Abstract
Globular proteins fold by minimizing the nonpolar surface that is exposed to water, while simultaneously providing hydrogen-bonding interactions for buried backbone groups, usually in the form of secondary structures such as α-helices, β-sheets, and tight turns. A primary thermodynamic driving force for the formation of globular structure is thus the sequestration of nonpolar groups, but the correlation between the parts of proteins that are observed to fold first (termed folding initiation sites) and the “hydrophobicity” (as customarily defined) of the amino acids in these regions has been quite weak. It has previously been noted that many amino acid side chains contain considerable nonpolar sections, even if they also contain polar or charged groups. For example, a lysine side chain contains four methylenes, which may undergo hydrophobic interactions if the charged ε-NH3+ group is salt-bridged or hydrogen-bonded. Folding initiation sites might therefore contain not only accepted “hydrophobic” amino acids, but also larger charged side chains. Recent experiments on the folding of mutant apomyoglobins provides corroboration for models based on the hypothesis that folding initiation sites arise from hydrophobic interactions. A near-perfect correlation was observed between the areas of the molecule that are present in the burst-phase kinetic intermediate and both the free energy of formation of hydrophobic initiation sites and the parameter “average area buried upon folding,” which pinpoints large side chains, even those containing charged or polar portions. These results provide a putative mechanism for the control of protein-folding initiation and growth by polar/nonpolar sequence propensity alone.
Keywords: folding pathways, myoglobin, ribonuclease
Freed of the ribosome, and in the absence of chaperones, a newly synthesized protein exists in an ensemble of states, the so-called statistical coil, the nature of which is a subject of much recent investigation (1). It subsequently folds to the native biologically active structure whose free energy is considered to be a minimum under appropriate solution conditions (2). Since the seminal work of Anfinsen (2), the kinetics of the folding process and the final structure have been considered to be determined by the physical interactions among the amino acid residues to attain the thermodynamically favored state.
A major question has involved the exact mechanism by which the interresidue interactions specify the initiation of folding in the unfolded polypeptide chain under folding conditions. Because nonpolar groups are not soluble in water, attention has been focused on the hydrophobic interactions between nonpolar side chains to achieve their sequestration from aqueous solvent. A model has been proposed in which nearby nonpolar groups in the chain participate in hydrophobic interactions with minimal loss of entropy of the chain because of the proximity of the interacting side chains in the amino acid sequence (3).
Yet the chemical nature of the polypeptide chain complicates the simple physical chemistry: each of the nonpolar amino acid side chains is attached to the polypeptide backbone, which is polar. The only way that the polar backbone can be sequestered from water in the nonpolar interior is for it to be involved in hydrogen bonds (4, 5). If exposed to water, such hydrogen bonds would be very weak because of competition from hydration of the polar C O and N
O and N H groups. However, polar amino acids such as lysine and glutamic acid have nonpolar necks that can interact with nonpolar side chains, as illustrated in Fig. 1 (6, 7). These interactions strengthen hydrogen bonds and electrostatic interactions between charged groups both by reducing the entropy of otherwise freely rotating polar or charged side chains and creating a water-diminished nonpolar environment. Thus, the final structure contains stable backbone hydrogen bonds in the form of secondary structure, namely α-helices, β-sheets, and β-turns.
H groups. However, polar amino acids such as lysine and glutamic acid have nonpolar necks that can interact with nonpolar side chains, as illustrated in Fig. 1 (6, 7). These interactions strengthen hydrogen bonds and electrostatic interactions between charged groups both by reducing the entropy of otherwise freely rotating polar or charged side chains and creating a water-diminished nonpolar environment. Thus, the final structure contains stable backbone hydrogen bonds in the form of secondary structure, namely α-helices, β-sheets, and β-turns.
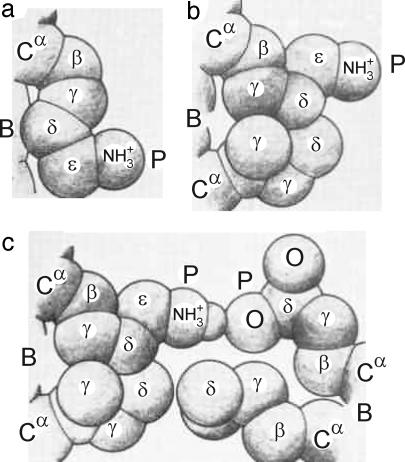
Fig. 1.
