Abstract
Aerial plant surfaces are colonized by diverse bacteria such as the ubiquitous Methylobacterium spp. The specific physiological traits as well as the underlying regulatory mechanisms for bacterial plant colonization are largely unknown. The purpose of this study was to identify proteins produced specifically in the phyllosphere by comparing the proteome of Methylobacterium extorquens colonizing the leaves either with that of bacteria colonizing the roots or with that of bacteria growing on synthetic medium. We identified 45 proteins that were more abundant in M. extorquens present on plant surfaces as compared with bacteria growing on synthetic medium, including 9 proteins that were more abundant on leaves compared with roots. Among the proteins induced during epiphytic growth, we found enzymes involved in methanol utilization, prominent stress proteins, and proteins of unknown function. In addition, we detected a previously undescribed type of two-domain response regulator, named PhyR, that consists of an N-terminal sigma factor (RpoE)-like domain and a C-terminal receiver domain and is predicted to be present in essentially all Alphaproteobacteria. The importance of PhyR was demonstrated through phenotypic tests of a deletion mutant strain shown to be deficient in plant colonization. Among PhyR-regulated gene products, we found a number of general stress proteins and, in particular, proteins known to be involved in the oxidative stress response such as KatE, SodA, AhpC, Ohr, Trx, and Dps. The PhyR-regulated gene products partially overlap with the bacterial in planta-induced proteome, suggesting that PhyR is a key regulator for adaptation to epiphytic life of M. extorquens.
Keywords: fitness, sigma factor, stress, two-component system
Molecular microbial ecology is often hampered by the difficulty of unraveling how the environment shapes bacterial physiology and allows microorganisms to multiply. One such habitat is the aerial parts of plants that are colonized by various microorganisms, mostly bacteria, which are often found in numbers averaging 106 to 107 cells per cm2. Epiphytes, defined as bacteria that are capable of multiplying on plant surfaces, encounter rather harsh conditions in the phyllosphere environment. This habitat is generally considered to be poor in nutrients. In addition, residing microorganisms are exposed to the atmosphere and radiation and are subjected to rapid changes with respect to their physical environment (1). Many plant-colonizing bacteria do no apparent harm to their hosts and might even be beneficial to the plant, whereas others are plant pathogens and can, after establishment of an epiphytic population, ultimately destroy the tissue on which they are living.
The chemical and physical features of leaf surfaces are not well known, and the same is true for the traits that allow these bacteria to multiply in the leaf habitat. Our current knowledge about bacterial physiology in the phyllosphere stems mainly from targeted approaches. Thus, phenotypes such as flagellar motility, UV-mediated mutagenic repair, and exopolysaccharide production contribute substantially to epiphytic fitness (2–4). In addition, random mutagenesis has been performed to identify novel targets important for phyllosphere colonization (5). Gene expression profiling is a good strategy for providing information about adaptations to specific conditions or environments. One powerful strategy for targeting gene expression in the natural context is through promoter trap analysis [i.e., in vivo expression technology (IVET)] (6). Variants of this approach have been successfully applied to identify genes induced during phyllosphere colonization of bacterial pathogens (7–9). However, by definition, the IVET strategy can only give an incomplete picture of the physiology of bacteria. Indeed, it is well known that changes at the protein level are not necessarily predictable from transcript levels because of differences in translation efficiency, proteolysis, and posttranslational modifications. In this study, we have therefore chosen a more direct way to gain insights into the physiology of bacteria in the phyllosphere through the analysis of the proteome of a bacterium in this ecosystem.
For this work, we have used the Alphaproteobacterium Methylobacterium extorquens AM1, a well studied model pink-pigmented facultative methylotroph (PPFM) (10), whose draft genome sequence is available (see www.integratedgenomics.com/genomereleases.html#6). Methylobacterium spp. are common leaf epiphytes that represent an important bacterial population on leaves (11, 12) and have been found on all analyzed plants (13). Methylobacterium spp. on plant surfaces benefit from methanol produced by plants (14) by means of methylotrophy (10, 15). However, methanol is not the only carbon substrate that these bacteria are able to consume in the phyllosphere (14). The presence of Methylobacterium may be beneficial to plants through the production of plant hormones (13, 16). The ubiquitous presence of Methylobacterium on plant surfaces makes them an interesting model for discovering the particular traits that these bacteria have acquired as successful epipytes. This work provides the identification of previously undescribed candidate proteins of Methylobacterium required for phyllosphere colonization and, in particular, the identification of a key regulator controlling adaptation to this habitat.
