Abstract
Factors influencing patterns in the distribution and abundance of plant and animal taxa modulate ecosystem function and ecosystem response to environmental change, which is often taken to infer low functional redundancy among such species, but such relationships are poorly known for microbial communities. Using high-resolution molecular fingerprinting, we demonstrate the existence of extraordinarily repeatable temporal patterns in the community composition of 171 operational taxonomic units of marine bacterioplankton over 4.5 years at our Microbial Observatory site, 20 km off the southern California coast. These patterns in distribution and abundance of microbial taxa were highly predictable and significantly influenced by a broad range of both abiotic and biotic factors. These findings provide statistically robust demonstration of temporal patterning in marine bacterial distribution and abundance, which suggests that the distribution and abundance of bacterial taxa may modulate ecosystem function and response and that a significant subset of the bacteria exhibit low levels of functional redundancy as documented for many plant and animal communities.
Keywords: marine, reoccurrence, automated ribosomal intergenic spacer analysis (ARISA)
The distribution and abundance of species and the factors that control them are central to understanding and predicting ecosystem function and response to environmental change (1). A central tenet of community ecology is that strong spatial and temporal patterns in the distribution and abundance of taxa are governed by environmental (e.g., abiotic, biotic, nutrient, or other) processes (2, 3), and that these processes link ecosystem function to biological diversity (4, 5), but the vast majority of studies have been done on animals and plants (macrobial species), not microbial species like bacteria. Recently, however, it has become increasingly evident that bacteria are often dominant species both in terms of relative abundance (densities or biomass) and in terms of contributing to ecosystem processes (6–8). Thus, the extent to which the interactions between abiotic and biotic processes govern ecosystem and biogeochemical processes remains difficult to assess in the absence of information about microbial biodiversity patterns of equivalent resolution to that of macrobial biodiversity. Although one might argue that microbial communities will mirror macrobial communities, recent analyses suggest that small organisms, such as protistan species, due to their huge population sizes and high dispersal capability, may have fundamentally different diversity patterns compared to larger organisms (9). Microbes are also thought to exhibit the potential for a high degree of functional redundancy, as shown in models like that of Curtis and Sloan (10), where several different bacterial species can fill a particular niche. Given the dominance of bacteria in communities and ecosystem processes, predicting ecosystem response to environmental change requires determining whether bacterial populations exhibit diversity patterns that are indeed fundamentally different from plants and animals.
Advances in molecular methods for quantifying microbial biodiversity, based mostly on 16S rRNA sequences (11), have recently provided mounting evidence that microbes may indeed exhibit spatial patterns in distribution and abundance akin to larger organisms, but temporal patterns remain relatively unexplored. Evidence for species turnover in terrestrial fungi (12), species area curves in sediment bacteria (13), and “island biogeography” in tree holes (14) and lakes (15) suggest that microbial communities may exhibit spatial or macroecological patterns well known for larger organisms in terrestrial ecosystems. A recent review concluded that some spatial patterns observed in microbes mirror those of larger organisms, although there may be some patterns unique to microbes (16).
Marine ecosystems in particular are less well studied, but here, too, these methods have provided evidence of broad-scale patterns in the distribution and abundance of dominant taxa of prokaryotic plankton. For example, bacterial groups like the SAR11 cluster are now known to be abundant through most of the ocean (17) and cyanobacteria and certain Roseobacters common only in certain habitats (18, 19). There is also evidence for spatial gradient patterns from rivers to the open sea (20, 21) and short-term (within a year) temporal changes (22). A new study by Morris et al. (23) examined temporal trends of various bacterial groups in the Sargasso Sea over multiple years, noting certain groups tended to be more common during certain seasons (some groups after seasonal deep mixing events, and others during summer periods). Another study by Crump and Hobbie (24) showed remarkably parallel development of bacterial communities in two adjacent rivers and related the composition to environmental parameters.
Such studies of trends and patterns in abundance, commonness, and specialization are suggestive of similarities to larger organisms. However, few, if any, studies demonstrate statistically robust, predictable patterns in microbial communities. That is, few studies demonstrate that it is possible to predict bacterial community taxonomic composition from environmental parameters. For example, are there annually repeating cyclical patterns for multiple microbial taxa in marine ecosystems that are predictable from environmental conditions? Such a study would more directly and convincingly show the extent to which bacterial communities may be deterministic. For example, one might predict changes in microbial communities and associated ecosystem processes in the face of climate change if such patterns existed.
We tested whether the distribution and abundance of marine bacterioplankton taxa exhibit strong temporal patterns in distribution and abundance over multiple years and at high resolution (e.g., >150 taxa at a monthly scale), and whether these patterns are associated with abiotic and biotic factors. We sampled monthly over 4.5 years for a suite of oceanographic environmental variables including temperature, salinity, dissolved oxygen, chlorophyll a, bacterial and viral abundance, bacterial heterotrophic production, and nutrients. For bacterial community composition, we used a PCR-based community fingerprinting method [automated ribosomal intergenic spacer analysis (ARISA); ref. 25]. ARISA classified bacteria into 171 different operational taxonomic units (OTUs) in our system, of 181 possible OTUs. ARISA analyzes the lengths of the intergenic spacers between 16S and 23S rRNA genes present in almost all bacteria. Our standardized version includes prefiltration to remove eukaryotes, and measured amounts of DNA at each step (21). Phylogenetic resolution is ≲98% 16S rRNA sequence identity, near the range widely considered to be bacterial “species” or “ecotypes” (26, 27). The assay does not miss any known major group of near-surface marine bacteria (27, 28); however, if it did, this would not alter our interpretation because they would be missed consistently throughout. Also, ARISA has been shown to be a reasonably quantitative measure of relative abundance of the only taxon we could independently measure (Prochlorococcus, by flow cytometry; ref. 27). Our interpretations here do not rely on accurately measuring the relative abundance of different taxa, but instead rely on ARISA being repeatable within taxa and with a response positively related to organism abundance; these criteria have been met when examined (27).
