Abstract
Based on functional studies in the bone marrow, it has been suggested that the ability to efflux Hoechst 33342 may represent a universal stem cell trait. In this phenotypic and functional characterization of the Hoechst side population (SP) in adult murine epidermis, we demonstrate that these cells are a rare subset of the keratinocyte stem cell-enriched α6briCD71dim fraction comprising SSClow/K14+/CD34−/Oil red O−/c-kit−/CD45− keratinocytes. Epidermal SPs have the smallest cell and nuclear size but exhibit the highest nuclear-to-cytoplasmic ratio of any fraction examined, consistent with a primitive cell type. Although SPs demonstrated poor cumulative in vitro proliferative output, they exhibited sustained epidermal tissue-regenerative activity in vivo compared with unfractionated and non-SP cells. Collectively, these results indicate that the epidermal SP contains the most potent keratinocyte stem cell population in skin epithelium.
Keywords: skin, self-renewal, tissue regeneration, cell cycle, proliferation
Because keratinocyte stem cells (KSCs) are the ultimate source of epidermal renewal in vivo, their isolation and characterization is of paramount importance. We have developed a method to enrich for KSCs based on the α6briCD71dim surface phenotype from both interfollicular and follicular epidermis (1, 2), confirmed by subsequent studies (3–6). Whether this population can be further enriched for the most potent KSCs by using Hoechst 33342 has not been determined. Although the existence of a side population (SP) resembling the Goodell bone marrow SP in disaggregated human and murine epidermis has been reported (7–11), its functional significance for epithelial tissue renewal remains unclear. Thus, although Montanaro et al. (7) demonstrated that SP cells isolated from total murine skin (i.e., epidermis and dermis) could engraft skeletal muscle, albeit with poor efficiency, whereas NSP cells could not, the capacity of these SPs to contribute to skin regeneration was not investigated. Interestingly, a recent study demonstrated that human SPs derived from passage 2 epidermal cultures exhibited greater long-term proliferative potential and skin regeneration in vitro (12). However, their relationship to the in vivo stem cell compartment was undetermined because of the paucity of markers for cultured KSCs (9).
Attempts have been made to infer the relatedness of epidermal SPs to known epidermal stem cell populations by phenotypic correlation with slowly cycling DNA label-retaining cells (LRCs) based on their α6briCD71dim expression (2). Terunuma et al. (8) concluded that, because epidermal SPs appeared to be α6dim, they must be distinct from LRCs (known to be α6bri) and, therefore, could not be stem cells. In another study, Triel et al. (10) concluded that SPs lacked stem cell characteristics, because the vast majority of LRCs in mouse epidermis were in the NSP fraction, and human epidermal SPs expressing the Hoechst efflux HSC marker Bcrp1 were also negative for the α6 and β1 integrins known to be highly expressed on KSCs. In stark contrast, Yano et al. (11) believed epidermal SPs were “closely related” to KSCs by virtue of their higher expression of α6 and β1 integrins and Bcrp1 as well as low levels of CD71 expression.
Presumably, the variations observed in the phenotype of the epidermal SP in these studies are due to species differences (murine vs. human), cell isolation techniques that may alter cell-surface markers, Hoechst staining techniques, and the anatomical origins of the epidermal cells (7–13). Importantly, the majority of conflicting reports have used phenotypic analysis or “guilt by association” as the major tool to infer whether stem cell status can be conferred on the epidermal SP, without directly testing these cells in functional assays for stem cell attributes. Furthermore, significant technical limitations exist in analyzing BrdU label retention directly in the SPs. For instance, the degree of difficulty in labeling 100% of epidermal stem cells (14) means that putative KSCs, like the SP, may evade BrdU labeling at the outset of the study. Thus, the apparent absence of LRCs in the SP region is subject to interpretation. It is therefore imperative to demonstrate actual functional differences between prospectively isolated epidermal SPs and NSPs directly in terms of proliferative output and tissue reconstitution (attributes considered to be hallmarks of KSCs), and define their relationship to previously described KSCs, to resolve the stem cell status of the epidermal SP. Here, we present data showing that resistance to Hoechst 33342 can indeed be exploited to further enrich for stem cells in murine epidermis.
Results
Murine Tail Epidermis Harbors a Rare SP of Keratinocytes.
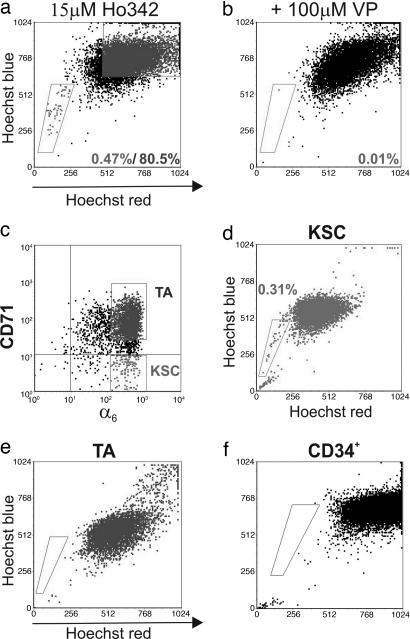
Given the rarity of SP incidence reported to date in various tissues (15–17), we chose adult murine tail epidermis for our studies because of the high density of clonogenic keratinocytes in this tissue and, therefore, its suitability as an important source of stem cells (4, 14, 18, 19). The frequency of SPs in primary adult tail keratinocyte preparations was 0.47 ± 0.17% (Fig. 1a) of viable cells, consistent with a recent study in mouse dorsal epidermis (8). The addition of 100 μM verapamil resulted in a reduction of the SP to background levels (Fig. 1b), verifying that the epidermal SP cells arise as a result of effluxing Hoechst.
