Abstract
Archaeal/eukaryotic initiation factor 2 (a/eIF2) consists of α-, β-, and γ-subunits and delivers initiator methionine tRNA (Met-tRNAi) to a small ribosomal subunit in a GTP-dependent manner. The structures of the aIF2βγ (archaeal initiation factor 2 βγ) heterodimeric complex in the apo and GDP forms were analyzed at 2.8- and 3.4-Å resolution, respectively. The results showed that the N-terminal helix and the central helix–turn–helix domain of the β-subunit bind to the G domain of the γ-subunit but are distant from domains 2 and 3, to which the α-subunit and Met-tRNAi bind. This result is consistent with most of the previous analyses of eukaryotic factors, and thus indicates that the binding mode is essentially conserved among a/eIF2. Comparison with the uncomplexed structure showed significant differences between the two forms of the β-subunit, particularly the C-terminal zinc-binding domain, which does not interact with the γ-subunit and was suggested previously to be involved in GTP hydrolysis. Furthermore, the switch 1 region in the γ-subunit, which is shown to be responsible for Met-tRNAi binding by mutational analysis, is moved away from the nucleotide through the interaction with highly conserved R87 in the β-subunit. These results implicate that conformational change of the β-subunit facilitates GTP hydrolysis by inducing the conformational change of the switch 1 region toward the off state.
Keywords: initiator methionine tRNA, ribosome
The fundamental steps in translation initiation are the binding of initiator methionine tRNA (Met-tRNAi) to a small ribosomal subunit and the positioning of them on the start codon of an mRNA. Eukaryotic initiation factor 2 (eIF2) is a heterotrimeric protein consisting of α-, β-, and γ-subunits that plays a pivotal role in these steps, delivering Met-tRNAi to a small ribosomal subunit as a ternary complex with GTP and facilitating start site selection through hydrolysis of GTP when the codon–anticodon base-pairing between Met-tRNAi and the start codon is formed (1). Recent analyses suggested that archaea also utilize the homolog of eIF2 [archaeal initiation factor 2 (aIF2)] for Met-tRNAi delivery during translation initiation (2, 3).
Many analyses have shown that each subunit of archaeal/eukaryotic initiation factor 2 (a/eIF2) has distinct activities. The γ-subunit is the structural core of a/eIF2 and binds primarily to GTP and Met-tRNAi. It is related structurally to translational EF1A (elongation factor 1A), although the rationale for GTP dependence of Met-tRNAi binding has not been fully understood (4–6). The N-terminal half of eIF2β is characteristic of eukaryotes and associates with RNA (7) and with the initiation factors eIF5 and eIF2B (8), which are also characteristic of eukaryotes. The remaining C-terminal half is common to a/eIF2β and encompasses the C2-C2 zinc-binding motif (9), which associates with RNA (7) and is implicated in the intrinsic GTPase activity (10). The α-subunit is phosphorylated by various stresses, resulting in general translation repression (11). The C terminus of aIF2α binds the γ-subunit and strongly enhances the binding of Met-tRNAi to the γ-subunit (2), although it is indirect (5). Such enhancement is mild in eukaryotes, and, indeed, the eIF2βγ complex facilitates efficient translation initiation in vitro (12).
Despite these studies that have revealed the individual functions of the subunits, it is still unclear how they cooperate to exert integrated functions, such as GTP hydrolysis upon start codon recognition, and the subsequent Met-tRNAi dissociation. In particular, the function of the a/eIF2β common region is less understood. Here, we describe the crystal structures of the aIF2βγ heterodimeric complex from Pyrococcus furiosus (PfIF2βγ) in the absence and presence of GDP at 2.8- and 3.4-Å resolution, respectively, which indicates that the β-subunit undergoes significant structural change in the vicinity of the guanine nucleotide and sequesters the switch 1 region (Sw1) away from the guanine nucleotide. Taken together with the previous implication of the β-subunit in GTP hydrolysis (10) and our mutational analysis that showed strong involvement of Sw1 in Met-tRNAi binding, we propose that the β-subunit promotes GTP hydrolysis and thereby the subsequent Met-tRNAi dissociation by modulating Sw1 to the off state.