To demonstrate predictable patterns in the relative distribution and abundance of bacterial species (or ecotypes) composition over time, three steps are necessary. First, demonstrate that the assemblage composition is patterned. Second, demonstrate that patterns in composition are repeatable. Third, demonstrate that abiotic, biotic, ecosystem function, nutrient, and biodiversity (i.e., environmental) factors can predict these particular temporal patterns. We accomplished the first step by using discriminant function analysis (DFA) to cluster samples (communities in each month) by month based only on ARISA data. We accomplished the second by using time series analyses (TSA) to identify significant autocorrelations among months as evidence of a repeating pattern, again based solely upon ARISA data. We accomplished the third by using multiple regression analyses (MRA) to test whether the temporally repeatable patterns in distribution and abundance are predictable by one or more environmental factors. We also posit that, when species repeat in predictable manner, it is evidence for a low degree of functional redundancy because substitutions would reduce repeatability and predictability.
Results
DFA generated discriminant functions that weighed the relative proportions of different OTUs based on ARISA. These DFA functions (i.e., linear mathematical functions in which coefficients are derived such that multivariate distances among samples are maximally dispersed) permit prediction of month from ARISA data. There are >1050 possible combinations of 171 OTUs we could explore by using DFA. For our data, preliminary exploration revealed that typically <20 OTUs could accurately classify months, and DFA tends to ignore the remaining OTUs once the months are already accurately classified. Therefore, if we simply performed DFA once with all of the 171 OTUs, most of the bacterial diversity would be ignored. Thus, to explore the majority of OTUs in our analysis, we ran DFA with 12 different, ecologically informative subsets of the OTUs. These included five sets based on fragment size, or more specifically the first, second, third, or fourth quarter of the 171 OTUs and all 171 OTUs, when ranked from smallest to largest (i.e., arbitrary sets of OTUs). They also included the OTUs that occurred on >10%, 33%, 50%, or ≥75% of the dates (i.e., commonness of OTUs). Finally, the OTU sets included those OTUs that each averaged 0.2, 1.1, or 1.6% or more of the total integrated DNA content in the amplified products in the samples over the duration of the study (i.e., abundance of OTUs). For example, only 16 OTUs were so abundant that they each accounted for 1.6% or more of the integrated DNA content of the amplified products in the 48 samples, whereas 62 OTUs were abundant enough to each account for at least 0.2% of the DNA in the 48 samples (Table 1). The percent of months correctly classified by DFA and the percent of dispersion explained by the first discriminant function are presented in Table 1.
Table 1.
Statistical analysis of ARISA profiles illustrating a repeating and predictable bacterioplankton community composition
| Statistic | OTU analyzed, sample size (n) | Abundance, OTU |
Commonness, frequency |
Arbitrary, OTUs |
All 171 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| >1.6% 16 | >1.1% 19 | >0.2% 62 | >75% 34 | >50% 63 | >33% 83 | >10% 133 | 399–528 44 | 531–657 43 | 660–844 43 | 849–1183 41 | ||||
| DFA | Dominant OTU | 1 | 719 | 739 | 633 | 739 | 704 | 687 | 477 | 447 | 546 | 699 | 919 | 447 |
| 2 | 675 | 704 | 477 | 687 | 739 | 666 | 408 | 444 | 531 | 769 | 914 | 444 | ||
| 3 | 681 | 687 | 624 | 699 | 519 | 534 | 516 | 441 | 555 | 687 | 441 | |||
| 4 | 402 | 600 | 704 | 734 | 1,040 | 534 | 465 | 621 | 690 | 465 | ||||
| 5 | 687 | 417 | 687 | 799 | 513 | 492 | 570 | 739 | 492 | |||||
| Percent correct | 85 | 57 | 98 | 57 | 64 | 89 | 98 | 94 | 100 | 70 | 19 | 93 | ||
| Percent dispersion | 47 | 51 | 61 | 46 | 44 | 46 | 58 | 52 | 40 | 50 | 59 | 46 | ||
| TSA | Lag, months | 1 | X | X | X | X | X | X | X | X | X | X | X | |
| 4 | X | X | X | X | X | |||||||||
| 5 | X | X | X | X | X | X | X | X | X | |||||
| 6 | X | X | X | X | X | X | ||||||||
| 10 | X | X | X | X | ||||||||||
| 20 | X | |||||||||||||
| MRA | Abiotic | Temp. | X | X | X | X | X | X | ||||||
| Oxygen | X | X | X | X | X | X | X | X | X | |||||
| Salinity | X | X | X | X | ||||||||||
| Biotic | Bacteria | X | ||||||||||||
| Virus | X | X | ||||||||||||
