Abstract
We develop a symbiogenetic concept of the origin of eukaryotic intracellular motility systems from anaerobic but aerotolerant spirochetes in sulfide-rich environments. The last eukaryotic common ancestors (LECAs) have extant archaeprotist descendants: motile nucleated cells with Embden-Meyerhof glycolysis and substrate-level phosphorylation that lack the α-proteobacterial symbiont that became the mitochondrion. Swimming and regulated O2-tolerance via sulfide oxidation already had been acquired by sulfidogenic wall-less archaebacteria (thermoplasmas) after aerotolerant cytoplasmic-tubule-containing spirochetes (eubacteria) attached to them. Increasing stability of sulfide-oxidizing/sulfur-reducing consortia analogous to extant sulfur syntrophies (Thiodendron) led to fusion. The eubacteria–archaebacteria symbiosis became permanent as the nucleus evolved by prokaryotic recombination with membrane hypertrophy, analogous to Gemmata obscuriglobus and other δ-proteobacteria with membrane-bounded nucleoids. Histone-coated DNA, protein-synthetic RNAs, amino-acylating, and other enzymes were contributed by the sulfidogen whereas most intracellular motility derives from the spirochete. From this redox syntrophy in anoxic and microoxic Proterozoic habitats LECA evolved. The nucleus originated by recombination of eu- and archaebacterial DNA that remained attached to eubacterial motility structures and became the microtubular cytoskeleton, including the mitotic apparatus. Direct LECA descendants include free-living archaeprotists in anoxic environments: archamoebae, metamonads, parabasalids, and some mammalian symbionts with mitosomes. LECA later acquired the fully aerobic Krebs cycle-oxidative phosphorylation-mitochondrial metabolism by integration of the protomitochondrion, a third α-proteobacterial symbiont from which the ancestors to most protoctists, all fungi, plants, and animals evolved. Secondarily anaerobic eukaryotes descended from LECA after integration of this oxygen-respiring eubacterium. Explanatory power and experimental predictions for molecular biology of the LECA concept are stated.
Keywords: karyomastigont, kinetosome-centriole, mitotic apparatus, nucleus origin, Thiodendron
The cytoskeleton in all eukaryotes comprises the mitotic spindle (often its kinetosome-centrioles within the centrosome), protist karyomastigonts (1), and neurotubules; the kinetome of 10,000 species of ciliates; microtubules of foram granuloreticulopodia; submembranous microtubules of red blood cells, trypanosomes, and many algae; and the undulipodium (cilium-eukaryotic “flagellum” or intrinsically motile 0.25-μm-diameter ninefold symmetrical intracellular “whip” generated from a kinetosome-centriole base). “This later structure is a highly complex organelle, composed of hundreds of proteins. It has no bacterial homolog, and undoubtedly evolved into its current form after the evolution of microtubules. However, it is a remarkably standardized organelle across diverse eukaryotic taxa, which indicates that it evolved once in the early evolution of eukaryotes” (2).
Movement inside cells, limited to eukaryotes, involves three classes of structures: actin filaments (6–8 nm in diameter) associated with myosin-ATPases, intermediate filaments (12–14 nm in diameter), and microtubules (24 nm) with microtubule-associated proteins (MAPs) (dynein, kinesin, and other ATP- or GTPases). We detail only the microtubular cytoskeleton, but a complete account of cell evolution must consider other motile proteins. Three possibilities for the origin of the cytoskeleton include (i) ancient origin from our genetically-not-yet-annealed ancestors (3), (ii) direct filiation from a prokaryotic ancestor, or (iii) symbiotic addition to a different eukaryotic ancestor. Lack of precision of “genetically-not-yet-annealed” precludes investigation of the first. Continuous reevaluation of the assumed second reveals slim evidence and terminology misused (2). Even the accepted concept of FtsZ (filament temperature-sensitive Z protein) as prokaryotic ancestor of tubulin protein is debatable (2). Symbiogenetic origin of intracellular motility including mitosis, via addition of Spirochaeta to a Thermoplasma-like archaebacterium (4–7), our concept, unlike other alternatives, explains superficially unrelated phenomena and generates details studiable by genomic, proteomic, cytologic, and geologic methods.
Results and Discussion
Our evolutionary scenario, developed independently of molecular sequence data, unlike others (8–10) reconstructs evolutionary history from extant organisms (Fig. 1). Taken as representative clues, many of the steps have been videographed in live microbes. We show how both claims that no eukaryote “primitively lacked mitochondria” and “the evolutionary gap between prokaryotes and eukaryotes is now deeper, and the nature of the host that acquired the mitochondrion more obscure, than ever before” are based on protistological ignorance but rely on the kind of molecular data that will support or disprove our model.
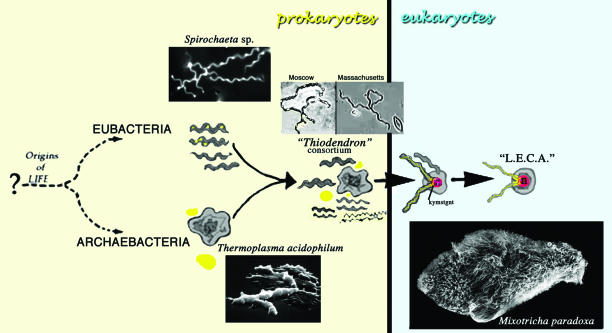
Fig. 1.