Schematic representation of various hydrophobic interactions of a polar side chain with its surroundings (B refers to the backbone, P to the polar head, and α to the α-carbon). (a) Interaction of a lysine side chain with the backbone. (b) Interaction of a lysine side chain with a nearby isoleucine side chain. (c) Interaction of two polar side chains (lysine and glutamic acid), engaged in hydrogen bonding, with two nonpolar side chains (isoleucine and leucine, respectively). A hydrophobic interaction is also formed between the two nonpolar side chains. [Reproduced with permission from ref. 6 (Copyright 1963, John Wiley & Sons, Inc.).]
The purpose of this article is to summarize recent experimental results that provide evidence in support of two related models of folding initiation (3, 8), one of which (8) also treats the subsequent propagation steps. Both models lead to similar conclusions.
Models for Initiation and Propagation
The first model (3), illustrated in Fig. 2, is based on hydrophobic interactions (6, 9, 10) and uses a running window of various numbers of residues along the amino acid sequence to calculate the free energy to convert an extended chain to a collapsed hydrophobic pocket; different windows are scored in terms of the most favorable stable pocket and other pocket sites of successively lower free energy of stabilization, i.e., indicating the possible presence of multiple initiation sites. The negative free energy of hydrophobic interactions compensates for the decrease in entropy to bring nonpolar residues into contact to form a hydrophobic pocket. Such a pocket is a folding initiation site, which is a specific conformation of a limited section of a polypeptide chain; its existence can significantly increase the rate of formation of the native structure from the unfolded form. However, as pointed out in refs. 3 and 11, such a conformation should not be considered as a static one but rather as one that fluctuates among stretches of nonpolar residues in many competing initiation sites of slightly lower stability, i.e., persistence of such a fluctuating site (with transitions through the unfolded conformation) in the ensemble of interconverting conformations in the unfolded state of the protein; usually the primary initation site is marginally stable by itself but acquires increased stability as it interacts with other pockets in nearby portions of the chain. When the initiation pocket is formed under conditions that permit folding of the remainder of the chain, it leads to rapid direct formation of the complete native structure; its absence, even under conditions in which folding may occur, prevents folding, independent of the state of the remaining nonpolar groups. Electrostatic interactions contribute to the stability of a hydrophobic pocket [and pH-dependent folding (3)] only when hydrophobic interactions lead to constraints that force charged or polar groups to be near each other, as illustrated in Fig. 1. A feature of an initiation site is the presence of a turn (not necessarily an ordered one), as illustrated in Fig. 2, that persists to the final native structure, despite small alterations in the initiation site during subsequent folding stages; even if such small alterations in structure occur, its nonpolar character is preserved. For those cases in which a turn is not found, the native conformation of all or most of the predicted initiation site is an α-helix; moreover, with the exception of the 103–115 pocket of myoglobin, the initiation sites are found very close to the ends of helical sequences (3). A pocket of the type shown in Fig. 2 can easily transform into a helix in subsequent folding stages. The model of ref. 3 suggests that the nonpolar character of an initiation site could also be important for hydrophobic interactions in the later stages of folding. A similar approach (10) is based on calculating buried surface area in a running window of nine residues.
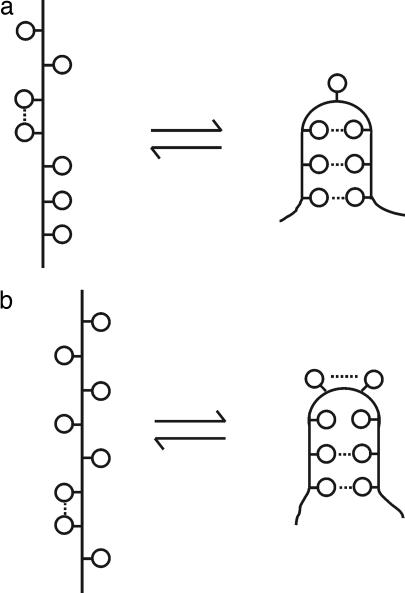
Fig. 2.