| Ecosystem funct. | ChlA | X | X | X | X | X | X | |||||||
| Phaeo | X | X | X | X | X | |||||||||
| Leu | X | |||||||||||||
| TdR | X | X | X | |||||||||||
| Nutrients | NO2 | X | X | X | X | X | X | |||||||
| NO3 | X | X | ||||||||||||
| SiO3 | X | X | X | |||||||||||
| PO4 | X | X | ||||||||||||
| Biodiversity | # OTUs | X | X | X | ||||||||||
| R2 | 0.41 | 0.48 | 0.56 | 0.39 | 0.72 | 0.22 | 0.28 | 0.71 | 0.12 | 0.54 | 0.42 | 0.2 | ||
| P value | <0.01 | <0.001 | <0.01 | <0.001 | <0.001 | <0.05 | <0.001 | <0.001 | <0.05 | <0.001 | <0.01 | <0.05 | ||
Results from OTUs that comprised 1.6, 1.1, or 0.2 % of the total integrated DNA content in the amplified products are shown under Abundance, results from OTUs that occurred in 75, 50, 33, or 10 % of the samples are shown under Commonness, results from arbitrarily selected OTUs are shown under Arbitrary, and results from all OTUs are shown in the column marked ALL. Sample size (n) indicates the number of OTUs that were used for a given set of DFA, TSA, and MRA. Dominant OTUs were OTUs that had the largest coefficients in a given DFA, presented in rank order, 1 being the highest rank. The length (bp) of the Dominant OTUs are shown above, and putative identities of the dominant OTUs (27) appear in Table 2. Percent Correct is the percentage of months correctly classified by DFA. Percent Dispersion is the percent dispersion of months explained by the first discriminant function. TSA that yielded a significant (P < 0.05) autocorrelation are indicated with an X; significant lags indicate the temporal span of repeated values of the first DFA score. Bacteria and Virus represent prokaryote and virus total abundance by direct counts, ChlA and Phaeo are concentrations of particulate chlorophyll a and phaeopigments, Leu and TdR are bacterial heterotrophic production as estimated by incorporation of tritiated leucine and thymidine, NO2, NO3, SiO3, and PO4 are concentrations of nitrite, nitrate, silicate, and phosphate, respectively, and # OTUs is the richness (n) of individual ARISA samples. Parameters that predict microbial diversity as identified by the MRA (see Materials and Methods) are indicated with an X, with a bold X indicating environmental variables with significant (P < 0.05) regression coefficients. Significant P values (P < 0.05) are indicated in bold. Illustrated examples in Fig. 1 were chosen based on MRA with highest R2 for abundance, commonness, and arbitrarily defined sets of OTUs. Ecosystem funct., ecosystem functionality.
Sampling months could be statistically predicted from the number and relative proportions of OTUs alone, indicating that bacterial composition is patterned. Of the 12 different subsamples of the bacterial OTUs tested by DFA, seven resulted in functions that could accurately classify 80% or more of the samples to their correct months based solely on the distribution and abundance of OTUs (Table 1). On average, the first DFA axis accounted for 50% of the dispersion of the samples in the multivariate space described by the OTUs used in the analyses. The DFA scores based on this first DFA axis therefore serve as a single multivariate index (DFA1) describing ≈50% of the variability in the distribution and abundance of bacterioplankton OTUs for a given sample. This single index of bacterioplankton diversity can be plotted against time (three examples are provided in Fig. 1) and analyzed for temporal patterns by TSA.
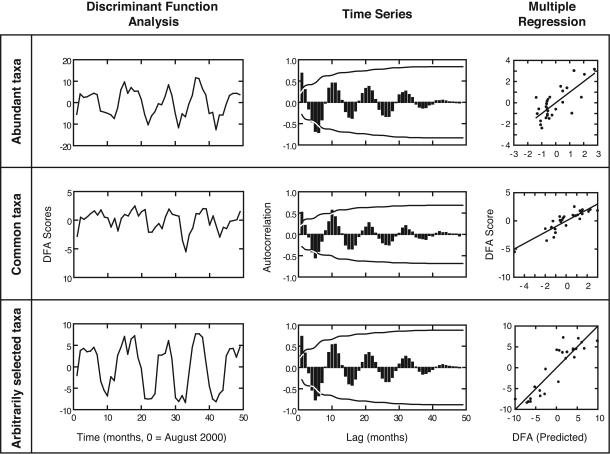
Fig. 1.
Multiple lines of evidence for temporal cyclicity in bacterial biodiversity at 5-m depths in open-ocean samples from coastal California waters. This figure provides three examples selected from Table 1. (Left) DFA scores that evidence annually repeated multivariate patterns in bacterioplankton. (Center) Autocorrelations from TSA (i.e., 95% confidence intervals illustrated by lines). Note evidence for strong, steady, seasonal patterns reflected by the sinusoidal pattern in autocorrelations as lag size increases. (Right) DFA scores and their predicted values based on MRA (see Table 1). (Top) DFA scores based on relatively high abundance (summed amplified DNA of OTUs over the 4.5 years, with OTUs representing >0.2% of the total amplified DNA). (Middle) DFA scores based on commonness (OTUs in 50% or more of the samples over the 4.5 years). (Bottom) DFA scores based on arbitrarily selected taxa, in this case, the first quarter (44 OTUs), i.e., shortest fragment lengths in ARISA analyses (Table 1).