Results
Structural Overview.
PfIF2βγ heterodimeric complex was formed by mixing the purified subunits and was separated by the following gel filtration. The protein was crystallized in apo form, and GDP was introduced by the soaking method, whereas the GTP analog could not be introduced successfully because of immediate cracking of the crystals. The structures in the apo and GDP forms are essentially identical, with a backbone rmsd of 0.3 Å. Differences between the two forms are limited to the local conformation around the guanine nucleotide-binding site (GBS), as shown in the uncomplexed γ-subunit (4). The middle region of Sw1 (residues 39–42), which is disordered in the apo form, as are other structures of the γ-subunit (4–6), could be modeled in the GDP form despite the lower resolution limit. Thus, we hereafter refer to the structure of GDP form except where indicated.
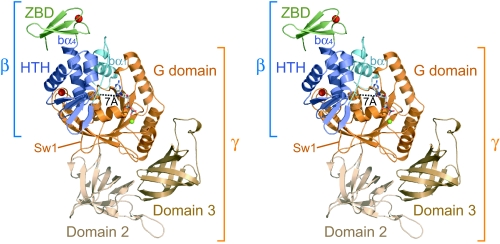
The overall structure of the γ-subunit in the heterodimeric complex consists of three consecutive domains, G, 2, and 3 (Fig. 1), and is closely similar to that of the uncomplexed form from Pyrococcus abyssi in the presence of GDP (4), which has 96% sequence identity with PfIF2γ, with a backbone rmsd of 0.7 Å. GDP-Mg2+ also interacts with the γ-subunit in essentially the same way as in the uncomplexed form. In contrast, the β-subunit shows significant differences from the uncomplexed structure (9), as described below. It consists of three parts: the N-terminal bα1-helix connected by loop (“b” and “g” in front of each secondary structural element or residue indicates that each element belongs to the β- or γ-subunit, respectively), the central helix–turn–helix domain (HTH), and the C-terminal zinc-binding domain (ZBD).
Fig. 1.
“Front” view of the aIF2βγ–GDP complex. Domains are color-coded as indicated. Zn2+, Mg2+, and GDP are shown as red balls, green balls, and stick models, respectively. The minimum distance between the β-subunit and GDP (bA35 CO–ribose 3′OH) is indicated as a dotted line.
In the complex, the N terminus and the central HTH of the β-subunit interact with the G domain of the γ-subunit, whereas the C-terminal ZBD is exposed to a solvent without making contact with the γ-subunit (Fig. 1). Upon complex formation, the N-terminal region of the β-subunit, which was unstructured or unstable in the uncomplexed form (9, 13), comprises the amphiphilic bα1-helix and interacts strongly with the “back side” of the GBS (where the “front” is defined as in Fig. 1). The subsequent loop region is then located near the ribose moiety of the bound GDP with a minimum distance of ≈7 Å, which is consistent with the affinity labeling of this region with the ribose moiety of the GTP analog (14), albeit without direct interaction. The β-subunit interacts only with the G domain and is distant from domains 2 and 3, to which the α-subunit and Met-tRNAi bind (5). This result is consistent with the absence of a stable interaction between the α- and β-subunits (4) but is in contrast to the cross-linking between the bβ1 region and Met-tRNAi in eukaryotes (15). Considering the significant conservation of the residues involved in the β–γ interactions among archaeal and eukaryotic factors (Fig. 2; see below) and the fact that the cross-linker used in the previous study specifically cross-links a peptide and a guanine base, this discrepancy can most likely be explained by the detection of an unintended GTP peptide instead of a desired tRNA peptide. However, biochemical analysis of the αγ complex still suggested the strong involvement of the eukaryotic β-subunit in Met-tRNAi binding (16), whereas that of the archaeal one did not (2), suggesting that a specific feature in eukaryotes, such as the N-terminal extension of the β-subunit, may participate in Met-tRNAi binding.