Schematic representation of folding initiation step in which a hydrophobic pocket is formed from an ensemble of unfolded species [which may involve neighbor-neighbor hydrophobic interactions (9) as indicated by the species on the left side of the equilibria]. (a) Only one side chain is involved in the “turn,” and it does not participate in hydrophobic interactions with nearby residues. (b) Two side chains (which themselves can involve a neighbor–neighbor hydrophobic interaction) are part of the turn, and the pocket is shown as an imperfect one, in that one pair of side chains is not sufficiently nonpolar to involve a hydrophobic interaction. Hydrophobic interactions are indicated by dotted lines. For pockets of types a and b, at least five and four residues, respectively, are assumed to be required, so that the pockets are large enough for the bends to be stereochemically feasible. [Reproduced with permission from ref. 3 (Copyright 1978, American Chemical Society).]
The second approach (8) makes use of the triangular contact map (12) of a native protein, illustrated in Fig. 3 (13), to trace a series of possible folding steps as initial clusters form among nearby residues in different multiple sites in the sequence and then coalesce in successive stages of folding. This folding model consists of three stages, A, B, and C. In stage A, short-range interactions lead to contacts among residues close to each other in the amino acid sequence, i.e., to form ordered backbone structures, (α-helices, β-sheets, or turns), at equilibrium above the denaturation temperature. Lowering of the temperature, or alteration of the solvent conditions in stage B, leads to medium-range interactions that form small fluctuating contact regions (nearby in amino acid sequence) between the structures formed in stage A, with possible rearrangements of the structures of stage A. The sizes of the pockets in the model of Fig. 2 are comparable to the sizes of the contact regions arising in stage B. The small contact regions formed in stage B can associate further in stage C in response to longer-range interactions, with possible small rearrangements of the structures formed in stages A and B. These structural features in stages A–C are reflected in the contact map of the native protein, i.e., the contacts close to the diagonal are those formed in stage A, and the medium- and long-range interactions are reflected in the contacts of stages B and C, respectively, as the distance of the contact region from the diagonal increases.
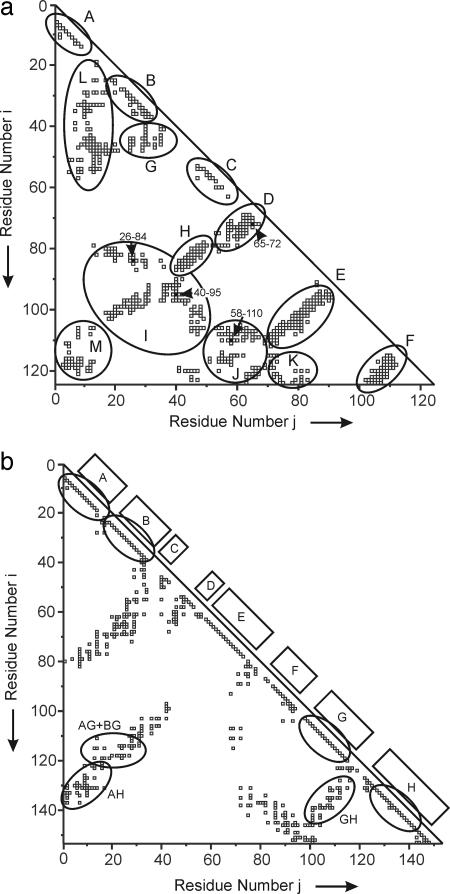
Fig. 3.
Triangular contact maps. (a) Contact map of RNase A constructed by the method of ref. 8. Each point of the map represents the presence (square) or absence (no marking) of a contact between two amino acid residues i and j. A contact is defined according to ref. 12. Contacts between residues are omitted whenever |i − j| ≤ 4. The pairs of half-cystine residues forming the disulfide bridges are denoted by black squares. The elliptical traces are the boundaries of contact regions A-M. (b) Contact map of apomyoglobin, constructed by the method of ref. 8.