Patterns in bacterial composition are highly repeatable over time, although slightly different depending on which OTUs are chosen. TSAs based on the DFA1 index of bacterioplankton diversity (for communities in each month) showed a significant autocorrelation at a lag of one, meaning that months were similar to temporally adjacent months (Table 1 and Fig. 1). Additional autocorrelations at higher lags (Table 1 and Fig. 1) indicate a strong sinusoidal pattern whereby composition (DFA1) is least similar in the “opposite” season half a year away, but reverts back to being very similar in the subsequent same season. This finding indicates strong, continuous seasonal fluctuations for several groups of OTUs (three examples are illustrated in Fig. 1).
Bacterioplankton diversity can be predicted by environmental factors. MRA showed that the bacterioplankton diversity, as indexed by DFA1, was consistently predictable from oceanographic environmental variables (Table 1). The particular environmental variables identified as significant contributors to the overall regression for the 12 sets of OTUs spanned the full range, from abiotic to biotic (Table 1).
Discussion
Our results demonstrate that, from the environmental conditions, one can predict the distribution and abundance of sets of bacterial OTUs (as indexed by DFA1), and the distribution and abundance of the OTUs can accurately predict the sampling month. Conversely, from the diversity of OTUs identified in DFA1, one cannot only tell the month, but also the likely environmental conditions. For example, the multiple regression with the best explanation of variance in bacterioplankton diversity (R2 = 0.72, or 72% of the variance diversity explained) was based on the set of OTUs occurring in >50% of the samples (Table 1). MRA for this case showed the DFA1 index of bacterioplankton diversity was predictable primarily from the environmental variables of temperature, oxygen, salinity, virus abundance, dissolved nitrite, and dissolved silicate (Table 1). Using the DFA coefficients to determine which OTUs dominated the calculation of the discriminant function, we can then use linked 16Sr RNA plus intergenic spacer clone data match sequences and identities to those OTUs (27). In this case, the most predictable OTUs were presumed to be from among the members of the Alteromonas, Bacteroidetes, α-Proteobacteria (including members of the SAR 11 and SAR 116 clusters), and Verrucomicrobium groups, although some were not identified (Table 2). Note that different combinations of parameters were significant contributors when different subsets of the bacterial community were examined (Table 1), as one would expect if the bacterial community consisted of species with highly diverse ecological niches.
Table 2.
Putative identification of dominant (most predictable) OTUs from DFA
| OTU | Group(s) (accession nos.) |
|---|---|
| 402 | γ-Proteobacteria - CHABI-7 subgroup (DQ009149) |
| 417 | Actinobacter (DQ157868) |
| 465 | SAR86 (AY552545) |
| 531 | γ-Proteobacteria - SPOTS121 subgroup (DQ009145) |
| 534 | γ-Proteobacteria - SPOTS121 subgroup (DQ009143) |
| 555 | Alteromonas (AF408841) |
| 621 | Bacteroidetes (DQ009091) |
| Fibrobacter (DQ009159) | |
| 624 | SAR406 (DQ009157) |
| 666 | SAR11 (DQ009203) |
| 675 | α-Proteobacteria (DQ009262) |
| 681 | SAR11 (DQ009194) |
| 687 | SAR116 (DQ009272) |
| SAR11 (DQ009253) | |
| Alteromonas (AF408829) | |
| 690 | Pelagibacter ubique str. 1062 (SAR 11) (NC_007205) |
| 699 | SAR11 (AY033325) |
| 704 | α-Proteobacteria (DQ009256) |
| SAR11 (AF151254) | |
| 719 | SAR11 (DQ009166) |
| 734 | Verrucomicrobia (AY033323) |
| 739 | Verrucomicrobia (DQ009368) |
| Bacteroidetes (DQ009104) | |
| 769 | Bacteroidetes (DQ157869) |
| 799 | Bacteroidetes (DQ646394) |
| 914 | Prochlorococcus (NC_007335) |
| 1040 | Synechococcus (DQ009365) |
| γ-Proteobacteria - OM182 subgroup (DQ009156) |
ARISA OTU numbers (ARISA fragment length, in base pairs) and group identities (phylogenetic groups of known marine sequences with matching ARISA lengths) for 22 of the 36 unique dominant OTUs from DFA listed in Table 1. OTUs not appearing here, but in Table 1, do not match any known marine sequence. GenBank accession numbers of identifying clones/cultures are also listed.
Collectively, these results demonstrate that the biodiversity of open ocean marine prokaryotic communities at this site exhibit distinctly repeatable temporal patterns that are predictable from a variety of abiotic, biotic, nutrient, and ecosystem function variables. Although we have explored only a limited number of sets of OTUs, as in the case of studies of larger organisms (e.g., based on abundance, commonness, or arbitrary subsets), a number of strong patterns are apparent. Significant associations with a variety of environmental variables that cover a complex range, from biotic to abiotic, suggest a large number of possible ecological mechanisms responsible for these patterns. Although experiments would be necessary to identify the particular ecological mechanisms, our results nevertheless suggest that these communities are likely to be structured by the environment, as are many plant and animal communities.
Note that our analysis does not imply that the distribution and abundance of all taxa in this system vary in lockstep in highly repeatable annual patterns. Rather, our analyses indicate that there are several highly predictable subsets of taxa as well as less predictable subsets of taxa (and visual inspection of the data shows some taxa occurred only a few times over the 4.5 years, others sporadically). However, this wide range of responses among taxa is a common feature of “macrobial” communities. Of interest is why some taxa show strong patterns and others do not.