Fig. 1.
The SP from adult mouse tail epidermis. Cells freshly harvested from the tail epidermis of 9-week-old C57/B6 mice harbor an SP (a) that was ablated with 100 μM verapamil (VP) (b). Fresh isolates subsetted on α6 (c, x axis) and CD71 (c, y axis) surface phenotype into fractions enriched for KSCs and TAs were subjected to Hoechst staining to demonstrate that SP enrichment occurs in the KSC subset (d), whereas TAs are devoid of SPs (e). (f) CD34+ epidermal cells did not harbor an SP.
To ascertain the lineage of the SPs in the epidermal isolates, cytospins of sorted SP cells were subjected to immunohistochemistry for the basal epidermal marker K14, revealing that the majority of this fraction contained small K14+ cells interspersed with some differentiated K14− cells (Fig. 6a, which is published as supporting information on the PNAS web site). Flow-cytometric analysis for K14 confirmed that this epithelial marker was highly expressed in 90–92% of the SP and NSP fractions (Fig. 6b). Interestingly, epidermal SPs exhibited slightly lower levels of K14 compared with NSPs, perhaps reflecting an undifferentiated status, as reported for putative mammary epithelial stem cells (20). This finding is also consistent with the observed low frequency of keratin filament bundles in putative KSC populations (14, 21, 22). We next subjected the Hoechst subsets to immunostaining for various markers to determine whether the SP contained any rare nonepidermal populations. The absence of c-kit+ melanocytes and CD45+ Langerhans cells/intraepithelial lymphocytes in the SP was established by flow-cytometric analysis. Thus, although 1.19 ± 0.05% c-kit+ cells were found in the unfractionated (Unf) tail epidermal preparations (Fig. 6c), these were all located in the NSP population (Fig. 6d). Similarly, the Unf sample contained 1.05 ± 0.18% CD45+ cells (Fig. 6e), which were also exclusive to the NSP population (Fig. 6f). Oil red O staining of Hoechst subsets revealed rare sebocytes at 1 in 10,000 Unf cells (Fig. 6g); a search among 20,000 SPs failed to locate any positive cells (Fig. 6h). These data confirm that the epidermal SP is enriched for keratinocytes.
Epidermal SPs Represent a Subset of the KSC-Enriched α6briCD71dim Compartment, Depleted of Mitotic Cells but Enriched for Small Cells with a High Nuclear-to-Cytoplasmic (N/C) Ratio.
We have reported that the α6briCD71dim subset contains cells exhibiting many attributes ascribed to KSCs, including slowly cycling, small cells with low complexity and high N/C ratio and the greatest long-term in vitro proliferative output (1, 2). To establish the relationship of the SP to that previously described KSC population, we set out to determine the SP phenotype with respect to α6 and CD71. Given that incubation at 37°C (required for Hoechst staining and efflux) alters surface expression of CD71 (our observations) (10), keratinocytes were sorted into KSC (α6briCD71dim) and transit-amplifying (TA) (α6briCD71bri) fractions (Fig. 1c) and then subjected to Hoechst staining. Our results demonstrate conclusively that the SP cofractionates with the α6briCD71dim KSC population (Fig. 1d), whereas the TA compartment (Fig. 1e) was devoid of any SP cells. Because both interfollicular and hair follicle KSCs express high levels of α6 and low levels of CD71, we sought to determine whether the SPs originated in the hair-follicle bulge, a known KSC repository (14), using CD34 as a marker for this region (23). Predictably, murine tail epidermis contained a CD34+ population with low side light scatter (SSC) comprising 7.78 ± 1.12% (n = 12) of Unf cells; however, all of the CD34+ cells were localized to the NSP population, demonstrating that the Hoechst SP cells are distinct from the hair-follicle bulge cells (Fig. 1f).