Experimental Verification of Models
The first approach (3) identified the primary initiation sites in 14 proteins, including bovine pancreatic RNase A and sperm whale myoglobin, and also other less-stable initiation sites (11) in RNase A. The primary initiation site for RNase A was identified in this model as residues 106–118. Application of the second approach (8) to RNase A (13) led to six sites (illustrated in Fig. 3a), which were consistent with those identified (11) by the first approach (3). These are residues 4–11 (region A), residues 25–34 (region B), residues 51–57 (region C), and antiparallel structures (not necessarily pleated sheets) formed by the association of residues 53–67 with residues 69–79 (region D), residues 71–90 with residues 91–111 (region E), and residues 103–111 with residues 115–124 (region F). The subsequent steps along the folding pathway consist of the growth or coalescence of these regions. Region G is formed when residues 36–48 fold against helix B. Region G, however, contains longer-range contacts than does helix B. Therefore, G is considered to form at a later stage than B. Region L is formed by association of region A with B, G, and C. This structural sequence is followed by region H, formed when regions C and D come into contact; by region K containing contacts between regions E and F; by region J in which regions C and D form contacts with regions E and F; and by region I containing contacts of regions E and H with G. Finally, region M is formed when region A comes into contact with E and F.
The predicted primary folding initiation site of RNase A is in agreement with the observed turn at residues 113 and 114 in the native molecule, with the very high degree of conservation of nonpolar residues in site 106–118 of 23 mammalian species of this protein (3), and with experimental information on intermediates detected in the thermal unfolding of RNase A (14). NMR evidence for local structure in unfolded RNase A under folding conditions (15) and in fragments thereof (16), and immunochemical studies on the formation of stable antigenic sites (on the surface of the protein, covering the buried primary hydrophobic initiation site when reduced RNase A is oxidized (ref. 17 and figure 8 of ref. 18), were also consistent with this picture. Other experimental evidence supporting the existence of initiation sites A-F is provided in ref. 11.
For apomyoglobin, the first approach (3) identified the primary folding initiation site as residues 103–115, which encompasses the major part of the G helix, with the sequence Tyr-Leu-Glu-Phe-Ile-Ser-Glu-Ala-Ile-Ile-His-Val-Leu. Application of the second approach (8), and conformational-energy calculations, led to the suggestion that interactions among the A, G, and H helices of apomyoglobin might be important for folding initiation (19), consistent with experimental results that implicated these regions in equilibrium intermediates of apomyoglobin (20) and in the earliest kinetic steps in the folding pathway (21). Recent experimental studies of mutants of myoglobin (22, 23) provide convincing support for models that incorporate hydrophobic initiation and propagation of folding.
Apomyoglobin has become a paradigm for research into folding pathways (24). Part of the apomyoglobin molecule, consisting of the A, G, and H helices and part of the B helix, folds rapidly to form an on-pathway intermediate, with the remainder of the polypeptide chain folding at a slower rate, of the order of seconds (21, 25). Peptide fragments of apomyoglobin corresponding to the G and H helices showed a propensity for helical structure (26–28), whereas peptides corresponding to the remainder of the protein showed far lower propensity for helix formation (29). Yet an experiment in which the H helix of myoglobin was mutated to decrease its helical propensity (30) showed very little influence on the folding rate of the protein. A much greater effect was observed when the distal histidine (H64) was changed to phenylalanine: the substitution of a nonpolar side chain for the buried polar histidine resulted in a significant increase in the rate of folding (31). These studies pointed toward the importance of side-chain packing, particularly in the contact surfaces of helices, for the rate of folding of apomyoglobin.