Past studies of planktonic bacteria have rarely examined this particular question, but have found some similar and some disparate results in comparison to those we report here. For example, Kent et al. (29), who used ARISA like we did, found little evidence for temporally repeating patterns in humic lake bacterioplankton. Earlier, Lindstrom (30) showed a slow steady change rather than annual or seasonal patterns over years in a boreal lake, and Yannarell et al. (31) found no clear temporal pattern within neighboring lake systems. Recently, Yannarell and Triplett (32) also found a decoupling between the community and environmental parameters in lake systems. The differences between our study and these lake based studies could be due to differences in taxa, sensitivity to environmental variables, the closed versus open nature of the systems, the differences in age of the systems, and other locally unique properties of these systems such as perturbation frequency, or in analytical approaches; further study is needed to evaluate which of these possibilities may best account for the observed differences.
In contrast to these reports on lake systems that appear to lack repeating annual cycles, two other studies have identified possible temporal patterns. Morris et al. (23) reported seasonal reappearance of certain groups related to large scale mixing events in the Sargasso Sea. Additionally, Crump and Hobbie (24) reported parallel and apparently deterministic bacterial community patterns in two adjacent rivers that drain into the same coastal area and reported evidence for seasonal repeatability and predictability from environmental parameters as judged by examination of multidimensional scaling plots. Their temporal pattern was most strongly correlated to parameters related to the strong seasonality of their site (e.g., large temperature shifts from 0 to 29°C and flow rate). However, neither Morris et al. nor Crump and Hobbie tested for the statistical significance of temporal trends (autocorrelation), nor did they test for the predictability of temporal patterns from nonbacterial parameters, so we do not know whether these studies show temporal patterning or predictability comparable to what we observed.
In an ocean environment, an obvious factor that can influence communities is the hydrographic regime. Our sample location off Southern California is within a system of currents that exhibit strong seasonality, with generally southward flow in spring, weak northward flow in early summer, and stronger northward flow in late summer and fall (33). Because the region north of our site has strong seasonal upwelling, especially in the spring, one might expect currents to bring more eutrophic conditions and associated organisms in spring, whereas summer–fall stratification and flow from the south would be associated with more oligotrophic conditions and associated organisms. This region also experiences storms during winter that can lead to moderately deep mixing that brings inorganic nutrients and deeper-dwelling organisms into the near-surface waters. It is likely that these seasonal hydrographic conditions and their influence on other organisms, such as eukaryotic phytoplankton species or other protists that we did not investigate here, contributed significantly to our observed patterns and predictability in bacterial community composition. Such variations in hydrography are reflected in several parameters we investigated, such as temperature, salinity, and nutrients.
Our results have important implications for understanding marine ecosystem function. If we had seen unpatterned distribution and abundance, then the implication would have been that bacterial species are well dispersed and to a large degree ecologically redundant, which would be consistent with the idea that much or most microbial diversity consists of functionally interchangeable taxa. Furthermore, if unpatterned, then ecosystem function (e.g., autotrophic or heterotrophic bacterioplankton production) would simply be a consequence of abiotic factors and nutrient availability, with the bacteria representing the equivalent of a black box multiplier associated with biomass that permits biogeochemical processes to occur; ecosystem models have not yet moved beyond that general portrayal of marine bacteria. However, our evidence of strong temporal patterns implies that the subsets of planktonic bacteria that showed the repeating predictable patterns are less functionally interchangeable (i.e., specialized), their distribution and abundance regulated by abiotic factors (e.g., temperature, salinity, dissolved oxygen), biotic factors (e.g., competition with other species, predation by viruses), and nutrients (e.g., types, supply rates, stoichiometric constraints). In other words, if the dominant bacterial niches in our area were all occupied by various combinations of functionally redundant bacteria, we would expect to see different combinations of OTUs occurring under similar conditions, but instead we see the repeating and predictable patterns of certain bacterial community subsets. Thus, this evidence of temporal patterns in bacterioplankton distribution and abundance implies that current marine ecosystem models that take a “black box” approach may oversimplify the true nature of microbial ecological functions.
A considerable number of well established theories in macrobial ecology [e.g., resource-based competition (34), trophic cascades (35), biodiversity and ecosystem functioning (5), and the metabolic theory of ecology (36)] apply to the kinds of patterns in distribution and abundance as we demonstrate here, but these theories are largely untested in bacteria. In situ experimental analyses designed to examine which theories are best supported by strong patterns in distribution and abundance represent a critical next step, but such approaches still face many challenges in microbial ecology. Overcoming these challenges would lead the way to the development of a more predictive open ocean microbial ecology, one in which the same level of understanding of the role of biodiversity in macrobial systems is achieved for prokaryotic microbial communities.
Materials and Methods
Sample Collection.
Samples from 5-m depth were collected at the San Pedro Ocean Time Series (SPOTS) Microbial Observatory site (33° 33′ N, 118° 24′ W) on board the R/V Sea Watch, using Niskin bottles approximately monthly from August 2000 to December 2004. Seawater (20 liters) was filtered through Gelman A/E glass fiber filters (nominal pore size, 1.2 μm) to remove eukaryotic cells (containing plastids that complicate the interpretation of ARISA fingerprints). The A/E-filtered seawater, containing the free-living bacterioplankton that have been shown to be ≈85% of the total bacteria (37), was then filtered through a 0.2-μm Durapore filter (Millipore) to collect the bacteria. Filters were frozen at −80°C before analysis at the University of Southern California (Los Angeles, CA).