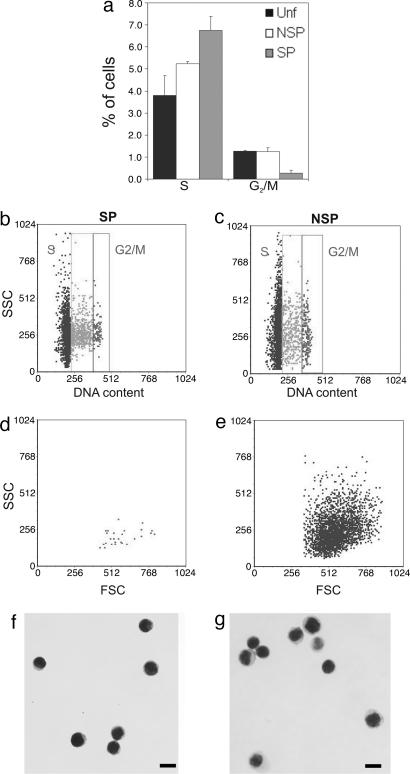
Given that the epidermal SPs were not part of the deeply quiescent hair-follicle bulge region, we sought to establish their cycling status to determine whether they belonged to the actively cycling TA compartment or whether they were a quiescent population as expected of a stem cell fraction. Unf epidermal cells, SPs, and NSPs were collected by FACS and subjected to propidium iodide staining for subsequent flow cytometric analysis for DNA content. No statistically significant differences were observed in the frequency of G0/G1 phase among total viable (94.93 ± 0.93%), NSP (93.51 ± 0.22%), and SP fractions (92.99 ± 0.49%, n = 3). The overall number of S-phase cells in the epidermis was similar to previous reports (2, 19, 24). However, the SPs contained a greater number of S-phase cells (Fig. 2a and b; 6.75 ± 0.64%) compared with NSPs (Fig. 2 a and c; 5.23 ± 0.11%, P = 0.0374) and Unf cells (3.8 ± 0.9%, P = 0.0589). Although this finding would suggest enrichment for cycling cells, examination of the G2/M phase revealed that the SPs contained a 5-fold decrease (0.26 ± 0.15%) in cells undergoing mitosis when compared with both NSP (1.24 ± 0.18%, P = 0.0209) and Unf (1.27 ± 0.04%, P = 0.0045) cells, suggesting that the SP contains fewer cycling cells.
Fig. 2.
Cell cycling status, light scattering, and morphology of SP and NSP subsets. Hoechst subsets were stained with 50 μg/ml propidium iodide to measure DNA content. (a) The proportion of cells in different phases of the cell cycle was estimated by using ModFit analysis. The DNA content vs. SSC (b and c) and forward scatter (FSC) vs. SSC profiles (d and e) of SP (b and d) and NSP (c and e) cells were compared to demonstrate the low G2/M frequency and homogeneously low intracellular complexity of SP cells vs. NSPs, irrespective of cycling status, consistent with a primitive cell type. Cytospins of SP (f) and NSP (g) stained with Giemsa.
It has been shown that a feature of stem cell populations, given their primitive state, is that they are homogeneously small, blast-like cells with a high N/C ratio (2, 25–27). Initial flow-cytometric analysis revealed that epidermal SPs exhibited homogeneously low intracellular complexity (i.e., low SSC) and formed a cluster in the low forward-scatter region (Fig. 2d), indicating enrichment for small, immature cells. In contrast, NSPs had very heterogeneous forward and SSC properties (Fig. 2e), indicative of a wide range of cell sizes and increased intracellular complexity consistent with maturing TA cells. Microscopic analysis of Giemsa-stained cytospins confirmed that SPs were uniformly small and blast-like (Fig. 2f), with a thin strip of cytoplasm around their nuclei, whereas NSPs (Fig. 2g) were larger, with increased cytoplasmic area. Importantly, quantitative analysis using morphometry software corroborated these visual observations, confirming that the SP cells were significantly smaller in both cell area and diameter compared with the NSP and Unf cells (Table 1). Smaller condensed nuclei are a hallmark of quiescent cells (28–30), and, notably, SPs exhibited the smallest nuclear area of all fractions, despite the lack of a statistically significant difference in G0/G1 frequency, suggesting that they are, indeed, enriched for quiescent cells. Notably, despite the significantly smaller nuclear diameter of SPs, they exhibited, by far, the highest N/C ratio at 1.83, whereas the NSP (1.394) and Unf (1.390) N/C ratios were virtually identical. This ratio compares with an N/C ratio of 1.54 for the KSC-enriched α6briCD71dim subset (2), demonstrating that the SP represents the most primitive blast-like subset of any epidermal population identified thus far.
Table 1.
Cell size characteristics of Hoechst-stained epidermal cell populations
| Characteristic | Unf | NSP | SP | SP vs. NSP |
P |
NSP vs. Unf |
|---|---|---|---|---|---|---|
| SP vs. Unf | ||||||
| Cell area, μm2 | 91.40 ± 18.310 | 91.50 ± 22.08 | 77.84 ± 16.21 | 7.28 × 10−14 | 1.04 × 10−8 | 0.0691 |
| Cell diameter, μm | 10.79 ± 1.06 | 10.72 ± 1.29 | 9.90 ± 1.04 | 2.29 × 10−13 | 1.26 × 10−8 | 0.7760 |
| Nuclear area, μm2 | 55.54 ± 12.77 | 54.29 ± 14.70 | 51.71 ± 11.32 | 0.0002 | 2.35 × 10−15 | 0.8025 |
Unf, n = 270; NSP, n = 336; SP, n = 347.
Epidermal SP Exhibits Inferior in Vitro Proliferative Output but Superior in Vivo Epidermal Tissue-Regenerative Activity.
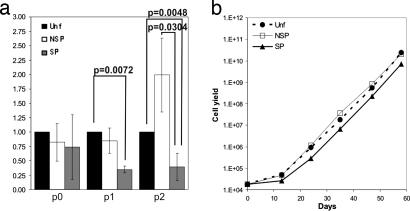
We have shown that short-term culture does not allow separation of KSCs from their immediate progeny in neonatal human foreskin epidermis (1). We therefore developed a method for long-term serial cultivation of adult murine keratinocytes (31), which we further optimized to determine the comparative proliferative output of the epidermal SP, NSP, and Unf populations. The data obtained showed that the SP displayed significantly lower cumulative cell yield at passages 1 and 2 (Fig. 3a). Beyond passage 2, there were no statistically significant differences in cell output between the fractions (Fig. 3b), presumably reflecting adaptation to culture and loss of intrinsic functional differences.