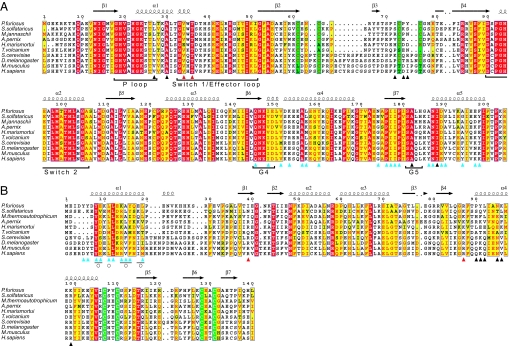
Fig. 2.
Sequence alignments of the G domains (A) and the β-subunits (B). The completely conserved residues are boxed in red, and conservative replacements are boxed in yellow. The zinc-binding motifs in each subunit are boxed in green. The secondary structural elements are designated as indicated. The residues marked with cyan, black, and red triangles indicate those involved in the bα1-γ, bα4-γ, and HTH-Sw1 contacts, respectively. Circles indicate the mutations in yeast that are defective in binding of the γ-subunit (17). Regions related to the guanine nucleotide are indicated at the bottom of the columns.
Dimer Interactions.
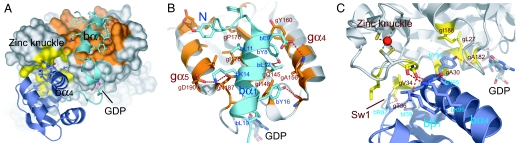
The β-subunit binds the γ-subunit mainly by means of two helices, bα1 at the N terminus and bα4 in the central HTH (Fig. 3A). The amphiphilic bα1-helix interacts with the hydrophobic surface, composed of gα4, gβ6, gβ7, and gα5 at the back side of the GBS, which comprises an extensive intersubunit hydrophobic core (Fig. 3B). Furthermore, the side chains of conservative residues, such as bY8, bE9, bK14, and bY16, additionally form hydrogen bonds with the γ-subunit to ensure the specificity. Particularly, bY8 and bY16 interact with gQ145 and gI148 in a QNKIE motif that recognizes the guanine base of the nucleotide. The contacting surface in this region accounts for ≈60% of the total interface (1,600 Å2), indicating that the specific binding of the two subunits is mainly attributable to this contact. Indeed, many of the residues involved in this contact are conserved or conservatively substituted in the both subunits of a/eIF2 (Fig. 2) and are correlated well with the mutants of the β-subunit that show defective association with the γ-subunit in yeast (17).
Fig. 3.
Dimer interfaces. (A) The topside view of the βγ complex. The γ-subunit is shown as a surface representation in which the residues interacting with bα1 (cyan) are colored orange and those interacting with HTH (dark blue) are colored yellow. ZBD is not shown. (B) The interactions between bα1 and the back side of the G domain. The residues involved in the contacts are shown as stick models, along with hydrogen bonds (dashed red lines). The conservative residues and the residues mentioned in the text are labeled. (C) The interactions between HTH and the topside of the G domain.
In contrast, the central HTH of the β-subunit shows only local contacts with the conserved motifs in the γ-subunit, mainly by means of the bα4 helix (Fig. 3C). That is, bN95, bK96, and bK99 in the middle region of the bα4 helix form hydrogen bonds with the main chain of the zinc knuckle of the γ-subunit, whereas bY91 in the N terminus of bα4 makes hydrophobic interactions with the concavity composed of the SALH motif and the P loop of the GBS. In addition, bI39 in bβ1 and the conservative bR87 in the loop between bβ4 and bα4 interact with gV34 and the strictly conserved gT36 in the N terminus of Sw1, respectively. However, most of these contacts between HTH and the γ-subunit are local and less specific, because they consist mainly of less conserved residues or main chains (Fig. 2), indicating that the core region of the β-subunit binds loosely to the γ-subunit.
Conformational Variability of the β-Subunit.