Apomyoglobin has the advantage that it can be studied at equilibrium under a number of solution conditions that approximate some of the observable stages on the kinetic folding pathway. Thus, it has been possible to define the conformational propensities of unfolded and partly folded apomyoglobin by NMR (32–35). Interestingly, acid-unfolded apomyoglobin shows a propensity for non-native helical structure in a region that connects the D and E helices, as well as the native-like structure observed in the A and H helices, which had been predicted in the kinetic studies (34). The NMR studies of the conformational propensities of the various forms of apomyoglobin (32–35) included determination of the relaxation parameters of the polypeptide chain to site-specifically determine the backbone dynamics in each of the states of the protein. Instead of the “model-free” analysis that is suitable for folded proteins, the relaxation data were analyzed by calculating reduced spectral density functions J(0), J(ωN), and J(ωH) (36). The function J(0) informs on μs–ms time-scale motions and suggested that, for apomyoglobin at pH 2.3, the A and G helices were making transient contacts (34). This exciting result, later validated by spin-label NMR studies (37), provided definitive evidence of native-like long-range contacts in an unfolded protein. Even more intriguingly, the function J(ωN), which informs on ps–ns time-scale motion, showed sequence-dependent variation, which could be correlated with variation in a calculated parameter, the “average area buried upon folding” (AABUF) (10). This parameter, which incorporates both residue size and hydrophobicity, is defined for individual amino acids, but is usually averaged over a window of five to nine residues in the sequence and provides an alternative quantitative estimate of the hydrophobicity defined by Matheson and Scheraga (3) in terms of free energy of formation. In the case of apomyoglobin at pH 2.3, peaks in the plot of J(ωN) vs. residue number, which indicate restriction of the sub-ns time-scale motion, corresponded extremely well with peaks in the AABUF plot, which indicate a higher than average number of large and/or nonpolar amino acids. This observation suggested that there is a pattern in the amino acid sequence, defined by groupings of large and/or nonpolar side chains, as in the model of ref. 3, that leads to restriction of the motion of the polypeptide backbone caused by local hydrophobic cluster formation.
Apomyoglobin at pH 2.3 retains a small propensity for helical structure in the A and H helices, as judged by the observed NMR chemical shifts (34). These propensities are abolished in the presence of 8 M urea (35), yet the sequence dependence of the relaxation rates of urea-denatured apomyoglobin also shows an intriguing correlation with the AABUF parameter. In this case, the correlation is between minima in J(ωH) and maxima of AABUF, suggesting that local hydrophobic interactions that restrict backbone motion on sub-ns time scales are persistent even in 8 M urea.
To test the hypothesis that the AABUF parameter, i.e., hydrophobicity, could be used to predict the folding initiation sites in the polypeptide chain, a specific set of apomyoglobin mutations were made (23). The A helix, which participates in the kinetic intermediate that is first formed upon folding of WT apomyoglobin, has a sequence that gives rise to a maximum in the AABUF parameter, whereas the E helix, which is in fact highly hydrophobic according to conventional hydrophobicity scales, has a rather low AABUF throughout its length. In folded apomyoglobin, the side chain of Trp-14, in the A helix, lies opposite the backbone of the E helix at Gly-73. A double-mutant myoglobin was prepared, in which Trp-14 was replaced with Gly, and Gly-73 was replaced with Trp. It was reasoned that the folded structure of the protein should be minimally perturbed by this swap, but that the AABUF parameter for the A and E helices should be drastically affected. The question was whether the folding pathway would also be affected. Indeed, folding of the mutant in which G73 and W14 are swapped occurs by a different pathway than the WT protein. All apomyoglobin variants that have so far been studied fold via a burst phase intermediate that contains helical structure. In the case of WT apomyoglobin, this intermediate contains the A, G, and H helices (and also part of the B helix). In the G73–W14 swap mutant, the intermediate contains the E, G, and H helices; the A helix is not protected in the initial phase of folding, but folds later. This result (illustrated in Fig. 4) was confirmed by spin-label studies showing contacts between the E, G, and H helices in the mutant, rather than the A, G, and H contacts observed in the WT protein.
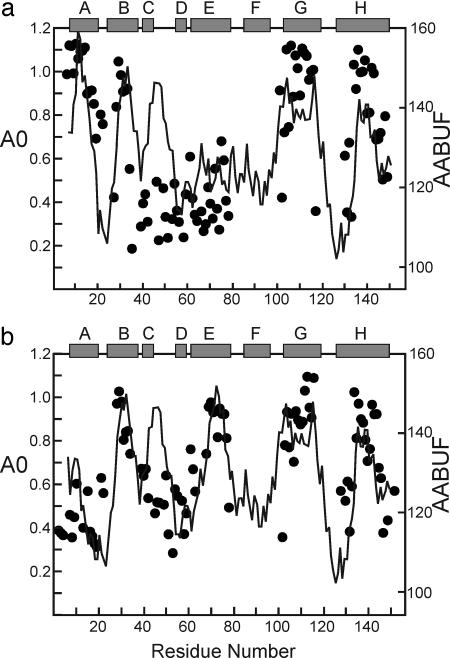
Fig. 4.