Fig. 3.
Cumulative in vitro proliferative output of Hoechst subsets. Hoechst subsets were plated at 104 cells per cm2 on col-IV in optimized AM-KGM and passaged at 103 cells per cm2 onto uncoated tissue culture plates. (a) The cumulative total cell output of SP and NSP subsets vs. Unf cells was compared up to passage 2. (b) The cell yield vs. time was monitored over four passages to reveal any differences in long-term output.
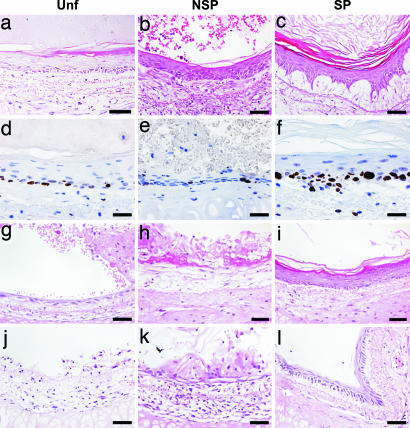
We then sought to determine the tissue-regenerative capacity of Hoechst subsetted keratinocytes, given that tissue reconstitution can rightly be regarded as a more rigorous assay for stem cells (32, 33). Denuded rat tracheas were inoculated with Hoechst fractions and transplanted s.c. into SCID recipients to assess their in vivo epidermal tissue regenerative capacity. This study revealed that all freshly isolated Hoechst fractions from adult murine epidermis were able to reconstitute a pluristratified epithelium for up to 6 weeks when transplanted without prior culture (Fig. 4a–c). Notably, only the SP subset gave rise to an epidermis exhibiting all of the hallmarks of the native tissue, including a polarized basal layer, multiple progressively flattening living cell layers, a granular layer, and thick stratum corneum sloughing from the surface in many contiguous strips (Fig. 4c), indicative of enrichment for tissue-regenerating cells in the SP fraction. This finding was further confirmed by analysis for Ki67 as a marker for the proliferative index of the epithelial sheets generated from primary cell transplants (Fig. 4 d–f). These data are consistent with the recent observation that human epidermal SPs isolated from cultures also exhibit sustained proliferative and organotypic potential (12).
Fig. 4.
In vivo epidermal tissue-regenerative activity of Hoechst subsets. Devitalized rat tracheas were inoculated with 5 × 105 irradiated p0 tail keratinocytes along with 5 × 104 total viable (a, g, and j), NSP (b, h, and k), and SP (c, i, and l) cells from adult murine tail epidermis freshly isolated (a–c) or cultured to passage 1 (g–i) and passage 2 (j–l). Epithelia regenerated from noncultured samples (a–c) were stained for Ki67 to assess the proliferative activity of Unf (d) vs. NSP (e) and SP (f) subsets in vivo. Transplants were harvested at 6 weeks (a–f) or 3 weeks (g–l). (Scale bars, 50 μm.)
It has been shown that the epidermal tissue-regenerative potential of human keratinocytes declines with serial culture (34), suggesting the uncoupling of in vitro cell-replicative capacity from tissue renewal. We therefore reasoned that partial exhaustion of this activity through serial culture would allow us to better distinguish between the Hoechst subsets in vivo. Thus, passage 1 (p1) Unf, SP, and NSP cells were assayed in transplants, and we observed that, although the Unf and NSP exhibited significant loss of tissue-regenerative ability, the SPs demonstrated sustained tissue-renewal capacity. Specifically, p1 Unf cells gave rise to a thin layer of epithelium with no progressive stratification and some cornified tissue harboring hyperkeratotic nuclei (Fig. 4g). The p1 NSP fraction fared worse still, appearing to differentiate almost immediately to produce dysplastic cornified tissue (Fig. 4h). In stark contrast, the p1 SP cells gave rise to a pluristratified epidermis with a densely populated basal layer and progressively flattening suprabasal layers, terminating in a granular layer with conspicuous keratohyalin granules and a stratum corneum (Fig. 4i). Thus, the SP cells outperformed the others in vivo, producing a pluristratified epithelium that more closely resembled their native tissue of origin. After two passages (Fig. 4 j–l), only the SP fraction was capable of sustained tissue regeneration (Fig. 4l). However, stratification was incomplete in all p2 fractions, perhaps reflecting the declining in vivo differentiation potential through successive in vitro passages. Collectively, the transplantation data demonstrate that the epidermal SPs are enriched for tissue-renewing cells compared with the Unf and NSP fractions, suggesting stem cell enrichment.