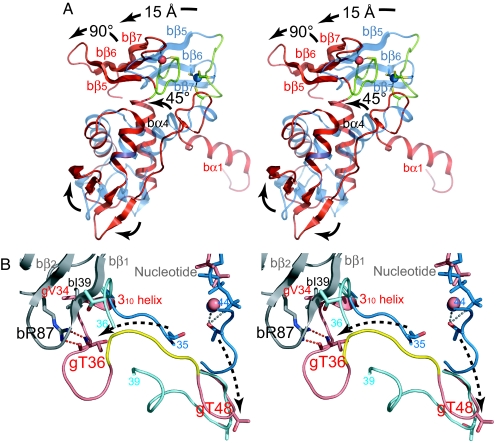
Consistent with the overall flexibility suggested by the uncomplexed structure of the β-subunit from Methanobacterium thermoautotrophicum (9), which shows 56% identity and 80% similarity with PfIF2β, the β-subunit in the βγ complex is significantly different from the uncomplexed form. The bα4 helix in the center of the molecule, which also interacts with the γ-subunit as described above, is rotated by ≈45° with respect to that of the uncomplexed form, entailing large movement of the connecting ZBD for ≈15 Å (Fig. 4A). Furthermore, ZBD is rotated by ≈90° about an axis perpendicular to the bα4 helix, which is due to the local conformational change of the loop between bα4 and bβ5 from the extended form in the uncomplexed subunit to the hairpin form in the βγ complex. Although this loop includes bC107 and bC110 of the C2-C2 zinc-binding motif, the tetrahedral coordination to the zinc ion is retained in the present conformation (Fig. 4A), which is consistent with the essentiality of the zinc ion in the stability of aIF2β (18). ZBD in the complex is packed closely to HTH, and an interdomain hydrogen bond is discernible between the conserved bT115 in ZBD and bR58 in HTH, in contrast to the uncomplexed form, which shows no obvious interaction between the domains. Along with these large domain movements, the antiparallel β-sheets in HTH (bβ1-bβ4) and ZBD (bβ5-bβ7) are twisted in the complex (Fig. 4A), which is necessary to maintain the integrity of the protein. In fact, severe internal steric hindrance would occur without such twisting, indicating that all of the structural changes of the β-subunit occur in a concerted manner. Considering the loose binding of HTH to the γ-subunit as described above, such conformational change of the β-subunit core region can occur in the intact a/eIF2 complex, whereas the N-terminal bα1-helix binds rigidly to the γ-subunit to anchor the core region. Indeed, we have observed the immediate cracking of the crystals when soaked in buffer containing GTP analog, which has never been observed in those soaked in GDP, suggesting significant conformational change of the βγ complex between the different nucleotide states.
Fig. 4.
Structural comparisons. (A) Comparison of the β-subunits in the βγ complex (red) and the uncomplexed form (blue) by superposing the Cα atoms of HTH (residues 35–89) with the program TOP3D (25). The loop between bα4 and bβ5 encompassing the N-terminal half of the C2-C2 zinc-binding motif is shown in green. Major conformational changes are indicated with arrows. (B) Comparison of the Sw1 of the γ-subunit in PfIF2βγ–GDP (pink), aIF2αγ–GDPNP (blue), and aIF2γ–GDP (cyan) by superposing the G domains with the program TOP3D. The β-subunit is shown in gray. The conserved EEΦ(R/K)R motif in the βγ complex is colored yellow. The residues between 35 and 44 in αγ–GDPNP and 36 and 39 in γ–GDP are disordered. Dashed arrows indicate the possible conformational change of Sw1 between the GDPNP and GDP forms.
Structural Change of Sw1 Induced by the β-Subunit.
In the present aIF2βγ complex in the GDP form, the β-subunit interacts directly with Sw1 as well as the conserved motifs in the GBS as described above (Fig. 3). Particularly, through the interaction between the conserved bR87 and gT36, the N-terminal half of Sw1 (residues 33–40) encompasses the 310 helix and is bent toward the zinc knuckle, ≈20 Å away from GBS, whereas the C-terminal half shows a similar conformation to that in the uncomplexed γ-subunit in GDP form (Fig. 4B).