Karyomastigont model of origin of nucleated cells. The LECA evolved from eubacterial-archaebacterial syntrophies in which sulfide-oxidizing spirochetes attached to sulfidogenic thermoplasmas to form a “Thiodendron”-like consortium. Archaeprotist (trichomonad Mixotricha paradoxa, lower right), LECA analogue in termite Mastotermes darwiniensis, swims via motility symbiosis with 200,000 Treponema sp. surface spirochetes. Four distinctive surface spirochetes are detected by morphological and molecular techniques (36). Chromatin appears first in the karyomastigont (“kymstgnt”), the precursor cytoskeletal organellar system from which the tethered nucleus (“n”) was released.
New results mandate that aspects of our putative evolutionary (chronological) karyomastigont model be reconsidered and the only propositions disputed in scientific literature [(i) symbiotic origin of archaebacterial cytoplasm from sulfidogens, not methanogens, and (ii) nucleus-associated microtubule cytoskeleton preceded acquisition of mitochondria] be defended.
Hydrogen sulfide (H2S)-oxygen (O2) interface habitats abounded during the Proterozoic Eon (2,500–541 mya) throughout the world ocean as inferred from sedimentary, isotopic, and other geologic observation (11, 12). Worldwide localities show preservation of ancient eukaryotes mostly as microfossils in thin-section of cherty rock; nucleated cells evolved before 1,000 mya in sulfidic, muddy habitats periodically well lit, dominated by cyanobacteria and therefore rich in organics. Spirochetes, an ancient cohesive group of helical, motile chemoheterotrophic eubacteria, had evolved, in response to cyanobacterial oxygenesis, from their strictly anaerobic origin through aerotolerant-facultative aerobes, to microaerophils like Leptospira (13, 14). Spirochetes, active chemotactic swimmers, thrived in marine microbial mats, pond scums, and fresh and saline hot springs, where many, as now, lived syntrophically.
-
The ancestors to which Spirochaeta were added were Thermoplasma-like sulfidogenic archaebacteria. This postulate, critical to our model, was argued by Searcy (15–21) (Table 1). Thermoplasma acidophilum, heat- and acid-tolerant pleiomorphs that lack cell walls, are amenable to study as co-descendants of conserved archaebacterial features in eukaryotes. Thermoplasma are predicted to enter metabolic associations more easily than walled relatives. Their DNA, unlike most prokaryotes, is protected from hot acid by “nucleosomes,” a coating of arginine- and lysine-rich histone-like proteins (20, 21), and thus, thermoplasmas in nature may constitute living refugia from the hydrolytic environmental habitat extremes in which they grow. Like eukaryotic cytoplasm Thermoplasma metabolizes glucose to pyruvate or lactate via Embden-Meyerhof enzymes anaerobically, but because its terminal electron acceptor is elemental sulfur (only oxygen at microoxic, <5% ambient concentrations), it tends to be sulfidogenic. Volcanoes, fumaroles, hot springs, and burning coal piles replete with elemental sulfur, H2S, or sulfide ion (HS−) provided Proterozoic sulfatara and sulfureta environmental selection pressures (high sulfur concentration, heat, low pH, and cyclically high toxic oxygen gas and organic food from cyanobacteria). These led hungry oxygen-intolerant spirochetes to cohabit and attach to sulfidogenic thermoplasmas. The merger of incessantly motile spirochetes with sluggish acid-mediating thermoplasmas formed the earliest protists: amitochondriate, anaerobic motile nucleated cells with low levels of oxygen tolerance and utilization.
Spirochetes detoxified fluctuating ambient gaseous oxygen by its conversion with sulfide to elemental sulfur usable by Thermoplasma as terminal electron acceptor. Conferrence of motility on this redox syntrophy permitted both partners access to organic-rich habitats that augmented survival and growth. Modern descendants of syntrophic consortia became archaeprotists, motile premitochondriates amenable to study. The Archaeprotista phylum, members riddled with endo-, epi-, and even nuclear symbionts (22), includes the archamoebae (pelomyxids, mastigoamoebae), metamonads Retortamonas and Giardia, and parabasalids such as trichomonads, devescovinids, hypermastigotes, and calonymphids (23, 24). Distinctive features of eukaryotic cells (e.g., archaebacterial-like transcription and translation in protein synthesis, histone-like proteins of nucleosomes and chromatin, glucose catabolism, sulfide generation, ATP- and GTP-based substrate-level energy transformation, details of ion regulation, membrane transport, and mechano-, chemo-, and photoreception), although recombined and refined, have been conserved such that their history can be reconstructed from ultrastructural, physiological, biochemical, and molecular data.