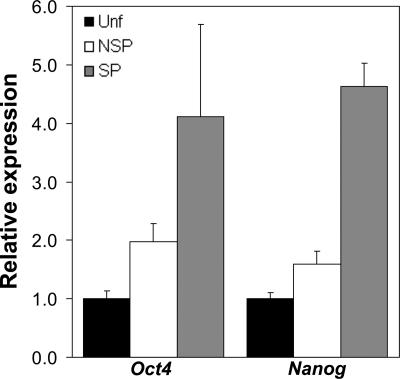
To further confirm the stem cell-like nature of the epidermal SP, we sought to investigate the expression of the transcription factors Oct4 and Nanog, critical mediators of self-renewal and pluripotency in embryonic stem cells (35, 36). Real-time PCR analysis of Unf, SP, and NSP fractions revealed that the epidermal SP was >4-fold enriched for both Oct4 and Nanog (Fig. 5), consistent with its greater tissue-renewal capacity. This finding is also consistent with observations reporting Oct4 in rare interfollicular basal cells of human epidermis in situ (37).
Fig. 5.
Relative expression of pluripotency genes Oct4 and Nanog in Hoechst subsets by quantitative real-time PCR. Quantitative real-time PCR was performed with cDNA made from Unf, NSP, and SP fractions by using optimized gene-specific primer sets to analyze expression levels of pluripotency markers Oct4 and Nanog normalized to EF-1α expression in corresponding samples.
Discussion
Although the existence of SPs in many tissues, including the epidermis, has been known for some time, their functional relevance to in vivo tissue renewal has been proven only in the hemopoietic system. The role of epidermal SPs has been unclear, with conflicting evidence for and against their representing a stem cell population largely based on correlative and phenotypic analysis. We have performed a functional and phenotypic analysis of murine epidermal SPs and demonstrate that this population satisfies many important criteria ascribed to KSCs, including low incidence, high N/C ratio, low complexity with blast-like morphology, high expression of Oct4 and Nanog, genes known to be highly expressed in self-renewing embryonic stem cells, and greatest tissue-renewal capacity after transplantation.
The comprehensive phenotypic characterization we undertook, particularly the absence of CD34, a marker for follicular-bulge stem cells, revealed that epidermal SPs are a bona fide interfollicular epidermal stem cell population not originating in the hair follicle. Interestingly, the epidermal SPs map to the KSC-enriched α6briCD71dim fraction and make up a minor subset of this population. Given that the NSP population contains the majority of the α6briCD71dim fraction and yet exhibited poorer tissue-renewal capacity, we suggest that epidermal SPs represent the most potent subset of KSCs within the α6briCD71dim fraction.
There is broad consensus that quiescence is a definitive KSC trait (4), and the SP subset was significantly depleted of mitotic G2/M cells. Similarly, bulge KSC-enriched cells exhibited only 1.36% G2/M cells as compared with 2.55% for total viable cells (23), and an elegant study using inducible histone H2B-driven GFP expression in the bulge demonstrated that only 0.5% of the GFPhigh LRCs were in G2/M-phase as compared with 2.7% for GFPlow nonstem cells (4). The latter study reported an ≈5-fold reduction in G2/M frequencies in KSCs, a finding that was mirrored in our analysis with respect to SPs vs. Unf keratinocytes. Although SP cells were significantly enriched for cells in S phase, the total lack of SPs in the proliferating α6briCD71bri TA fraction and paucity of G2/M cells is consistent with a very slow or arrested S phase for the majority of SPs. However, the relationship, if any, between SP functionality and cell cycling status remains to be determined. Indeed, bone marrow SPs in S-G2M were just as potent in reconstituting lethally irradiated recipients as those SPs in G0/G1 (38).
We have used relative long-term in vitro proliferative output to distinguish candidate stem cells from TA cells isolated from human epidermis. However, murine epidermal SPs exhibited poor proliferative activity in vitro compared with NSPs. It is possible that the in vitro SP results merely reflect their initial slow cycling status. This possibility is supported by observations of human KSCs that are initially slow to replicate in organotypic cultures (33) and has also been suggested recently for ocular SP cells (39). Another possibility is that not all SPs are recruited to proliferate in the culture conditions we used. Indeed, quiescent HSCs exhibit minimal in vitro clonogenicity unless specific cytokines are present (40), and germinative epidermal stem cells realize their immense in vitro proliferative potential only when combined with dermal papilla cells (22). Perhaps the more surprising observation is the uncoupling of proliferative ability in vitro from tissue reconstitution ability in vivo. Thus, although murine SP and NSP cells could be propagated in vitro quite efficiently over many passages, this propagation was not accompanied by the ability to reconstitute epithelial tissue. The hemopoietic system is once again instructive in interpreting this paradox, where it was shown several decades ago that cultivating hemopoietic stem cells led to loss of bone marrow reconstitution–cocultivation in Dexter cultures incorporating a stromal element was critical in restoring this activity (41). These data and our own previous (33) and current work strengthen the argument for placing candidate stem cells in the appropriate microenvironment to permit their tissue-regenerative abilities to be realized. Increasingly, it is becoming evident that epidermal stem cell self-renewal does not occur indefinitely in vitro, because the cell-replicative capacity of keratinocyte cultures is exhausted over time (33, 34). The loss of tissue-regenerative ability over time from Unf, (p1) NSP (p1), and even SP cells by p2 supports this notion but could also be attributed to the well documented chromosomal instability of murine keratinocytes in culture (42). Clearly, the lesser regenerative potential of cultured murine epidermal SPs (this study) vs. those of human origin (12) assists in more rapid discrimination of stem vs. TA cells in vivo. Collectively, these observations demonstrate that stem cell populations may lack critical proliferative stimuli in vitro and, therefore, caution against assigning stem cell status based solely on in vitro surrogate assays, as suggested in ref. 9. The most definitive evidence that the Hoechst SP represent a stem cell population of the epidermis is the detection of elevated levels of the self-renewal genes Oct4 and Nanog and the demonstrable superior in vivo epidermal tissue-reconstitution ability exhibited by these cells. In conclusion, Hoechst efflux ability may represent a universal stem cell trait, permitting isolation of viable stem cells from many tissues.
