leaves by using TRIzol reagent (Invitrogen, Carlsbad, CA). Fifteen micrograms of total RNA was used for RNA blot analysis. Total DNA was extracted from T0 transgenic plants by CTAB method, and four micrograms of total DNA was digested by EcoRI for Southern blot. Hybridization was performed with 32P-labeled gene-specific cDNA, and the results were detected by autoradiography.
DNA Chip Analysis.
A custom DNA chip of ≈60,000 oligos (70 mer) representing all putative genes of rice genome was purchased from the Beijing Genomic Institute. For the DNA chip analysis, each transgenic plant and control mRNA samples were reverse transcribed and labeled with Cy3 and Cy5, respectively. Each pair of samples was reversely labeled with transgenic sample labeled by Cy5 and control sample labeled by Cy3, which served as a technical repeat. Hybridization and data processing were carried out as described (33). The genes with the ratios exceeding 2.1-fold in all of the repeated hybridizations were selected for further analysis.
GFP Imaging and SEM.
Root slices of transgenic rice were stained with propidium iodide (10 μg/ml), and the GFP fluorescence was observed by confocal microscopy (TCS SP2, Leica). Leaves of the transgenic and WT plants at booting stage with the same period of dehydration-stress were fixed by glutaraldehyde (2.5%), and the aperture of stomata on upper surface was visualized in a SEM (S-570; Hitachi).
Biochemical Assay in Yeast.
For transactivation assay, the full ORF and a series of deleted SNAC1 were generated by PCR (see Table 5, which is published as supporting information on the PNAS web site, for primer information) and fused in frame to the yeast GAL4 DNA-binding domain in pDEST32 by recombination reactions (Invitrogen). Fusion proteins of GAL4 DNA-binding domain with different portions of SNAC1 were expressed in yeast cells MV203. The colony-lift filter assay (β-gal assay) was performed as described by the manufacturer (Invitrogen). For the one-hybrid assay, SNAC1 was fused to the GAL4 activation domain in the vector pGADT7-Rec2 (Clontech, Palo Alto, CA) and cotransformed with the reporter vector (pHIS2-cis) into yeast cell Y187 for determination of the DNA–protein interactions.
Acknowledgments
This research work was supported by grants from the National Program on the Development of Basic Research, the National Special Key Project on Functional Genomics and Biochips, and the National Natural Science Foundation of China.
Abbreviations
- NAC
NAM, ATAF and CUC transcription factor
- NACRS
NAC recognized sequence
- SNAC1
stress-induced NAC transcription factor gene 1
- PVC
poly(vinyl chloride)
- ABA
abscisic acid
- ERD1
early responsive to drought 1
- RWC
relative water content
- mRWC
minimum RWC.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The sequence of SNAC1 cDNA reported in this paper has been deposited in the GenBank database (accession no. DQ394702).
References
- 1.Deng N. Rev. China Agric. Sci. Technol. 1999;1:3–8. [Google Scholar]
- 2.Shinozaki K., Yamaguchi-Shinozaki K. Curr. Opin. Plant Biol. 2000;3:217–223. [PubMed] [Google Scholar]
- 3.Pastori G. M., Foyer C. H. Plant Physiol. 2002;129:460–468. doi: 10.1104/pp.011021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J. K. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shinozaki K., Yamaguchi-Shinozaki K., Seki M. Curr. Opin. Plant Biol. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- 6.Seki M., Kamei A., Yamaguchi-Shinozaki K., Shinozaki K. Curr. Opin. Biotechnol. 2003;14:194–199. doi: 10.1016/s0958-1669(03)00030-2. [DOI] [PubMed] [Google Scholar]
- 7.Tamura T., Hara K., Yamaguchi Y., Koizumi N., Sano H. Plant Physiol. 2003;131:454–462. doi: 10.1104/pp.102.011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward J. M., Schroeder J. I. Plant Cell. 1994;6:669–683. doi: 10.1105/tpc.6.5.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein M., Geisler M., Suh S. J., Kolukisaoglu H. U., Azevedo L., Plaza S., Curtis M. D., Richter A., Weder B., Schulz B., Martinoia E. Plant J. 2004;39:219–236. doi: 10.1111/j.1365-313X.2004.02125.x. [DOI] [PubMed] [Google Scholar]
- 10.Bartels D. Trends Plant Sci. 2001;6:284–286. doi: 10.1016/s1360-1385(01)01983-5. [DOI] [PubMed] [Google Scholar]
- 11.Singh K., Foley R. C., Onate-Sanchez L. Curr. Opin. Plant Biol. 2002;5:430–436. doi: 10.1016/s1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 12.Riechmann J. L., Heard J., Martin G., Reuber L., Jiang C., Keddie J., Adam L., Pineda O., Ratcliffe O. J., Samaha R. R., et al. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 13.Thomashow M. F. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- 14.Mukhopadhyay A., Vij S., Tyagi A. K. Proc. Natl. Acad. Sci. USA. 2004;101:6309–6314. doi: 10.1073/pnas.0401572101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaguchi-Shinozaki K., Shinozaki K. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oeda K., Salinas J., Chua N. H. EMBO J. 1991;10:1793–1802. doi: 10.1002/j.1460-2075.1991.tb07704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi H., Hong J., Ha J., Kang J., Kim S. Y. J. Biol. Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- 18.Uno Y., Furihata T., Abe H., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. Proc. Natl. Acad. Sci. USA. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. Nat. Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- 20.Haake V., Cook D., Riechmann J. L., Pineda O., Thomashow M. F., Zhang J. Z. Plant Physiol. 2002;130:639–648. doi: 10.1104/pp.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J. C., Lee S. H., Cheong Y. H., Yoo C. M., Lee S. I., Chun H. J., Yun D. J., Hong J. C., Lee S. Y., Lim C. O., Cho M. J. Plant J. 2001;25:247–259. doi: 10.1046/j.1365-313x.2001.00947.x. [DOI] [PubMed] [Google Scholar]
- 22.Park J. M., Park C. J., Lee S. B., Ham B. K., Shin R., Paek K. H. Plant Cell. 2001;13:1035–1046. doi: 10.1105/tpc.13.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ooka H., Satoh K., Doi K., Nagata T., Otomo Y., Murakami K., Matsubara K., Osato N., Kawai J., Carninci P., et al. DNA Res. 2003;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 24.Tran L. S., Nakashima K., Sakuma Y., Simpson S. D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Souer E., van Houwelingen A., Kloos D., Mol J., Koes R. Cell. 1996;85:159–170. doi: 10.1016/s0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 26.Aida M., Ishida T., Fukaki H., Fujisawa H., Tasaka M. Plant Cell. 1997;9:841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sablowski R. W., Meyerowitz E. M. Cell. 1998;92:93–103. doi: 10.1016/s0092-8674(00)80902-2. [DOI] [PubMed] [Google Scholar]
- 28.Xie Q., Frugis G., Colgan D., Chua N. H. Genes Dev. 2000;14:3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collinge M., Boller T. Plant Mol. Biol. 2001;46:521–529. doi: 10.1023/a:1010639225091. [DOI] [PubMed] [Google Scholar]
- 30.Fujita M., Fujita Y., Maruyama K., Seki M., Hiratsu K., Ohme-Takagi M., Tran L. S., Yamaguchi-Shinozaki K., Shinozaki K. Plant J. 2004;39:863–876. doi: 10.1111/j.1365-313X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- 31.Hegedus D., Yu M., Baldwin D., Gruber M., Sharpe A., Parkin I., Whitwill S., Lydiate D. Plant Mol. Biol. 2003;53:383–397. doi: 10.1023/b:plan.0000006944.61384.11. [DOI] [PubMed] [Google Scholar]
- 32.Kikuchi K., Ueguchi-Tanaka M., Yoshida K. T., Nagato Y., Matsusoka M., Hirano H. Y. Mol. Gen. Genet. 2000;262:1047–1051. doi: 10.1007/pl00008647. [DOI] [PubMed] [Google Scholar]
- 33.Ma L., Chen C., Liu X., Jiao Y., Su N., Li L., Wang X., Cao M., Sun N., Zhang X., et al. Genome Res. 2005;15:1274–1283. doi: 10.1101/gr.3657405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., Ying J., Kuzma M., Chalifoux M., Sample A., McArthur C., Uchacz T., Sarvas C., Wan J., Dennis D. T., et al. Plant J. 2005;43:413–424. doi: 10.1111/j.1365-313X.2005.02463.x. [DOI] [PubMed] [Google Scholar]
- 35.Lafitte H. R., Price A. H., Courtois B. Theor. Appl. Genet. 2004;109:1237–1246. doi: 10.1007/s00122-004-1731-8. [DOI] [PubMed] [Google Scholar]
- 36.Yue B., Xue W., Xiong L., Yu X., Luo L., Cui K., Jin D., Xing Y., Zhang Q. Genetics. 2006;172:1213–1228. doi: 10.1534/genetics.105.045062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen Y. G., Zhang W. K., He S. J., Zhang J. S., Liu Q., Chen S. Y. Theor. Appl. Genet. 2003;106:923–930. doi: 10.1007/s00122-002-1131-x. [DOI] [PubMed] [Google Scholar]
- 38.Price A. H., Townend J., Jones M. P., Audebert A., Courtois B. Plant Mol. Biol. 2002;48:683–695. doi: 10.1023/a:1014805625790. [DOI] [PubMed] [Google Scholar]
- 39.Sarda X., Tousch D., Ferrare K., Legrand E., Dupuis J. M., Casse-Delbart F., Lamaze T. Plant J. 1997;12:1103–1111. doi: 10.1046/j.1365-313x.1997.12051103.x. [DOI] [PubMed] [Google Scholar]
- 40.Chen Y. L., Huang R., Xiao Y. M., Lu P., Chen J., Wang X. C. Plant Physiol. 2004;136:4096–4103. doi: 10.1104/pp.104.047837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Assmann S. M. Plant Cell. 2002;14(Suppl.):S355–S373. doi: 10.1105/tpc.001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahlfors R., Lang S., Overmyer K., Jaspers P., Brosche M., Tauriainen A., Kollist H., Tuominen H., Belles-Boix E., Piippo M., et al. Plant Cell. 2004;16:1925–1937. doi: 10.1105/tpc.021832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cominelli E., Galbiati M., Vavasseur A., Conti L., Sala T., Vuylsteke M., Leonhardt N., Dellaporta S. L., Tonelli C. Curr. Biol. 2005;15:1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 44.Liang Y. K., Dubos C., Dodd I. C., Holroyd G. H., Hetherington A. M., Campbell M. M. Curr. Biol. 2005;15:1201–1206. doi: 10.1016/j.cub.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 45.Shi H., Lee B. H., Wu S. J., Zhu J. K. Nat. Biotechnol. 2003;21:81–85. doi: 10.1038/nbt766. [DOI] [PubMed] [Google Scholar]
- 46.Hiei Y., Ohta S., Komari T., Kumashiro T. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 47.Barr H. D., Weatherley P. E. Aust. J. Biol. Sci. 1962;15:413–428. [Google Scholar]