Extrapolated proton occupancy A0 for the burst phase intermediate, calculated as described (25), plotted as a function of residue number (black circles), and AABUF, averaged over a nine-residue moving window (gray trace). (a) WT apomyoglobin. (b) Quadruple mutant Leu-11 → Gly, Trp-14 → Gly, Ala-71 → Leu, Gly-73 → Trp. [Reproduced with permission from ref. 23 (Copyright 2005, Elsevier).]
Discussion
These experiments validate the apparent connection between the empirical AABUF parameter (10) [or hydrophobicity of Matheson and Scheraga (3)] and those parts of the polypeptide chain that fold first. Thus, if it is accepted that the amino acid sequence alone is capable of folding reliably into the correct 3D structure, then it appears that the best predictor of the sites of initiation of this process is the local pattern of amino acids that are ultimately most likely to be buried in the interior of the protein. This mechanism does make sense, but the point has been obscured somewhat in the past by the accepted ideas of hydrophobicity. As pointed out above, amino acids such as lysine have not traditionally been classed as nonpolar, yet they are frequently found in the interior of proteins. As long as the ε-NH3+ group is neutralized by hydrogen bonding or salt-bridge formation, the lysine side chain may be considered as nonpolar as methionine, and more so than leucine or isoleucine, whose branched side chains may not be ideal for optimum hydrophobic packing.
For apomyoglobin, the simple paradigm that works for ribonuclease (primary initiation sites close to the diagonal, with contacts further from the diagonal formed later) is complicated by two factors. First, myoglobin is a heme protein, and the absence of the prosthetic group causes specific changes in the final, folded structure of the apoprotein, in particular the lack of a unique structure in the F helix, which forms a gate over the heme pocket. Second, apomyoglobin is an all-helix protein, which results in a lower density of contacts in the contact map of the folded protein, particularly in the region of the diagonal (compare Fig. 3 a and b). This explanation renders the sequence of events in the folding process much more fluid than those for ribonuclease: the differences in the propensity of the various helices to fold early, as part of the molten globule intermediate, or later during the observable folding phase, are quite small, and can rather easily be perturbed by single-site mutations such as those described above. Thus, although contact regions D and E appear near the diagonal of Fig. 3b, they are not observed to form in the initial folding stages. [Interestingly, a propensity for non-native helical structure is observed in the D/E helix region of the acid-unfolded state of apomyoglobin (34).] During the growth process, in which contacts are formed in the kinetic burst phase of the WT protein, medium- and long-range interactions come into play in the formation of the GH, AG + BG, and AH contact regions. These are further shown in Fig. 5 as a model of one of the earliest hydrophobic cores to be formed along the folding pathway (22, 23); this model confirms the predicted folding interaction site (3) at residues 103–115. In the GW mutants (23), replacement of the tryptophan side chain from the A helix to the E helix preserves these contacts in the final folded form of the protein, but alters the sequence of formation of helices that are solvent-protected in the first steps of the folding pathway.
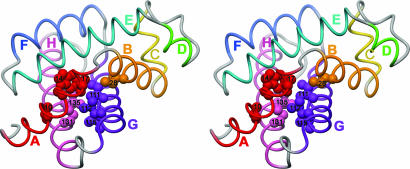
Fig. 5.
Stereoview of the backbone structure of sperm whale myoglobin (CO complex; Protein Data Bank ID code 1MBC) (38). Helices are labeled and colored red (A helix), orange (B helix), yellow (C helix), green (D helix), turquoise (E helix), blue (F helix), purple (G helix), and pink (H helix) with loop regions shown in gray. Heavy atoms of the side chains of some of the residues that participate in the earliest folded forms of WT apomyoglobin, corresponding to the elliptically traced contact regions in Fig. 3b, are shown as spheres, red for the A-helix residues Val-10, Trp-14, and Val-17; orange for the B-helix residue Ile-28; purple for the G-helix residues Ile-111, Ile-112, and Leu-115; and pink for the H-helix residues Met-131 and Leu-135, corresponding to the backbone color of the corresponding helix.