DNA Extraction and ARISA.
DNA was extracted from frozen filters by hot SDS lysis followed by phenol-chloroform purification of nucleic acids (38), and DNA was stored frozen at −80°C in TE buffer or dry. ARISA (25) was conducted on 5 ng of DNA as measured by PICO Green fluorescence. A standard amount of template genomic DNA was used in each PCR, with the intention of analyzing the same amount of bacteria from each sample. PCRs (50 μl) contained 1× PCR buffer, 2.5 mM MgCl2, 250 μM of each deoxynucleotide, 200 nM each of universal primer 16s [1392F (5′-G[C/T]ACACACCGCCCGT-3′)] and bacterial primer 23s [125R labeled with a 5′ TET (5′-GGGTT[C/G/T]CCCCATTC(A/G)G-3′)], 2.5 units Taq polymerase (Promega), and BSA (Sigma, St. Louis, MO, catalog no. A-7030; 40 ng/μl final concentration). These primers target specifically bacteria; therefore, archaea are not included in our analysis, and we know of no significant group of common near-surface marine bacteria whose DNA these primers fail to amplify (39). Thermocycling was preceded by a 3-min heating step at 94°C, followed by 30 cycles of denature at 94°C for 30 s, anneal at 56°C for 30 s, extend at 72°C for 45s, with a final extension step of 7 min at 72°C. Amplification products were cleaned by using Clean & Concentrator-5 (Zymo Research, Orange, CA) and DNA in purified products were measured by PICO Green fluorescence. Purified products were then diluted to 5 ng/μl so that we could load a standardized amount in the fragment analysis and prevent differences arising from different amounts of loaded DNA (such variations can occur with different loaded amounts because as more DNA is loaded, more small peaks can exceed the threshold of detectability). Products were then run for 5.5 h on an ABI 377XL automated sequencer operating as a fragment analyzer (40) with a custom-made ROX-labeled 1,500-bp standards (Bioventures, Murfreesboro, TN). The sequencer electropherograms were then analyzed by using ABI Genescan software.
Outputs from the ABI Genescan software were transferred to Microsoft Excel (Seattle, WA) for subsequent analysis. Peaks less than five times baseline fluorescence were discarded because they were judged not clearly distinguishable from instrument noise (21). With this criterion, the practical detection limit for one OTU is ≈0.09% of the total amplified DNA (21). Due to the nature of fragment analysis, there is some uncertainty in the estimates of ARISA fragment length, which led us to “bin” the OTUs. This uncertainty increases with increasing fragment length (largely from diffusive band spreading), and bins were 3 bp wide up to 700 bp, then 5 bp wide from 700–1,000 bp, and 10 bp wide >1,000 bp; reported OTU lengths in the tables are the mean value of the bins, and bin width was accounted for in determination of putative OTU identification (hence, some of the ambiguous identifications). Further analysis was then conducted as outlined under Statistical Analysis.
The standardization we use is comparable to animal and plant studies because ecologists have long recognized that counting more individuals in a sample can make diversity appear larger, so standardization of sample size is needed.
Ancillary Parameters.
Bacterioplankton production was estimated by incorporation of [3H]thymidine and [3H]leucine into DNA and protein, respectively, as described (41, 42). Although both are measures of growth, thymidine incorporation is thought to primarily follow cell division, whereas leucine incorporation also indicates changing growth rates. Briefly, 10-ml seawater samples in sterile, sample-rinsed polypropylene centrifuge tubes were inoculated with 5 nM (final concentration) [3H]thymidine or [3H]leucine and incubated 1 h at ambient temperature. After incubation, samples were filtered onto 25-mm-diameter 0.45-μm Millipore (type HA) nitrocellulose filters to dryness (Millipore, Billerica, MA). Small molecules were then extracted by incubation for 2 min with 2 ml of ice-cold trichloroacetic acid (TCA). Afterward, TCA was vacuum filtered through, and filters were placed in 5 ml of Ultima Gold scintillation fluid. Radioactivity was measured in a Beckman Coulter (Fullerton, CA) 6500 SC scintillation counter after 1 h to allow clearing of filter membranes. We used a conversion factor of 1.5 × 1017 cells per mol of leucine and 2 × 1018 cells per mol of thymidine incorporated to estimate bacterial production (41, 42).
Bacterial and viral abundance was determined by SYBR Green I staining and epifluorescence microscopy (43). Briefly, samples (50 ml) were fixed with 2% 0.02-μm-filtered formaldehyde and kept at 4°C in the dark until processing, which occurred within 24 h of sampling. Samples were processed by firstly filtering aliquots (3–20 ml, depending on depth) onto 0.02-μm Anodisc Al2O3 filters, drying the filter on tissue paper, then staining on a 100-μl 1:2,500-diluted drop of SYBR Green I. After staining, the filters were redried on tissue paper and mounted on a glass slide with a solution of 50:50:0.01 glycerol/PBS/p-phenylenediamine as mountant. Slides were observed under blue light excitation at ×1,000 magnification on an Olympus BH-60 microscope. More than 200 particles of each of viruses and bacteria were counted in 20 fields.
Hydrocast CTD instrument data included in vivo chlorophyll a fluorescence, salinity, oxygen, and temperature. Additionally, samples for the determination of extracted chlorophyll a (44), conductivity, and oxygen (Winkler titration; ref. 44) were taken corresponding to the target depths of our microbial analyses.