Because Sw1 in the structure of aIF2αγ in GTP form showed a similar conformation to that of EF1A in GTP form, it was suggested that Sw1 participates in the binding of both Met-tRNAi and GTP-Mg2+ (5). Indeed, two mutations in Sw1 have been identified in yeast that cause defects in the function of eIF2 (10, 19), further emphasizing importance of Sw1. However, it has not been evident whether Sw1 in the γ-subunit is actually involved in ligand binding as in EF1A. Therefore, we mutated the conserved 41EEΦ(R/K)R45 motif (in which “Φ” indicates a hydrophobic residue) and gT36 in Sw1, and we tested whether they affect the Met-tRNAi affinity. However, because the α-subunit of PfIF2 is insoluble when produced in Escherichia coli, we used aIF2 from Sulfolobus solfataricus (SsIF2) for the mutational assay, which is one of the most well characterized aIF2s and shares 35%, 31%, and 56% identity and 64%, 60%, and 76% similarity with the PfIF2 α-, β-, and γ-subunits, respectively. As a result, it was clearly shown that the former mutations result in a severe decrease in Met-tRNAi affinity and that the latter mutation also impairs slightly Met-tRNAi binding in the presence of GTP (Table 1; see also Fig. 5 A–H, which is published as supporting information on the PNAS web site). Because the region around the former mutations corresponds to the A′ helix in EF1A, which interacts directly with aminoacyl-tRNA and GTP-Mg2+, and the latter corresponds to the A” helix, which indirectly interacts with GTP-Mg2+, the effects of the mutations indicate that Sw1 in the γ-subunit is strongly involved in binding of Met-tRNAi in a manner similar to EF1A. In sum, the current structure reveals that the β-subunit sequesters gT36, which interacts indirectly with the γ-phosphate and Mg2+ as a part of the A” helix in GTP form from ligand binding through interaction with the conserved bR87 in GDP form.
Table 1.
Summary of Met-tRNAi affinities
| Protein | Kd, nM |
|---|---|
| Wild type | 9.1 ± 0.5 |
| gT34V (gT36) | 22 ± 4.1 |
| g39–43A (g41–45) | >20,000 |
| gE40A (gE42) | 380 ± 64 |
Values are shown as average ± SD of three independent experiments. The residue numbers refer to SsIF2; those in parentheses refer to PfIF2. In the g39–43A mutant, all residues in the 39EEΦ (R/K)R43 motif are mutated to alanine.
Discussion
We have analyzed the structures of aIF2βγ complex in the apo and GDP forms, which show binding of the β-subunit to the G domain of the γ-subunit by means of two helices, the N-terminal bα1 and the central bα4. The structural comparison indicates the large conformational change of the overall β-subunit, particularly the C-terminal ZBD that does not interact with the γ-subunit. Furthermore, cracking of the crystals by soaking in GTP analog also suggests significant conformational change of the βγ complex between the different nucleotide states. In contrast, the previous study showed that one of the SUI3 suppressor mutants in yeast (S264Y, corresponding to the mutation at A133 in ZBD of PfIF2β) confers a significant increase in intrinsic GTP hydrolysis that is independent of any other factors (10). Taking these facts together, it is likely that the involvement of ZBD in intrinsic GTP hydrolysis is structurally related to the conformational change of ZBD between the different nucleotide states. Moreover, because ZBD was shown to interact with RNA (7), it is possible that the conformational change of ZBD is triggered by mRNA or rRNA, by which the start codon signal can be mediated.
In the current structure, gT36 in Sw1 is far away from GBS through the interaction with the conserved bR87 in the β-subunit. Considering that our mutational analyses of Sw1 and the previous structure of aIF2αγ in GTP form (5) strongly suggest that Sw1 binds with Met-tRNAi and the γ-phosphate-Mg2+ in a manner similar to EF1A, Sw1 in the current GDP form seems no longer to participate in ligand binding. The simplest explanation of this Sw1 sequestering is that the β-subunit stabilizes Sw1 off conformation after GTP hydrolysis, thereby facilitating Met-tRNAi dissociation. However, this explanation is unlikely, because the mutation of bR87 or the absence of the β-subunit did not affect Met-tRNAi affinity in the presence of GDP (Fig. 5 I–K). Assuming that GTP hydrolysis is related to the structural change of the β-subunit as discussed above, another fascinating explanation is that the sequestering of gT36 by the β-subunit precedes or is concurrent with GTP hydrolysis and causes an environmental change around the γ-phosphate and Mg2+, which leads to GTP hydrolysis and a release of the γ-phosphate with the help of other factors, such as eIF1 and eIF5 (in the case of eukaryotes). In this context, the current structure in GDP form reflects that the β-subunit facilitates conformational change of Sw1 toward the off state.