Although it is axiomatic that the amino acid sequence controls the kinetics and thermodynamics of formation of globular protein structure, it has not been clear in detail why some parts of the protein fold early in the process, whereas others fold only later on. The key to folding initiation was early recognized to be the presence of nonpolar amino acid side chains and the possibility of formation of hydrogen-bonded secondary structure to satisfy buried charges and polar backbone groups. A key concept in our present understanding of the initiation process was the recognition (3, 6, 10) that large polar or charged amino acid side chains could be included in the groups of hydrophobic amino acids, and could thus contribute to the hydrophobic stabilization of the initial folded structures, through interactions of the nonpolar CH2 groups between the backbone and the heteroatom functional groups. Experimental tests of this hypothesis (22, 23) have shown definitively that, at least in the case of the helical protein apomyoglobin, the earliest events in the folding of the protein can be re-engineered precisely by making simple changes in the amino acid sequence based on the positions of key nonpolar groups.
Abbreviation
- AABUF
average area buried upon folding
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Rose G. D., editor. Advances in Protein Chemistry. Vol. 62. San Diego: Academic; 2002. Unfolded Proteins. [Google Scholar]
- 2.Anfinsen C. B. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 3.Matheson R. R., Jr., Scheraga H. A. Macromolecules. 1978;11:819–829. [Google Scholar]
- 4.Yang A. S., Honig B. J. Mol. Biol. 1995;252:351–365. doi: 10.1006/jmbi.1995.0502. [DOI] [PubMed] [Google Scholar]
- 5.Yang A. S., Honig B. J. Mol. Biol. 1995;252:366–376. doi: 10.1006/jmbi.1995.0503. [DOI] [PubMed] [Google Scholar]
- 6.Némethy G., Steinberg I. Z., Scheraga H. A. Biopolymers. 1963;1:43–69. [Google Scholar]
- 7.Fernandez A., Scheraga H. A. Proc. Natl. Acad. Sci. USA. 2003;100:113–118. doi: 10.1073/pnas.0136888100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka S., Scheraga H. A. Macromolecules. 1977;10:291–304. doi: 10.1021/ma60056a015. [DOI] [PubMed] [Google Scholar]
- 9.Poland D. C., Scheraga H. A. Biopolymers. 1965;3:283–304. [Google Scholar]
- 10.Rose G. D., Roy S. Proc. Natl. Acad. Sci. USA. 1980;77:4643–4647. doi: 10.1073/pnas.77.8.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montelione G. T., Scheraga H. A. Acc. Chem. Res. 1989;22:70–76. [Google Scholar]
- 12.Tanaka S., Scheraga H. A. Proc. Natl. Acad. Sci. USA. 1975;72:3802–3806. doi: 10.1073/pnas.72.10.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Némethy G., Scheraga H. A. Proc. Natl. Acad. Sci. USA. 1979;76:6050–6054. doi: 10.1073/pnas.76.12.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matheson R. R., Jr., Scheraga H. A. Biochemistry. 1979;18:2437–2445. doi: 10.1021/bi00579a001. [DOI] [PubMed] [Google Scholar]
- 15.Swadesh J. K., Montelione G. T., Thannhauser T. W., Scheraga H. A. Proc. Natl. Acad. Sci. USA. 1984;81:4606–4610. doi: 10.1073/pnas.81.14.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montelione G. T., Arnold E., Meinwald Y. C., Stimson E. R., Denton J. B., Huang S.-G., Clardy J., Scheraga H. A. J. Am. Chem. Soc. 1984;106:7946–7958. and erratum (1985) 107, 1457. [Google Scholar]