Phylogenetic Identification of ARISA OTUs.
16S–ITS–23S sequences obtained from clone libraries constructed previously from the San Pedro Ocean Time Series (SPOTS) station and 16S–ITS–23S sequences available in GenBank were used to assign putative identities to the 36 unique ARISA bins presented in Table 2. Twenty-two of 36 unique bins were identified. Occasionally, more than one possible identification could be assigned to an individual OTU (five of 36), whereas, conversely, 14 ARISA bins did not have a corresponding clone of the same length and hence could not be identified.
Statistical Analysis.
DFA is a multivariate statistical method that identifies functions that maximize distances (dispersion) among groups in multivariate space. The discriminant functions yield scores that reflect differences among groups better than individual variables. The method uses eigenvalue analysis to solve for the coefficients in the discriminant functions. In our case, the groups are months and the variables are OTU proportions derived from the ARISA analyses. Because the discriminant scores of the first function represents the best multivariate metric for distinguishing months based on OTUs, we use DFA1 scores as our measure of microbial diversity that best distinguishes one month from another. DFA is described in Manly (45). Time series (46) and multiple regression analyses (47) are described in the main text. We used SYSTAT version 11 for these analyses.
Acknowledgments
We thank Ximena Hernandez, Sheila O’Brien, Anand Patel, Xiaolin Liang, Reni Smith, Michael Neumann, Tony Michaels, and the crew of the R/V Sea Watch. This work was supported by U.S. National Science Foundation (NSF) Microbial Observatories Program Grant MCB0084231, and NSF Grant OCE0527034.
Abbreviations:
- ARISA
automated ribosomal intergenic spacer analysis
- OTU
operational taxonomic unit
- DFA
discriminant function analysis
- TSA
time series analysis
- MRA
multiple regression analyses.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ009149, DQ157868, AY552545, DQ009145, DQ009143, AF408841, DQ009091, DQ009159, DQ009157, DQ009203, DQ009262, DQ009194, DQ009272, DQ009253, AF408829, NC_007205, AY033325, DQ009256, AF151254, DQ009166, AY033323, DQ009368, DQ009104, DQ157869, DQ646394, NC_007335, DQ009365, and DQ009156).
References
- 1.Chapin F. S., III, Zavaleta E. S., Eviner V. T., Naylor R. L., Vitousek P. M., Reynolds H. L., Hooper D. U., Lavorel S., Sala O. E., Hobbie S. E., et al. Nature. 2000;405:234–242. doi: 10.1038/35012241. [DOI] [PubMed] [Google Scholar]
- 2.Morin P. J. Community Ecology. Malden, MA: Blackwell Science; 1999. [Google Scholar]
- 3.Elser J. J., Sterner R. Ecological Stoichiometry: The Biology of Elements from Molecules to the Biosphere. Princeton: Princeton Univ. Press; 2002. [Google Scholar]
- 4.Loreau M., Naeem S., Inchausti P., Bengtsson J., Grime J. P., Hector A., Hooper D. U., Huston M. A., Raffaelli D., Schmid B., et al. Science. 2001;294:804–808. doi: 10.1126/science.1064088. [DOI] [PubMed] [Google Scholar]
- 5.Hooper D. U., Chapin F. S., III, Ewel J. J., Hector A., Inchausti P., Lavorel S., Lawton J. H., Lodge D. M., Loreau M., Naeem S., et al. Ecol. Monogr. 2005;75:3–35. [Google Scholar]
- 6.Fuhrman J. A., Sleeter T. D., Carlson C. A., Proctor L. M. Mar. Ecol. Prog. Ser. 1989;57:207–217. [Google Scholar]
- 7.Whitman W. B., Coleman D. C., Wiebe W. J. Proc. Natl. Acad. Sci. USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azam F., Worden A. Z. Science. 2004;303:1622–1624. doi: 10.1126/science.1093892. [DOI] [PubMed] [Google Scholar]
- 9.Fenchel T., Finlay B. J. Bioscience. 2004;54:777–784. [Google Scholar]
- 10.Curtis T. P., Sloan W. T. Curr. Opin. Microbiol. 2004;7:221–226. doi: 10.1016/j.mib.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Olsen G. J., Lane D. L., Giovannoni S. J., Pace N. R. Annu. Rev. Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 12.Green J. L., Holmes A. J., Westoby M., Oliver I., Briscoe D., Dangerfield M., Gillings M., Beattie A. J. Nature. 2004;432:747–750. doi: 10.1038/nature03034. [DOI] [PubMed] [Google Scholar]
- 13.Horner-Devine M. C., Lage M., Hughes J. B., Bohannan B. J. Nature. 2004;432:750–753. doi: 10.1038/nature03073. [DOI] [PubMed] [Google Scholar]
- 14.Bell T., Ager D., Song J. I., Newman J. A., Thompson I. P., Lilley A. K., van der Gast C. J. Science. 2005;308:1884. doi: 10.1126/science.1111318. [DOI] [PubMed] [Google Scholar]