The earliest symbiogenetic fusion that integrated thermoacidophilic archaebacterial thermoplasmas with aerotolerant spirochetes produced the first protist, a swimming chimera that evolved into a stable nucleated protist cell: the last eukaryotic common ancestor, LECA. The nucleus in the karyomastigont was generated by recombination of eu- and archaebacterial DNA that remained attached to membrane and to spirochete motility proteins. This three-part microtubular-mitotic apparatus-cytoskeleton-organellar system, the karyomastigont includes: (i) undulipodium connected to the (ii) nucleus by (iii) nuclear connector (rhizoplast). A “parabasal body” (= golgi) is often also associated. Many forms of motility including cytoskeletal-based ingestion originated from the fusion. The eukaryotic cell née motility-sulfur syntrophy was selected for during the Archean or lower Proterozoic Eon by variables that fluctuated diurnally and seasonally: temperature, light, water abundance, pH, salinity, organics, and oxygen in chemically reduced, sulfide-rich anoxic habitats. The mitotic nucleus, evolved as part of the karyomastigont, preceded obligate aerobiosis because the acquisition of oxygen respiration occurred only after the endosymbiotic α-proteobacterium became the mitochondrion. Eukaryotic-style evolution where entire genomes are ingestible preceded both the symbiotic acquisition of mitochondria-mediated aerobiosis and cyanobacteria that conferred photosynthesis on algal and plant ancestors.
Origin of the cytoskeletal intracellular motility requires so many coordinated genes and biochemical activities that it can only be explained by symbiogenesis. Motile phenomena, visible in live cells by light microscopy, include undulipodial beating; “raft” transport along ciliary axonemes; chromosome movement by MAP motors on spindle microtubules; nuclear rotation; cyclosis, exocytosis, endocytosis, phagocytosis, and other particle translocation; evacuation of vacuoles, formation of vesicles, and cell process (pseudopod, axopod, nerve growth cone) extension; and fusion of cells in fertilization, myogenesis, and myriad other locomotion exclusive to eukaryotes.
The formation and function of the kinetosome-centriole-based undulipodium requires >360 different proteins (25). By contrast, large prokaryotic cells (e.g., Lyngbya, Thiomargarita, and gliding myxobacteria) show no directed internal motility at highest magnifications (×1,000) with differential interference or phase contrast microscopy. Sexual life history cycles require syngamic or karyogamic intracellular movement followed by meiosis. Microtubules and MAPs are so intrinsic to the eukaryotes that any hypothesis for the origin of the nucleus is fatally deficient unless it includes an account of the cytoskeleton (2).
Histone orthologues have been isolated from methanogens and other archaebacteria (26). However, the paucity of relict methanogenesis or its coenzyme biochemistry requires rejection of methanogens in eukaryotic ancestry. Methanogenesis, like oxygenic photosynthesis, is correlated with secondary acquisition of symbiotic bacteria in anoxic environments (27, 28). The absence of primary methanogenesis in anaerobic protists contrasts strikingly with the widespread, if not universal, detection of cytoplasmic sulfidogenesis (17) in all four eukaryotic lineages [protoctists, plants, fungi, and animals including human erythrocytes (16)] consistent with the concept that sulfidogenesis was introduced by the Thermoplasma archaebacterium symbiont (19).
Table 1.
Microtubular semes and their selective advantage
| Seme and its source | Putative selective advantage |
|---|---|
Condensation reaction of acetate (carboxylic acid, from Spirochaeta fermentation) and thiol to form thioester [R1 C( C( O)S O)S R2] from Thermoplasma R2] from Thermoplasma
|
Soluble energy transfer (14 kC/g molecule) and proton (H+) generator as source of ATP via pyrophosphate phosphorylation (50) |
| Karyomastigont: kinetosome-centriole from spirochete attachment structure connected by protein (nuclear connector, rhizoplast) to recombined nucleic acid of syntrophic eubacterial-archaebacterial partners | Swimming, other cell locomotion; chemo- and mechanosensitivity in feeding and defense; assurance of joint heritability of merged symbiont genomes |
| Nucleus (from syntrophic-motility integration) permanent metabolic and genetic integration of symbionts: sulfide-oxidizing Spirochaeta and sulfidogenic Thermoplasma | Sulfide oxidation by Spirochaeta for oxygen-hydrogen peroxide detoxification produces elemental sulfur for Thermoplasma’s terminal electron acceptor. Endomembrane system to distribute ion channels (ER, golgi). Energy generation by aerotolerant glycolysis to acetyl CoA, hydrogen, and oxidizable 3-C compounds (pre-adaptation for acquisition of mitochondrial ancestors) |
| Nuclear membrane (from motile syntrophic partnership integration) by fusion and proliferation of composite endomembrane system | Insure joint stable and permanent integration followed by segregation of symbiont DNA (Spirochaeta + Thermoplasma) each generation to produce offspring |
| Phagocytosis, cyclosis, pinocytosis, endocytosis, exocytosis (from spirochete innards) particulate feeding (bacteriovory), intracellular motility and transport, facultative fertilization, and other forms of cell fusion | Feeding, cell “drinking,” locomotion, prerequisite for mitotic reproduction and meiotic sexuality, complex sexual life histories, neuron-based sensory systems, ciliated retinal, olfactory, and auditory epithelial cells, and other eukaryotic features that require cytoskeleton |
| Synaptonemal complex (from bacterial conjugation protein structures) | Enzymatic and gene redundancy reduction, accurate recombination that guarantees euploidy: precursor to plant and animal tissue differentiation |
Mitochondria Evolved After Nuclei.
That hundreds of species of anaerobic eukaryotes lost mitochondria on return to anoxic habitats and/or that all eukaryotic lineages began with hydrogenosome-mitochondrial ancestry is more assumption than conclusion based on evidence (9).