Materials and Methods
Culture of Adult Murine Tail Epidermal Keratinocytes.
Keratinocytes were isolated from adult C57B6/J mice and cultured as described (31), with some modifications. Briefly, adult mouse-keratinocyte growth medium (AM-KGM) was further optimized, consisting of KBM (Cambrex, Mt. Waverly, Australia) supplemented with 70 μg/ml bovine pituitary extract (Hammond Cell Technologies, Alameda, CA), 500 ng/ml hydrocortisone, 10 ng/ml EGF, 2 mg/ml BSA, 5 μg/ml insulin and transferrin, 40 pM triiodothyronine, and 100 ng/ml cholera toxin (Sigma, St. Louis, MO) with penicillin (1.2 μg/ml)/gentamycin (17 μg/ml), and diflucan (6 μg/ml). Cultures were initiated in triplicate at 104 cells per cm2 on human collagen-IV (Sigma) and replated at 103 per cm2 on tissue-culture plastic thereafter. Cumulative cell yields were calculated by assuming all cells from previous passages were replated.
Hoechst 33342 Staining and Phenotypic Characterization.
The method of Goodell et al. (38) was optimized to resolve an epidermal SP that was verapamil-sensitive and homogeneously SSClow, consistent with a pure population of actively effluxing undifferentiated cells. Cells were prewarmed for 15 min and then stained for 90 min at 37°C with 15 μM Hoechst 33342 at 106 cells per ml in PBS/2% FCS. Hoechst exclusion was inhibited by adding 100 μM verapamil. Cells were resuspended at 4 × 106 per ml with 100 μg/ml viability dye 7-amino-actinomycin D (Molecular Probes, Invitrogen, Melbourne, Australia) to exclude dead cells and sorted on a FACSDiva (BD Biosciences, Franklin Lakes, NJ) at 50 psi. Hoechst-DNA binding was detected by excitation with a UV laser (355 nm at 50 mW), followed by a 610-nm short-pass dichroic mirror and 670 LP and 450 BP 20 detection filters for red and blue emission wavelengths, respectively. Freshly isolated epidermal cells were immunostained for α6 (GoH3, 1:100) and CD71 (C2, 1:100) or CD34 (RAM34, 1:100) with antibodies from BD Biosciences as described (43) and sorted before Hoechst staining. Immunophenotyping for K14, c-kit, CD45, and Oil red O staining was performed as described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Cell-Cycle Analysis.
Hoechst subsets were resuspended in 400 μl of PBS with 50 μg/ml propidium iodide, 100 μg/ml RNase A, and 0.1% (vol/vol) IGEPAL CA-630 nonionic detergent (Sigma). Samples were incubated at 37°C for 30 min and immediately analyzed on a FACScan flow cytometer (BD Biosciences). The proportion of cells in each phase of the cell cycle was estimated by using ModFit software (Verity Software House, Topsham, ME) supplied with the flow cytometer.
Measurement of Cell Size and Cytoplasmic and Nuclear Area.
Cytospins of Hoechst fractions were Giemsa-stained and photographed microscopically for automated morphometry software image analysis to determine nuclear and total cell areas and to calculate the N/C ratio for at least 270 cells per fraction. Statistical significance was calculated with Student’s paired t test.
In Vivo Epidermal Tissue Regeneration.
Epidermal tissue regeneration in rat trachea transplants was performed as described (44). Briefly, tracheas aseptically removed from adult Sprague–Dawley rats were denuded by multiple cycles of freeze–thawing and inoculated with 5 × 104 to 1 × 105 Hoechst-stained cells along with 5 × 105 p0 lethally irradiated (12 Gy) adult murine support keratinocytes in a 1:1 mixture of optimized AM-KGM and DMEM with 10% (vol/vol) FCS, 20 ng/ml EGF, 400 ng/ml hydrocortisone, and 10 ng/ml cholera toxin (Sigma). For each condition, two SCID mice were transplanted s.c. with two tracheas each for harvest at 3–6 weeks, and each experiment was replicated two to three times. Tracheas excised from recipients were fixed in 4% (wt/vol) buffered formalin and decalcified in 20% (wt/vol) EDTA, pH 8.5, for 24 h at 4°C and processed for routine histology.