In eukaryotes, intrinsic GTP hydrolysis of eIF2 is facilitated by GTPase-activating protein (GAP) eIF5, which interacts stably with the β-subunit (8). Because the active domain of eIF5 shows homology with the a/eIF2β common region, it was proposed that eIF5 displaces the β-subunit during GTPase activation (20). Therefore, taken together with our hypothesis above, the conformational change of Sw1 by the β-subunit may lead to recruitment of the eIF5 arginine finger, the proposed catalytic residue of GAP activity, in the vicinity of the γ-phosphate. However, in the case of archaea, a homolog of eIF5 is apparently not coded in their genomes (21), except for aIF2β. Because the current knowledge of the archaeal system is mainly based on in silico analyses, further study is needed to elucidate whether the GAP activity is assigned to an unknown factor or is complemented by known aIFs. In any case, the evolutional relationship between the archaeal and eukaryotic systems will be of interest.
In summary, in the present study, we determined the structures of aIF2βγ in the apo and GDP forms, which implicate a structural role of the β-subunit in GTP hydrolysis. We hope this work will stimulate further studies on both archaeal and eukaryotic systems.
Materials and Methods
Sample Preparation.
Each gene encoding PfIF2γ and PfIF2β was cloned into pET-26b and was expressed in E. coli strain BL21-Codonplus-(DE3)-RIL as C-terminal His-6-tagged protein, which was purified independently as follows. To ensure the yield, the point mutation (G236D) was introduced into the γ-subunit (4). The cells were disrupted and heat-treated for 30 min at 80°C at pH 8.0. The extracted protein was purified by Ni-affinity, gel-filtration, and cation exchange chromatography at pH 6.0. The purified subunits were mixed together in equimolar amounts to form the intact βγ complex, which was then further purified by gel filtration with buffer containing 20 mM Tris·HCl (pH 8.0) and 0.2 M NaCl and concentrated to 22 mg/ml.
Each gene encoding SsIF2α, SsIF2β, and SsIF2γ was cloned into pET-22b or pET-28b and was expressed in BL21-Codonplus-(DE3)-RIL. The cells were disrupted and heat-treated at 70°C for 30 min. SsIF2γ was further purified by anion exchange and gel-filtration chromatography at pH 9.0, whereas SsIF2α and -β were purified by cation exchange and gel-filtration chromatography at pH 6.0. All of the SsIF2 variants were constructed by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. All of the mutants were purified essentially the same as the wild-type protein and were confirmed to be thermostable and have no defect in the formation of the heterotrimeric complex, which was analyzed by nondenaturing PAGE as described in ref. 3. The concentration of each protein was estimated by standard Bradford assay, using BSA as a standard.
Met-tRNAi from S. solfataricus was prepared as described in ref. 3 by using [3H]-labeled methionine [specific activity of 78 Ci/mmol (1 Ci = 37 GBq)].
Crystallization and Structural Analysis.
The crystals of apo-PfIF2βγ appeared within 2 months by the hanging-drop method at 18°C with a reservoir containing 0.2 M MgCl2, 26–30% (wt/vol) PEG 8000, and 0.1 M Tris·HCl (pH 8.5). The crystal containing GDP was obtained by soaking the crystal in the mother liquor containing 1.5 mM GDP for 1 h. In contrast, soaking with the buffer containing 1.5 mM GDPNP resulted in cracking of the crystal within 1 min. The diffraction data were collected at beamline BL41XU or BL44B2 (at SPring-8) and were analyzed and processed by using the program HKL2000 (22).