“The genomes of E. [Entamoeba] histolytica and the amitochondrial protist pathogens Giardia lamblia and Trichomonas vaginalis share several metabolic adaptations. These include reduced or eliminated mitochondrial metabolic pathways. Indeed the genome data are consistent with the lack of a mitochondrial genome. Tricarboxylic acid cycle and mitochondrial electron transport chain enzymes are lacking …” (29).
Yet many kinds of eubacterial genes are known in anaerobic eukaryotes (30, 31). The best available comparative genomic analysis suggests that mitochondria, hydrogenosomes, and related organelles evolved independently several times in various anaerobic ancestral eukaryotic lineages through evolutionary tinkering (32). The claim of Martin and Koonin (10) that the nucleus evolved in response to symbiotic acquisition of the α-proteobacterial protomitochondrion ignores cytoskeletal data, protoctist biology, and comparative organellar genetics. The abundance of introns in α-proteobacterial relative to other eubacterial DNA required separation of the genome from the cytoplasm. But following the logic of those authors, initial acquisition of this intron-rich DNA would have been lethal to the cell (10).
So-called mitochondria “relict genes” in anaerobic protists (Trichomonas, Giardia) do not involve direct use of oxygen gas, the cytochrome a/a3 (cytochrome-oxidase) terminal electron transport or other enzymes unique to mitochondria. The speciose amitochondriate protist taxa (i.e., trichomonads, calonymphids, and hypermastigotes) have no mitochondriate relatives; it is more likely they are descendants of Proterozoic evolution before the acquisition of protomitochondria than that all evidence of mitochondria has disappeared. Many, if not all eubacterial enzymes asserted to derive from relict mitochondria are common to eubacteria. Loss and/or dedifferentiation of mitochondria in isolated species is well known: the rumen protist Neocallimastix, a chytrid; plagiopylid ciliates with stripped bodies in the cytoplasm; and many sapropel ciliates (27, 28). Some fungi (glucose-repressed yeast) and marine worms have dedifferentiated mitochondria interpretable as legacies of local evolution: nearly all fungi and animals are mitochondriates in oxic habitats. Secondarily amitochondriate taxa are analogous to nongreen plants (e.g., Monotropa, Cuscuta, and Corallorhiza) whose cells contain proplastids or chromoplasts derived from plastidic ancestors. Nongreen plants, isolated rarities in the overwhelmingly green Kingdom Plantae, evolved where specialized habitat led to chloroplast loss. In stark contrast, hundreds of protists including archaeamoebae (e.g., Pelomyxa and Mastigoamoeba), metamonads (Retortamonas and Hexamitus) and parabasalids (Family Devescovinidae: Devescovina, Mixotricha, Oxymonas, Pyrsonympha, and Trichomonas; the Calonymphids, e.g., Coronympha and Stephanonympha), Snyderella and all hypermastigotes (including genera such as Barbulanympha, Staurojoenina, and Trichonympha), lack mitochondrial relict organelles and mitochondriate relatives (33).
Epi-, endo-, and intranuclear bacterial symbionts of protists tend to be eubacterial Gram-negative rods and coccoids. Whereas genus identification is rarely available, distinctive eubacteria regularly adorn the same anaerobic protist cell. In Caduceia versatilis, a devescovinid, more than four different bacterial types, including intranuclear, were detected (34); in the giant trichomonad, Mixotricha paradoxa, at least six different eubacteria were verified in Radek’s electron microscopic analysis (35, 36), and more than seven were detected with molecular techniques (36). Staurojoenina associates with at least three distinguishable bacterial symbionts (37). Study of protists in low-oxygen insect tissue led Kirby to his masterpiece on bacteria living inside and on protists (38). Pervasive bacterial associations, including ones mistaken for mitochondria, is marked under anoxic relative to oxic conditions. Kirby’s observations have been confirmed by transmission electron microscopy (39, 40).
Only if uniquely mitochondrial (not general eubacterial) genes are proved present in protists with taxon-specific cytoskeletons in which all species live anaerobically in anoxia and lack mitochondria at all stages must we reevaluate the archaeprotist concept. We explain the eubacterial enzymes [acetate kinase, phosphoglucomutase, NADH oxidase, and NADH peroxidase (41)] as legacy of the aerotolerant spirochete not from the protomitochondrion.
Increasingly stable sulfide-oxidizing/sulfur-reducing consortia, analogous to “Thiodendron” syntrophies, were precursors to the LECA. As the Thermoplasma/Spirochaeta syntrophy became permanent, the nucleus evolved by prokaryotic recombination and intracellular membrane hypertrophy that led to a membrane-bounded nucleus analogous to the nucleoid of Gemmata obscuriglobus (42).
Inadequacy of Other Nuclear Origin Hypotheses.
Three other symbiogenetic origin-of-eukaryotic cell hypotheses include the following: (i) the methanogen syntrophy (43), which postulates an original methanogen-proteobacteria symbiosis under anoxic conditions before mitochondria acquisition; (ii) the hydrogen hypothesis (9), where the nucleus and mitochondria originated concurrently—facultatively anaerobic mitochondrial metabolism provides the hydrogen (H2) and carbon dioxide (CO2) for methanogenic syntrophy; and (iii) overwhelming evidence, nearly exclusively drawn from amino acid residue sequence comparisons in many proteins, supports the chimera hypothesis—eukaryotes simultaneously have archaebacterial and eubacterial ancestors (44, 45). Sequence analysis in anaerobes (Giardia, Trichomonas) suggests to Gupta that some nucleated lineages never acquired mitochondria, but he fails to specify which genera represent archae- and eubacteria that formed LECA. He depicts the endoplasmic reticulum origin by membrane motility yet postulates no origin for the intracellular capacity required for endocytotic in-folding. Even Rizzotti (46), who understands the importance of the cytoskeleton in his “cilium from peduncle” hypothesis, fails to account for the nuclear membrane, other endomembranes, or sulfidogenesis. Unlike the karyomastigont model (7), no previous hypothesis was documented by videography (35).