Quantitative Real-Time PCR.
mRNA was isolated from sorted cells by using the RNeasy kit (Qiagen, Valencia, CA) and reverse-transcribed into cDNA with SuperScript III (Invitrogen) as per the manufacturer’s instructions. Gene-specific primers for Oct4 (forward, 5′-GAAGTTGGAGAAGGTGGAACCA-3′; reverse, 5′-GCTTCAGCAGCTTGGCAAA-3′, 1 μM) and Nanog (forward, 5′-TCTTCCTGGTCCCCACAGTTT-3′; reverse, 5′-GCAAGAATAGTTCTCGGGATGAA-3′, 100 nM) were optimized for expression analyses by real-time PCR on the ABI 7500 thermocycler (Applied Biosystems, Foster City, CA) using ABsolute QPCR SYBR Green ROX mix (ABgene, Epsom, U.K.) with 2 min at 50°C, 15 min at 95°C, 40 cycles of 15 s at 95°C, and 1 min at 60°C, followed by dissociation-curve analysis to confirm specificity. Mouse embryonic stem cells and H2O were used as positive and negative controls, respectively. EF-1α (forward, 5′-ATTCGAGACCAGCAAATACTATGTGA-3′; reverse, 5′-AGCCTGGGATGTGCCTGTAA-3′, 200 nM) was used for normalization, and relative expression was calculated by using the comparative CT method (45).
Supplementary Material
Acknowledgments
We thank Ralph Rossi and Andrew Fryga for invaluable FACS expertise, Stewart Fabb and Makoto Tanaka for technical assistance with real-time PCR, and Brenda Aisbett for histology work. R.P.R. was supported by scholarships from the Cancer Council of Victoria and the Australian Stem Cell Centre. This work was supported by grants from the National Health and Medical Research Council of Australia (to P.K.).
Abbreviations
- KSC
keratinocyte stem cell
- LRC
label-retaining cell
- N/C
nuclear/cytoplasmic
- NSP
non-SP
- SSC
side scatter
- SP
side population
- TA
transit-amplifying
- Unf
unfractionated.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Li A., Simmons P. J., Kaur P. Proc. Natl. Acad. Sci. USA. 1998;95:3902–3907. doi: 10.1073/pnas.95.7.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tani H., Morris R. J., Kaur P. Proc. Natl. Acad. Sci. USA. 2000;97:10960–10965. doi: 10.1073/pnas.97.20.10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun K. M., Niemann C., Jensen U. B., Sundberg J. P., Silva-Vargas V., Watt F. M. Development (Cambridge, U.K.) 2003;130:5241–5255. doi: 10.1242/dev.00703. [DOI] [PubMed] [Google Scholar]
- 4.Tumbar T., Guasch G., Greco V., Blanpain C., Lowry W. E., Rendl M., Fuchs E. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris R. J., Liu Y., Marles L., Yang Z., Trempus C., Li S., Lin J. S., Sawicki J. A., Cotsarelis G. Nat. Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 6.Ohyama M., Terunuma A., Tock C. L., Radonovich M. F., Pise-Masison C. A., Hopping S. B., Brady J. N., Udey M. C., Vogel J. C. J. Clin. Invest. 2006;116:249–260. doi: 10.1172/JCI26043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montanaro F., Liadaki K., Volinski J., Flint A., Kunkel L. M. Proc. Natl. Acad. Sci. USA. 2003;100:9336–9341. doi: 10.1073/pnas.1133179100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terunuma A., Jackson K. L., Kapoor V., Telford W. G., Vogel J. C. J. Invest. Dermatol. 2003;121:1095–1103. doi: 10.1046/j.1523-1747.2003.12531.x. [DOI] [PubMed] [Google Scholar]
- 9.Kaur P., Li A., Redvers R., Bertoncello I. J. Invest. Dermatol. Symp. Proc.; 2004. pp. 238–247. [DOI] [PubMed] [Google Scholar]
- 10.Triel C., Vestergaard M. E., Bolund L., Jensen T. G., Jensen U. B. Exp. Cell Res. 2004;295:79–90. doi: 10.1016/j.yexcr.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 11.Yano S., Ito Y., Fujimoto M., Hamazaki T. S., Tamaki K., Okochi H. Stem Cells. 2005;23:834–841. doi: 10.1634/stemcells.2004-0226. [DOI] [PubMed] [Google Scholar]
- 12.Larderet G., Fortunel N. O., Vaigot P., Cegalerba M., Maltere P., Zobiri O., Gidrol X., Waksman G., Martin M. T. Stem Cells. 2006;24:965–974. doi: 10.1634/stemcells.2005-0196. [DOI] [PubMed] [Google Scholar]
- 13.Montanaro F., Liadaki K., Schienda J., Flint A., Gussoni E., Kunkel L. M. Exp. Cell Res. 2004;298:144–154. doi: 10.1016/j.yexcr.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Cotsarelis G., Sun T. T., Lavker R. M. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 15.Hulspas R., Quesenberry P. J. Cytometry. 2000;40:245–250. [PubMed] [Google Scholar]
- 16.Uchida N., Fujisaki T., Eaves A. C., Eaves C. J. J. Clin. Invest. 2001;108:1071–1077. doi: 10.1172/JCI13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asakura A., Rudnicki M. A. Exp. Hematol. 2002;30:1339–1345. doi: 10.1016/s0301-472x(02)00954-2. [DOI] [PubMed] [Google Scholar]