The structure of apo-PfIF2βγ complex was solved by molecular replacement by using the program AMoRe (23) with the structure of uncomplexed aIF2γ (4) as the search model. The model of the β-subunit was built manually, based on an omit map. The structure was refined by rigid-body refinement, simulated annealing, and cycles of positional and B factor refinement by using the program CNS (24) with manual fittings. Particularly, Sw1 was completely rebuilt with an omit map to minimize model bias. Finally, the model was refined to 2.8-Å resolution with an R factor of 24.4% and an Rfree factor of 29.0% and containing one heterodimeric complex, a zinc ion in each subunit, and 180 water molecules in an asymmetric unit. The structure of PfIF2βγ–GDP-Mg2+ was determined by positioning the apo structure with rigid-body refinement. The model was refined to 3.4-Å resolution in a manner similar to the apo structure, with an R factor of 24.9% and an Rfree factor of 29.8%. Residues 1, 2, and 140 in the β-subunit and residues 1–5 in the γ-subunit are disordered and could not be modeled in both structures, whereas residues 39–42 in the γ-subunit are disordered only in the apo form. The data collection and refinement statistics are summarized in Table 2.
Table 2.
Data collection and refinement statistics
| Data set | Apo | GDP complex |
|---|---|---|
| Data collection | ||
| Beamline | SPring-8 BL41XU | SPring-8 BL44B2 |
| Wavelength, Å | 0.9000 | 0.9788 |
| Space group | P212121 | P212121 |
| Unit cell, Å | 68.4, 76.2, 98.2 | 69.2, 76.6, 97.9 |
| Unique reflections | 12,894 (1,172) | 8,036 (651) |
| Resolution, Å | 50.0–2.80 (2.90–2.80) | 50.0–3.30 (3.42–3.30) |
| Completeness, % | 98.3 (91.1) | 97.3 (82.4) |
| Redundancy | 5.3 (3.2) | 4.4 (3.0) |
| I/σ(I) | 16.7 (2.0) | 7.0 (1.4) |
| Rmerge, % | 10.2 (31.4) | 11.9 (49.7) |
| Refinement | ||
| Resolution range, Å | 20.0–2.80 | 20.0–3.4 |
| R factor/Rfree, % | 24.4/29.0 | 24.9/29.8 |
| No. of atoms (protein, water, and others) | 4,216/180/2 | 4,250/108/31 |
| rmsd bond length, Å/angles, degrees | 0.0067/1.85 | 0.0071/1.81 |
| Ramachandran plot, % | ||
| Favored | 85.0 | 80.7 |
| Allowed | 14.1 | 17.6 |
| Generous | 0.9 | 1.7 |
| Average B factor, Å2 | 61.9 | 53.7 |
Values in parentheses are for the highest-resolution shell. Rfree factors were calculated for 7.5% randomly selected test sets that were not used in the refinement.
Met-tRNAi Affinity Measurement.
The affinity to Met-tRNAi was estimated by deacylation protection assay as described in ref. 2. The concentrations of SsIF2 variants were varied from 2 nM to 20 μM according to the Kd value to be measured. Reaction mixtures (100 μl) containing the buffer, the protein, 1 mM GTP or GDP, and 30 nM [3H]Met-tRNAi were incubated at 62°C. Aliquots (20 μl) were withdrawn at 6-min intervals and spotted onto 3-MM filter disks (Whatman, Clifton, NJ) saturated with 5% (wt/vol) trichloroacetic acid (TCA); the disks were washed with 5% TCA and ethanol, dried, and subjected to liquid scintillation counting (Fig. 5).
Supplementary Material
Acknowledgments
We thank Prof. M. Kimura for kind advice on this work and the staff of the SPring-8 BL41XU and BL44B2 beamlines for help with the x-ray diffraction experiments. This work was supported by a Grant-in-Aid for Scientific Research (16570089); a grant for the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; and by the Human Frontier Science Program.
Abbreviations
- Met-tRNAi
initiator methionine tRNA
- eIF2
eukaryotic initiation factor 2
- aIF2
archaeal initiation factor 2
- a/eIF2
archaeal/eukaryotic initiation factor 2
- GBS
guanine nucleotide-binding site
- HTH
helix–turn–helix domain
- ZBD
zinc-binding domain
- Sw1
switch 1 region
- PfIF2
aIF2 from Pyrococcus furiosus
- SsIF2
aIF2 from Sulfolobus solfataricus
- EF1A
elongation factor 1A
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2D74 and 2DCU).