Semes.
Although frequently inexplicit about methods, “traits in common” are used to reconstruct phyletic lineages by great evolutionists, e.g., Darwin, Haeckel, and Mayr (47). This is seme analysis (48) as used by Brogniart and many paleobotanists (49), Romer, Simpson, and other paleontologists. Semes, always determined by more than a single or even a few genes, are characteristics of clear selective advantage in given environments at specified times (tabulated in ref. 24, p. 106). A single mutation may cause a seme loss (of photosynthesis in plants or vision in cave animals) but never a gain. Semes, in order of evolution, include the following: heat-resistant spores, magnetosomes, dinitrogen gas fixation, oxygenic photosynthesis, cellulosic cell walls, desmosomes, actomyosin muscles, chitinous exoskeletons, amniote eggs, feathers, and speech. The karyomastigont, a seme that preadapted eukaryotes for mitosis, generated a descendant neoseme: the microtubular ninefold symmetrical shaft [9 (2) + 2] of the undulipodium that grows from a [9 (3) + 0] kinetosome-centriole. Selective advantages of semes from the symbiogenetic Spirochaeta/Thermoplasma syntrophy fusion are listed in evolutionary order (Table 1). The earliest probably is substrate-level phosphorylation that provided electron transfer and ATP for biosynthesis and motility since the beginning of the Archean Eon’s anoxic world (50).
Karyomastigont Evolved from Attached Symbiotic Aerotolerant Spirochetes.
The karyomastigont we place in its evolutionary context (4) was described in 1915 (51). Because a review of our model that emphasizes our protistological predecessors (H. J. Kirby, Jr., and L. R. Cleveland) appeared in Paleobiology [dedicated to S. J. Gould (7)], only newer work is presented here. Examples of explanatory power and experimental predictions of superficially unrelated phenomena include the fact that epitopes of gamma tubulin and a scleroderma antigen (anti-pericentrin serum) localized in the rotary motor zone in the archaeprotist (e.g., Caduceia versatilis) (5, 23).
Isolation of a filamentous bacterium from an abundant white sulfurous slime in marine coastal habitats associated with Fucus (rockweed, brown algae) led Perfiliev (52) to introduce the genus Thiodendron latens (“lazy sulfur tree”) to bacteriology. Thirty-five years of study by Dubinina (53, 54) proved that what Perfiliev had identified as a single bacterium with alternating motile-by-flagella with unicellular-stringy filamentous sulfur-ridden life-history stages was a syntrophic consortium. “Thiodendron” evolved convergently in at least six aquatic locations; it is a heterotrophic sulfate-reducing sulfidogen associated with a sulfur-oxidizing spirochete. The sulfate-reducing physiology suggested the spirochete’s partner was a Desulfovibrio. Further work identified at least two new genera (i.e., Desulfobacter and Dethiosulfovibrio) as the sulfidogens (41). In all “Thiodendrons” studied, the spirochete partners, based on swimming behavior, electron microscopic morphology, metabolism, and 16S rRNA, classify as Spirochaeta sp. However, the strains of Spirochaeta from distant sites (e.g., the White Sea, two Pacific Islands, and Moscow Starayiya hot springs) differ in detail including 16S rRNA (5), which supports convergent origins of the partnership.
Dubinina’s work illuminates our karyomastigont model by provision of analogous spirochete syntrophies for field and laboratory investigation. The hypothetical syntrophic Spirochaeta sp. are physiologically identical to Dubinina’s aerotolerant ones: substrate-level phosphorylation generated by glycolysis pathways was enhanced by ambient oxygen in the spirochetes with their minimal oxygen metabolism—acetogenesis by pyruvate oxidation and exopolysaccharide production (41). The microxic, more efficient glucose oxidation preadapted spirochetes for association with Thermoplasma. However, the marine sulfidogens (Desulfobacter and Dethiosulfovibrio) (55) differ from hypothetical archaebacterial associates: Thermoplasma acidophilum (18) is found in fresher, hotter, and more acidic waters. Sulfide-sulfur redox metabolism was retained as intracellular physiological signal that enabled environmental expansion by LECA.