- 18.Al-Barwari S. E., Potten C. S. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1976;30:201–216. doi: 10.1080/09553007614550981. [DOI] [PubMed] [Google Scholar]
- 19.Morris R., Argyris T. S. Cancer Res. 1983;43:4935–4942. [PubMed] [Google Scholar]
- 20.Alvi A. J., Clayton H., Joshi C., Enver T., Ashworth A., Vivanco M. M., Dale T. C., Smalley M. J. Breast Cancer Res. 2003;5:R1–R8. doi: 10.1186/bcr547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavker R. M., Sun T. T. Science. 1982;215:1239–1241. doi: 10.1126/science.7058342. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds A. J., Jahoda C. A. J. Cell Sci. 1991;99:373–385. doi: 10.1242/jcs.99.2.373. [DOI] [PubMed] [Google Scholar]
- 23.Trempus C. S., Morris R. J., Bortner C. D., Cotsarelis G., Faircloth R. S., Reece J. M., Tennant R. W. J. Invest. Dermatol. 2003;120:501–511. doi: 10.1046/j.1523-1747.2003.12088.x. [DOI] [PubMed] [Google Scholar]
- 24.Potten C. S. J. Invest. Dermatol. 1975;65:488–500. doi: 10.1111/1523-1747.ep12610194. [DOI] [PubMed] [Google Scholar]
- 25.Barrandon Y., Green H. Proc. Natl. Acad. Sci. USA. 1985;82:5390–5394. doi: 10.1073/pnas.82.16.5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pavlovitch J. H., Rizk-Rabin M., Gervaise M., Metezeau P., Grunwald D. Am. J. Physiol. 1989;256:C977–C986. doi: 10.1152/ajpcell.1989.256.5.C977. [DOI] [PubMed] [Google Scholar]
- 27.Webb A., Li A., Kaur P. Differentiation (Berlin) 2004;72:387–395. doi: 10.1111/j.1432-0436.2004.07208005.x. [DOI] [PubMed] [Google Scholar]
- 28.Darzynkiewicz Z., Traganos F. In: Genetic Expression in the Cell Cycle. McCarthy K. S., Padilla G. M., editors. Vol. 1. New York: Academic; 1982. pp. 103–128. [Google Scholar]
- 29.Wheeler K. T., Nelson G. B., Terrell K. E., Wallen C. A. Int. J. Radiat. Biol. 1988;54:245–255. doi: 10.1080/09553008814551681. [DOI] [PubMed] [Google Scholar]
- 30.Mares V., Giordano P. A., Pellicciari C., Scherini E., Lisa V., Bottone M. G., Bottiroli G. Cell Prolif. 1991;24:569–577. doi: 10.1111/j.1365-2184.1991.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 31.Redvers R. P., Kaur P. In: Epidermal Cells: Methods and Protocols, Methods in Molecular Biology. Turksen K., editor. Vol. 289. Totowa, NJ: Humana; 2005. pp. 15–22. [Google Scholar]
- 32.Potten C. S., Loeffler M. Development (Cambridge, U.K.) 1990;110:1001–1020. doi: 10.1242/dev.110.4.1001. [DOI] [PubMed] [Google Scholar]
- 33.Li A., Pouliot N., Redvers R., Kaur P. J. Clin. Invest. 2004;113:390–400. doi: 10.1172/JCI19140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fortunel N. O., Hatzfeld J. A., Rosemary P. A., Ferraris C., Monier M. N., Haydont V., Longuet J., Brethon B., Lim B., Castiel I., et al. J. Cell Sci. 2003;116:4043–4052. doi: 10.1242/jcs.00702. [DOI] [PubMed] [Google Scholar]
- 35.Pesce M., Scholer H. R. Stem Cells. 2001;19:271–278. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- 36.Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 37.Tai M. H., Chang C. C., Kiupel M., Webster J. D., Olson L. K., Trosko J. E. Carcinogenesis. 2005;26:495–502. doi: 10.1093/carcin/bgh321. [DOI] [PubMed] [Google Scholar]
- 38.Goodell M. A., Brose K., Paradis G., Conner A. S., Mulligan R. C. J. Exp. Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Budak M. T., Alpdogan O. S., Zhou M., Lavker R. M., Akinci M. A., Wolosin J. M. J. Cell Sci. 2005;118:1715–1724. doi: 10.1242/jcs.02279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haylock D. N., To L. B., Dowse T. L., Juttner C. A., Simmons P. J. Blood. 1992;80:1405–1412. [PubMed] [Google Scholar]
- 41.Dexter T. M., Allen T. D., Lajtha L. G. J. Cell Physiol. 1977;91:335–344. doi: 10.1002/jcp.1040910303. [DOI] [PubMed] [Google Scholar]
- 42.Bruegel Sanchez V. L., Zhou J., LaCivita D., Milstone L. M. J. Invest. Dermatol. 2004;123:403–404. doi: 10.1111/j.0022-202X.2004.23218.x. [DOI] [PubMed] [Google Scholar]
- 43.Li A., Kaur P. In: Epidermal Cells: Methods and Protocols, Methods in Molecular Biology. Turksen K., editor. Vol. 289. Totowa, NJ: Humana; 2005. pp. 87–96. [Google Scholar]
- 44.Pouliot N., Redvers R. P., Ellis S., Saunders N. A., Kaur P. Exp. Dermatol. 2005;14:60–69. doi: 10.1111/j.0906-6705.2005.00252.x. [DOI] [PubMed] [Google Scholar]
- 45.Livak K. J., Schmittgen T. D. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.