References
- 1.Kapp L. D., Lorsch J. R. Annu. Rev. Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 2.Yatime L., Schmitt E., Blanquet S., Mechulam Y. J. Biol. Chem. 2004;279:15984–15993. doi: 10.1074/jbc.M311561200. [DOI] [PubMed] [Google Scholar]
- 3.Pedullà N., Palermo R., Hasenöhrl D., Bläsi U., Cammarano P., Londei P. Nucleic Acids Res. 2005;33:1804–1812. doi: 10.1093/nar/gki321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitt E., Blanquet S., Mechulam Y. EMBO J. 2002;21:1821–1832. doi: 10.1093/emboj/21.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yatime L., Mechulam Y., Blanquet S., Schmitt E. Structure (London) 2006;14:119–128. doi: 10.1016/j.str.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 6.Roll-Mecak A., Alone P., Cao C., Dever T. E., Burley S. K. J. Biol. Chem. 2004;279:10634–10642. doi: 10.1074/jbc.M310418200. [DOI] [PubMed] [Google Scholar]
- 7.Laurino J. P., Thompson G. M., Pacheco E., Castilho B. A. Mol. Cell. Biol. 1999;19:173–181. doi: 10.1128/mcb.19.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asano K., Krishnamoorthy T., Phan L., Pavitt G. D., Hinnebusch A. G. EMBO J. 1999;18:1673–1688. doi: 10.1093/emboj/18.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutiérrez P., Osborne M. J., Siddiqui N., Trempe J. F., Arrowsmith C., Gehring K. Protein Sci. 2004;13:659–667. doi: 10.1110/ps.03506604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang H. K., Yoon H., Hannig E. M., Donahue T. F. Genes Dev. 1997;11:2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Proud C. G. Semin. Cell Dev. Biol. 2005;16:3–12. doi: 10.1016/j.semcdb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Nika J., Rippel S., Hannig E. M. J. Biol. Chem. 2001;276:1051–1056. doi: 10.1074/jbc.M007398200. [DOI] [PubMed] [Google Scholar]
- 13.Cho S., Hoffman D. W. Biochemistry. 2002;41:5730–5742. doi: 10.1021/bi011984n. [DOI] [PubMed] [Google Scholar]
- 14.Bommer U. A., Kraft R., Kurzchalia T. V., Price N. T., Proud C. G. Biochim. Biophys. Acta. 1991;1079:308–315. doi: 10.1016/0167-4838(91)90074-a. [DOI] [PubMed] [Google Scholar]
- 15.Gaspar N. J., Kinzy T. G., Scherer B. J., Hümbelin M., Hershey J. W., Merrick W. C. J. Biol. Chem. 1994;269:3415–3422. [PubMed] [Google Scholar]
- 16.Flynn A., Oldfield S., Proud C. G. Biochim. Biophys. Acta. 1993;1174:117–121. doi: 10.1016/0167-4781(93)90105-m. [DOI] [PubMed] [Google Scholar]
- 17.Hashimoto N. N., Carnevalli L. S., Castilho B. A. Biochem J. 2002;367:359–368. doi: 10.1042/BJ20020556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutiérrez P., Coillet-Matillon S., Arrowsmith C., Gehring K. FEBS Lett. 2002;517:155–158. doi: 10.1016/s0014-5793(02)02610-8. [DOI] [PubMed] [Google Scholar]
- 19.Dorris D. R., Erickson F. L., Hannig E. M. EMBO J. 1995;14:2239–2249. doi: 10.1002/j.1460-2075.1995.tb07218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson G. M., Pacheco E., Melo E. O., Castilho B. A. Biochem J. 2000;347:703–709. [PMC free article] [PubMed] [Google Scholar]
- 21.Londei P. FEMS Microbiol. Rev. 2005;29:185–200. doi: 10.1016/j.femsre.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Navaza J. Acta Crystallogr. A. 1994;50:157–163. [Google Scholar]
- 24.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Lu G. J. Appl. Crystallogr. 2000;33:176–183. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.