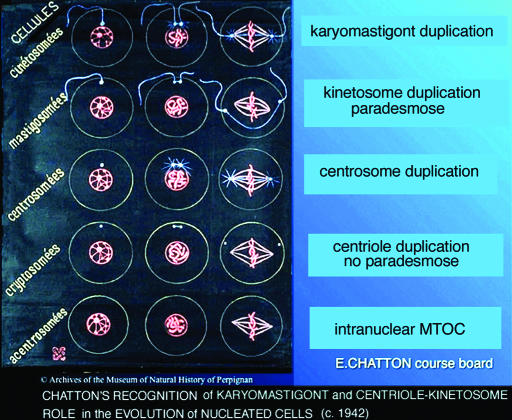
Homology of mitotic-microtubule variation was impossible to recognize until the development of glutaraldehyde fixatives after 1963, when by electron microscopy, microtubules became visible. Yet the prescient protistologist Edouard Chatton (56) anticipated this analysis. On “course boards” for students, he depicted cell evolution by centrosome-centriole-mitotic spindle morphology (“cellules cinetosomées” and “cellules mastigonemées”; Fig. 2). An expert marine protistologist and director of Laboratoire Arago, Banyuls sur Mer, France, he was first to tabulate all organisms as either “procariotíque” or “eucariotíque” (57, 58). His classification of “cells by centrosome behavior” genealogically organizes protoctist taxa (phyla, classes, and orders) in a valid manner consistent with our model.
Fig. 2.
Importance of the karyomastigont in the evolution of mitosis {Chatton’s 1938 course board (Left) Classification of cell types by the presence and localization of their centrosomes [Archives of the Museum of Natural History, Perpignan, France, bequest of André Lwoff (56)] corresponding to major taxa (Right)}. First row, mitosis including karyomastigont duplication, e.g., Chlamydomonas and Trypanosoma; second row, mitosis including “paradesmose” (pole-to-pole thin spindle) parabasalids—Trichomonas, devescovinids, and some hypermastigotes; third row, mitosis including centrosome duplication, animal cells; fourth row, mitosis includes duplication of intranuclear membrane-attached spindle-microtubule-organizing center (MTOC) of ciliates, red algae, conjugating green algae, and fungi (in ciliates and fungi with closed mitosis, the MTOC is attached to inner nuclear membrane); fifth row, acentrosomal mitosis typical of plants. [Reproduced with permission from Marie-Odile Soyer-Gobillard (56).]
The Test.
A definitive proof of our origin-of-the-nucleus hypothesis requires complete genome sequence comparison of appropriate Dubinina “Thiodendron”-Spirochaeta with other hypothetical prokaryotic-ancestor-of-eukaryote co-descendants. An aerotolerant spirochete the size of undulipodia (0.25-μm diameter by 10- to 14-μm length) that oxidizes sulfide to intracellular elemental sulfur globules [that contains, as does Hollandina (14, 59), 24-nm-diameter cytoplasmic tubules] is predicted to contain DNA and protein sequences with greater homology to genes that code for cytoskeletal nucleic acid and proteins (e.g., MAPs) than do other prokaryotes. The identification of unique centrosome-specific RNA molecules in Spisula surf clams (60) helps identify potentially relevant homologous sequences. Indeed, all 30 million species of eukaryotes should have retained cytoskeletal nucleic acid and protein (MAPs) sequences. Appropriate “control” genome comparisons, in addition to any arbitrarily chosen bacterium, should include thermoacidophilic archaebacteria (cytoplasm homologue), α-proteobacteria (mitochondrial homologue), and cyanobacteria (plastid homologue). The full oxidation of elemental sulfur to sulfate should correlate with later acquisition of the α-proteobacterium that became the mitochondrion. New techniques (e.g., genomics, proteomics, microbial physiology, geochronology, and geochemistry) are powerful enough to resolve this century-old evolutionary problem.
Materials and Methods
The methods, mostly traditional, are published but ignored by molecular and microbiologists, namely seme analysis (48). The unit of evolutionary analysis is the “seme”: newly gained “neosemes” and changed “aposemes”; increase or decrease in the number or size of an existing seme (hyper-/hyposeme, respectively) (48). Other methods include microbial ecological and laboratory techniques (53, 58), standard electron microscopy (23), the fluorescent immunocytology (5), and sulfidogen analysis by sulfidometer (16). To fill the largest evolutionary gap in the living world, we use knowledge of the organisms that bridged it: the unicellular eukaryotes in response to relevant environmental variables (fluctuating temperatures, salinities, pH, organic matter concentration, desiccation–rewetting cycles, and oxic-anoxic-sulfidic diurnal and seasonal variation). We attempt to recover an immense protistological literature: meticulous study of nucleated microbes in nature, their developmental life histories, and their morphological, cell biological, and ecological relationships.
Acknowledgments
Dr. Michael Dolan aided in all aspects. We are grateful to Mark Alliegro, Celeste Asikainen, David Bermudes, Christian de Duve, Johannes Hackstein, Susan Leschine, Harold J. Morowitz, Kenneth H. Nealson, Gemma Reguera, Dennis Searcy, Werner Schwemmler, Marie-Odile Soyer-Gobillard, Andrew Wier, and Elizabeth Stephens for critical aid in manuscript preparation. Andrew Wier, Dean Soulia, and Galena Dubinina helped supply the Spirochaeta, Mixotricha, and Thiodendron photographs in Fig. 1. We thank the Tauber Fund, Abraham Gomel, the University of Massachusetts at Amherst, and Alexander von Humboldt-Stiftung for financial support.
Abbreviations
- LECA
last eukaryotic common ancestor
- MAP
microtubule-associated protein.
Notes Added in Proof.
Dubinina’s aerotolerant spirochetes differ significantly enough from Spirochaeta to warrant a new genus description. The two articles (i) that place at least six strains of these spirochetes in a new genus named for B. V. Perfiliev and (ii) that report DNA sequence for one of them (White Sea strain) await the deposition of the pure cultures in two international culture collections (G. A. Dubinina, personal communication).