- 17.Chavez L. G., Scheraga H. A. Biochemistry. 1977;16:1849–1856. doi: 10.1021/bi00628a014. [DOI] [PubMed] [Google Scholar]
- 18.Scheraga H. A. In: Protein Folding. Jaenicke R., editor. Amsterdam: Elsevier/North–Holland; 1980. pp. 261–288. [Google Scholar]
- 19.Gerritsen M., Chou K.-C., Némethy G., Scheraga H. A. Biopolymers. 1985;24:1271–1291. doi: 10.1002/bip.360240714. and erratum (1985) 24, 2177. [DOI] [PubMed] [Google Scholar]
- 20.Hughson F. M., Wright P. E., Baldwin R. L. Science. 1990;249:1544–1548. doi: 10.1126/science.2218495. [DOI] [PubMed] [Google Scholar]
- 21.Jennings P. A., Wright P. E. Science. 1993;262:892–896. doi: 10.1126/science.8235610. [DOI] [PubMed] [Google Scholar]
- 22.Nishimura C., Wright P. E., Dyson H. J. J. Mol. Biol. 2003;334:293–307. doi: 10.1016/j.jmb.2003.09.042. [DOI] [PubMed] [Google Scholar]
- 23.Nishimura C., Lietzow M. A., Dyson H. J., Wright P. E. J. Mol. Biol. 2005;351:383–392. doi: 10.1016/j.jmb.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 24.Wright P. E., Baldwin R. L. In: Mechanisms of Protein Folding. Pain R. H., editor. Oxford: Oxford Univ. Press; 2000. pp. 309–329. [Google Scholar]
- 25.Nishimura C., Dyson H. J., Wright P. E. J. Mol. Biol. 2002;322:483–489. doi: 10.1016/s0022-2836(02)00810-0. [DOI] [PubMed] [Google Scholar]
- 26.Waltho J. P., Feher V. A., Merutka G., Dyson H. J., Wright P. E. Biochemistry. 1993;32:6337–6347. doi: 10.1021/bi00076a006. [DOI] [PubMed] [Google Scholar]
- 27.Shin H.-C., Merutka G., Waltho J. P., Wright P. E., Dyson H. J. Biochemistry. 1993;32:6348–6355. doi: 10.1021/bi00076a007. [DOI] [PubMed] [Google Scholar]
- 28.Shin H.-C., Merutka G., Waltho J. P., Tennant L. L., Dyson H. J., Wright P. E. Biochemistry. 1993;32:6356–6364. doi: 10.1021/bi00076a008. [DOI] [PubMed] [Google Scholar]
- 29.Reymond M. T., Merutka G., Dyson H. J., Wright P. E. Protein Sci. 1997;6:706–716. doi: 10.1002/pro.5560060320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavagnero S., Dyson H. J., Wright P. E. J. Mol. Biol. 1999;285:269–282. doi: 10.1006/jmbi.1998.2273. [DOI] [PubMed] [Google Scholar]
- 31.Garcia C., Nishimura C., Cavagnero S., Dyson H. J., Wright P. E. Biochemistry. 2000;39:11227–11237. doi: 10.1021/bi0010266. [DOI] [PubMed] [Google Scholar]
- 32.Eliezer D., Yao J., Dyson H. J., Wright P. E. Nat. Struct. Biol. 1998;5:148–155. doi: 10.1038/nsb0298-148. [DOI] [PubMed] [Google Scholar]
- 33.Eliezer D., Chung J., Dyson H. J., Wright P. E. Biochemistry. 2000;39:2894–2901. doi: 10.1021/bi992545f. [DOI] [PubMed] [Google Scholar]
- 34.Yao J., Chung J., Eliezer D., Wright P. E., Dyson H. J. Biochemistry. 2001;40:3561–3571. doi: 10.1021/bi002776i. [DOI] [PubMed] [Google Scholar]
- 35.Schwarzinger S., Wright P. E., Dyson H. J. Biochemistry. 2002;41:12681–12686. doi: 10.1021/bi020381o. [DOI] [PubMed] [Google Scholar]
- 36.Farrow N. A., Zhang O., Szabo A., Torchia D. A., Kay L. E. J. Biomol. NMR. 1995;6:153–162. doi: 10.1007/BF00211779. [DOI] [PubMed] [Google Scholar]
- 37.Lietzow M. A., Jamin M., Dyson H. J., Wright P. E. J. Mol. Biol. 2002;322:655–662. doi: 10.1016/s0022-2836(02)00847-1. [DOI] [PubMed] [Google Scholar]
- 38.Kuriyan J., Wilz S., Karplus M., Petsko G. A. J. Mol. Biol. 1986;192:133–154. doi: 10.1016/0022-2836(86)90470-5. [DOI] [PubMed] [Google Scholar]