- 15.Reche I., Pulido-Villena E., Morales-Baquero R., Casamayor E. O. Ecology. 2005;86:1715–1722. [Google Scholar]
- 16.Martiny J. B. H., Bohannan B. J. M., Brown J. H., Colwell R. K., Fuhrman J. A., Green J. L., Horner-Devine M. C., Kane M., Krumins J. A., Kuske C. R., et al. Nat. Rev. Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 17.Morris R. M., Rappe M. S., Connon S. A., Vergin K. L., Siebold W. A., Carlson C. A., Giovannoni S. J. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- 18.Rocap G., Distel D. L., Waterbury J. B., Chisholm S. W. Appl. Environ. Microbiol. 2002;68:1180–1191. doi: 10.1128/AEM.68.3.1180-1191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selje N., Simon M., Brinkhoff T. Nature. 2004;427:445–448. doi: 10.1038/nature02272. [DOI] [PubMed] [Google Scholar]
- 20.Troussellier M., Schafer H., Batailler N., Bernard L., Courties C., Lebaron P., Muyzer G., Servais P., Vives-Rego J. Aquat. Microbiol. Ecol. 2002;28:13–24. [Google Scholar]
- 21.Hewson I., Fuhrman J. A. Appl. Environ. Microbiol. 2004;70:3425–3433. doi: 10.1128/AEM.70.6.3425-3433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinhassi J., Hagstrom A. Aquat. Microbiol. Ecol. 2000;21:245–256. [Google Scholar]
- 23.Morris R. M., Vergin K. L., Cho J. C., Rappe M. S., Carlson C. A., Giovannoni S. J. Limnol. Oceanogr. 2005;50:1687–1696. [Google Scholar]
- 24.Crump B. C., Hobbie J. E. Limnol. Oceanogr. 2005;50:1718–1729. [Google Scholar]
- 25.Fisher M. M., Triplett E. W. Appl. Environ. Microbiol. 1999;65:4630–4636. doi: 10.1128/aem.65.10.4630-4636.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown M. V., Fuhrman J. A. Aquat. Microb. Ecol. 2005;41:15–23. [Google Scholar]
- 27.Brown M. V., Schwalbach M. S., Hewson I., Fuhrman J. A. Environ. Microbiol. 2005;7:1466–1479. doi: 10.1111/j.1462-2920.2005.00835.x. [DOI] [PubMed] [Google Scholar]
- 28.Giovannoni S. J., Rappe M. In: Microbial Ecology of the Oceans. Kirchman D. L., editor. New York: Wiley; 2000. pp. 47–84. [Google Scholar]
- 29.Kent A. D., Jones S. E., Yannarell A. C., Graham J. M., Lauster1 G. H., Kratz T. K., Triplett E. W. Microbiol. Ecol. 2004;48:550–560. doi: 10.1007/s00248-004-0244-y. [DOI] [PubMed] [Google Scholar]
- 30.Lindstrom E. S. FEMS Microbiol. Ecol. 1998;27:163–174. [Google Scholar]
- 31.Yannarell A. C., Kent A. D., Lauster G. H., Kratz T. K., Triplett E. W. Microbiol. Ecol. 2003;46:391–405. doi: 10.1007/s00248-003-1008-9. [DOI] [PubMed] [Google Scholar]
- 32.Yannarell A. C., Triplett E. W. Appl. Environ. Microbiol. 2005;71:227–239. doi: 10.1128/AEM.71.1.227-239.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Lorenzo E. Deep Sea Res. II. 2003;50:2371–2388. [Google Scholar]
- 34.Tilman D. Resource Competition and Community Structure. Princeton: Princeton Univ. Press; 1982. [PubMed] [Google Scholar]
- 35.Polis G. A., Strong D. R. Am. Nat. 1996;147:813–846. [Google Scholar]
- 36.Brown J. H., Gillooly J. F., Allen A. P., Savage V. M., West G. B. Ecology. 2004;85:1771–1789. [Google Scholar]
- 37.Lee S., Kang Y. C., Fuhrman J. A. Mar. Ecol. Progr. Ser. 1995;119:285–290. [Google Scholar]
- 38.Fuhrman J. A., Comeau D. E., Hagstrom A., Chan A. M. Appl. Environ. Microbiol. 1988;54:1426–1429. doi: 10.1128/aem.54.6.1426-1429.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown M. V., Fuhrman J. A. Aquat. Microb. Ecol. 2005;41:15–23. [Google Scholar]
- 40.Avaniss-Aghajani E., Jones K., Chapman D., Brunk C. BioTechniques. 1994;17:144–149. [PubMed] [Google Scholar]
- 41.Simon M., Azam F. Mar. Ecol. Prog. Ser. 1989;51:201–213. [Google Scholar]
- 42.Fuhrman J. A., Azam F. Mar. Biol. 1982;66:109–120. [Google Scholar]
- 43.Noble R. T., Fuhrman J. A. Aquat. Microb. Ecol. 1998;14:113–118. [Google Scholar]
- 44.Parsons T. R., Maita Y., Lalli C. M. A Manual of Chemical and Biological Methods for Seawater Analysis. New York: Pergamon; 1984. [Google Scholar]
- 45.Manly B. F. J. Multivariate Statistical Methods, A Primer. Boca Raton, FL: Chapman & Hall/CRC; 2005. [Google Scholar]
- 46.Rasmussen P. W., Heissey D. M., Nordheim E. V., Frost T. M. In: Design and Analysis of Ecological Experiments. Scheiner S. M., Gurevitch J., editors. Oxford: Oxford Univ. Press; 2001. pp. 158–177. [Google Scholar]
- 47.Philippi T. E. In: Design and Analysis of Ecological Experiments. Scheiner S. M., Gurevitch J., editors. New York: Chapman & Hall; 1993. pp. 183–210. [Google Scholar]