A first example of retention of microtubule-associated proteins (MAPs) has been published (61).
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Dolan M. F., Kirby H., Jr Eur. J. Protistol. 2002;38:73–81. [Google Scholar]
- 2.Dolan M. F. In: Microbial Phylogeny and Evolution: Concepts and Controversies. Sapp J., editor. New York: Oxford Univ. Press; 2005. pp. 281–289. [Google Scholar]
- 3.Woese C. Proc. Natl. Acad. Sci. USA. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dolan M., Melnitsky H., Margulis L., Kolnicki R. Anat. Rec. 2002;268:290–301. doi: 10.1002/ar.10161. [DOI] [PubMed] [Google Scholar]
- 5.Melnitsky H., Rainey F., Margulis L. In: Microbial Phylogeny and Evolution: Concepts and Controversies. Sapp J., editor. New York: Oxford Univ. Press; 2005. pp. 261–280. [Google Scholar]
- 6.Margulis L., Dolan M. F., Guerrero R. Proc. Natl. Acad. Sci. USA. 2000;97:6954–6959. doi: 10.1073/pnas.97.13.6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Margulis L., Dolan M. F., Whiteside J. Paleobiology. 2005;31:175–191. [Google Scholar]
- 8.Embley T. M., Martin W. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 9.Martin W., Müller M. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 10.Martin W., Koonin E. V. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 11.Canfield D. E. Nature. 1998;396:450–453. [Google Scholar]
- 12.Knoll A. H. Life on a Young Planet: Three Billion Years of Evolution on Earth. Princeton, NJ: Princeton Univ. Press; 2003. [Google Scholar]
- 13.Canale-Parola E. Bergey’s Manual of Systematic Bacteriology. Vol. 1. Baltimore/London: Williams & Wilkins; 1984. pp. 38–46. [Google Scholar]
- 14.Margulis L. In: Encyclopedia of Microbiology. 2nd Ed. Lederberg J., editor. Vol. 4. New York: Academic; 2000. pp. 353–363. [Google Scholar]
- 15.Searcy D. G., Hixon W. G. BioSystems. 1991;25:1–11. doi: 10.1016/0303-2647(91)90008-9. [DOI] [PubMed] [Google Scholar]
- 16.Searcy D. G., Lee S. H. J. Exp. Zool. 1998;282:310–322. doi: 10.1002/(sici)1097-010x(19981015)282:3<310::aid-jez4>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 17.Searcy D. G., Lee S. H., Gleeson D., Yong R., Abderazzaq K., Dowd G. In: From Symbiosis to Eukaryotism—Endocytobiology VII. Wagner E., Normann J., Greppin H., Hackstein J. H. P., Herrmann R. G., Kowallik K. V., Schenk H. E. A., Seckbach J., editors. Geneva: Geneva Univ. Press; 1999. pp. 43–51. [Google Scholar]
- 18.Searcy D. G. In: Symbiosis. Seckbach J., editor. Dordrecht, The Netherlands: Kluwer Academic; 2001. pp. 163–183. [Google Scholar]
- 19.Searcy D. G. Cell Res. 2003;13:229–238. doi: 10.1038/sj.cr.7290168. [DOI] [PubMed] [Google Scholar]
- 20.Searcy D. G., Delange R. J. Biochim. Biophys. Acta. 1980;609:197–200. doi: 10.1016/0005-2787(80)90212-9. [DOI] [PubMed] [Google Scholar]
- 21.Searcy D. G., Stein D. B. Biochim. Biophys. Acta. 1980;609:180–195. doi: 10.1016/0005-2787(80)90211-7. [DOI] [PubMed] [Google Scholar]
- 22.Dolan M. F. Int. Microbiol. 2001;4:203–208. doi: 10.1007/s10123-001-0038-8. [DOI] [PubMed] [Google Scholar]
- 23.Dolan M. F., d’Ambrosio U., Wier A. M., Margulis L. Acta Protozool. 2001;39:135–141. [Google Scholar]
- 24.Margulis L. Symbiosis in Cell Evolution. 2nd Ed. New York: Freeman; 1993. [Google Scholar]
- 25.Pazour G. J., Agrin N., Leszyk J., Witman G. B. J. Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slesarev A. I., Belova G. I., Kozyavkin S. A., Lake J. A. Nucleic Acids Res. 1997;26:427–430. doi: 10.1093/nar/26.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Bruggen J. A., Stumm C. K., Vogels G. D. Arch. Microbiol. 1983;136:89–95. [Google Scholar]
- 28.Hackstein J. H. P., Yarlett N. In: Molecular Basis of Symbiosis: Progress in Molecular and Subcellular Biology. Overmann J., editor. Berlin/Heidelberg: Springer; 2005. pp. 118–142. [Google Scholar]
- 29.Hertz-Fowler C., Berriman M., Pain A. Nat. Rev. Microbiol. 2005;3:670–672. doi: 10.1038/nrmicro1237. [DOI] [PubMed] [Google Scholar]
- 30.Golding G. B., Gupta R. S. Mol. Biol. Evol. 1995;12:1–6. doi: 10.1093/oxfordjournals.molbev.a040178. [DOI] [PubMed] [Google Scholar]
- 31.Gupta R. S. Mol. Microbiol. 1998;29:695–708. doi: 10.1046/j.1365-2958.1998.00978.x. [DOI] [PubMed] [Google Scholar]
- 32.Hackstein J. H. P., Tjaden J., Huynen M. Curr. Genet. 2006;43 doi: 10.1007/s00294-006-0088-8. in press. [DOI] [PubMed] [Google Scholar]
- 33.Margulis L., McKhann H. I., Olendzenski L., editors. Illustrated Glossary of the Protoctista. Boston: Jones and Bartlett; 1993. [Google Scholar]
- 34.d’Ambrosio U., Dolan M., Wier A. M., Margulis L. Eur. J. Protistol. 1999;35:327–337. doi: 10.1016/S0932-4739(99)80011-X. [DOI] [PubMed] [Google Scholar]
- 35.Margulis L., MacAllister J. Eukaryosis. 2005 digital video, 14 min, available on loan upon written request. [Google Scholar]
- 36.Wenzel M., Radek R., Brugerolle G., König H. Eur. J. Protistol. 2003;39:11–23. [Google Scholar]
- 37.Wier A. M., Dolan M. F., Margulis L. Symbiosis. 2004;36:153–168. [Google Scholar]
- 38.Kirby H., Jr . In: Protozoa in Biological Research. Calkins G. N., Summers F. M., editors. New York: Hafner; 1941. pp. 1009–1113. [Google Scholar]
- 39.Hollande A., Gharagozlou I. C. R. Acad. Sci. Ser. D. 1967;265:1309–1312. [PubMed] [Google Scholar]
- 40.Hollande A., Caruette-Valentin J. Protistologica. 1971;7:5–100. [Google Scholar]
- 41.Eprintsev A. T., Falaleeva M. I., Grabovich M. Y., Parfenova N. V., Kashirskaya N. N., Dubinina G. A. Mikrobiologiya. 2004;73:367–371. [PubMed] [Google Scholar]
- 42.Fuerst J. A., Webb R. I. Proc. Natl. Acad. Sci. USA. 1991;88:8184–8188. doi: 10.1073/pnas.88.18.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.López-Garcia P., Moreira D. J. Mol. Evol. 2002;47:517–530. doi: 10.1007/pl00006408. [DOI] [PubMed] [Google Scholar]
- 44.Gupta R. S. Crit. Rev. Microbiol. 2000;26:111–131. doi: 10.1080/10408410091154219. [DOI] [PubMed] [Google Scholar]
- 45.Gupta R. S. In: Microbial Phylogeny and Evolution: Concepts and Controversies. Sapp J., editor. New York: Oxford Univ. Press; 2005. pp. 261–280. [Google Scholar]
- 46.Rizzotti M. Acta Biotheor. 1995;43:227–240. [Google Scholar]
- 47.Mayr E. Science. 1972;176:981–989. doi: 10.1126/science.176.4038.981. [DOI] [PubMed] [Google Scholar]
- 48.Hanson E. D. The Origin and Early Evolution of Animals. London: Wesleyan Univ. Press/Pitman; 1977. pp. 68–122. [Google Scholar]
- 49.Andrews H. N. The Fossil Hunters: In Search of Ancient Plants. Ithaca, NY: Cornell Univ. Press; 1980. [Google Scholar]
- 50.de Duve C. Singularities: Landmarks On the Pathways of Life. New York: Cambridge Univ. Press; 2005. [Google Scholar]
- 51.Janicki C. Z. Wiss. Zool. 1915;112:573–691. [Google Scholar]
- 52.Perfiliev B. V. Izv. Akad. Nauk. SSSR Ser. Biol. 1969:181–198. [Google Scholar]
- 53.Dubinina G. A., Leshcheva N. V., Grabovich M. Yu. Mikrobiologiya. 1993;62:432–444. [Google Scholar]
- 54.Dubinina G. A., Grabovich M. Yu., Leshcheva N. V. Mikrobiologiya. 1993;62:450–456. [Google Scholar]
- 55.Surkov A. V., Dubinina G. A., Lysenko A. M., Glöckner F. O., Kuever J. Int. J. Syst. Evol. Microbiol. 2001;51:327–337. doi: 10.1099/00207713-51-2-327. [DOI] [PubMed] [Google Scholar]
- 56.Soyer-Gobillard M.-O., Schrevel J. Edouard Chatton (1883–1947)—Life, Discoveries, and Complete Publication List of a Great Scientist. 2007 (DVD of course boards and booklet), in press. [Google Scholar]
- 57.Sapp J. In: Microbial Phylogeny and Evolution: Concepts and Controversies. Sapp J., editor. New York: Oxford Univ. Press; 2005. p. 21. [Google Scholar]
- 58.Sapp J. Microbiol. Mol. Biol. Rev. 2005;69:292–305. doi: 10.1128/MMBR.69.2.292-305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bermudes D., Hinkle G., Margulis L. Microbiol. Mol. Biol. Rev. 1995;58:387–400. doi: 10.1128/mr.58.3.387-400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alliegro M. C., Alliegro M. A., Palazzo R. E. Proc. Natl. Acad. Sci. USA. 2006;103:9034–9038. doi: 10.1073/pnas.0602859103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Melnitsky H., Margulis L. Symbiosis. 2004;37:323–333. [Google Scholar]