Results and Discussion
Proteome Analysis of M. extorquens During Phyllosphere Colonization.
With the aim to identify proteins that are specifically induced when M. extorquens AM1 colonizes the phyllosphere of Arabidopsis thaliana ColO plants, we performed a differential analysis of the proteome of M. extorquens that had colonized plants under gnotobiotic conditions after seed inoculation by comparison with the proteome of bacteria that were cultivated on the surface of synthetic minimal medium (MM) under the same conditions of light and temperature. We used succinate as a carbon source, because it enters directly into the tricarboxylic acid (TCA) cycle and allows us to observe the induction of methylotrophy markers (17). Proteins were separated by 2D gel electrophoresis (2-DE), and we identified those that were induced at least 3-fold based on image analysis (Fig. 1). In total, 40 proteins were identified as up-regulated during phyllosphere colonization (Table 1). To distinguish between proteins specific for phyllosphere colonization with respect to more general epiphytic adaptation, we compared the proteome of bacteria from the aerial parts with that of rhizosphere-colonizing bacteria. We identified 9 proteins that were >3-fold induced relative to the rhizosphere proteome (Table 1), out of which 5 were not identified in the earlier comparison. The high similarity between the phyllosphere and rhizosphere proteomes suggests a similar adaptation to the epiphytic state in both plant environments. Relatively few down-regulated proteins were identified (see Table 3, which is published as supporting information on the PNAS web site), and these generally corresponded to housekeeping proteins, which could reflect a general down-regulation of metabolism during epiphytic growth compared with in vitro conditions.
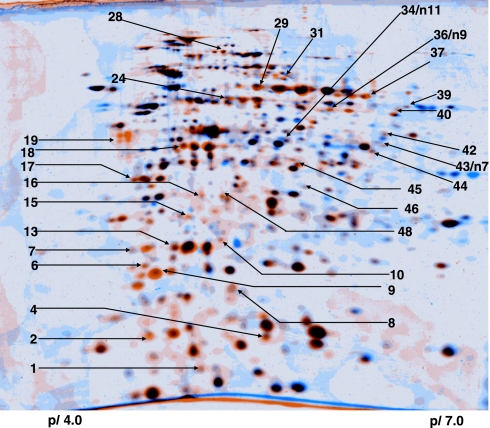
Fig. 1.
Dual-channel image analysis of 2-DE protein pattern of M. extorquens AM1 to reveal proteins induced during phyllosphere colonization. Proteins from cells harvested from the phyllosphere of A. thaliana ColO are colored in orange, and those of cells harvested from the surface of solid MM containing succinate as the only carbon source are colored in blue. Spots of identified proteins are marked (see Table 1).
Table 1.
List of proteins from M. extorquens AM1 found to be induced during phyllosphere (P) colonization relative to MM and rhizosphere (R) colonization
| Spot no.* | RMQ no. | Gene product(s)†† | CD search | Mr | pl | Ratio,‡ P/MM | Ratio,‡ P/R |
|---|---|---|---|---|---|---|---|
| Metabolism | |||||||
| 29 | RMQ05966 | MxaF, methanol dehydrogenase, large subunit (M31108) | pfam01011 | 67.2 | 5.8 | + | |
| 48 | RMQ00044 | MxaJ protein (M31108) | pfam00497 | 27.4 | 6.0 | ∞ | |
| 4 | RMQ09682 | Fae, formaldehyde activating enzyme (L43136) | 20.7 | 7.0 | + | ||
| 16 | RMQ08765 | PqqB, PQQ biosynthesis polypeptide (L25889) | 30.6 | 5.4 | ∞ | ||
| 34 n11 | RMQ03830 | PhaA, β-ketothiolase (AF287907) | pfam00108 | 44.1 | 6.7 | + | + |
| 42 | RMQ09548 | Crr, crotonyl-CoA reductase (L48340) | pfam00107 | 47.5 | 6.3 | ∞ | |
| 2 | RMQ01365 | Gap20 (AF442749) | 19.0 | 5.8 | ∞ | ||
| 45 | RMQ05381 | Malyl-CoA lyase-like protein | pfam03328 | 37.9 | 5.8 | ∞ | |
| 24 n10 | RMQ03452 | Aldehyde dehydrogenase | pfam00171 | 62.8 | 6.4 | + | ∞ |
| 28 | RMQ07560 | Xanthine oxidase-related aldehyde oxidoreductase | pfam02738 | 80.6 | 5.3 | + | |
| 44 | RMQ07805 | Putative NADP-dependent oxidoreductase | pfam00107 | 35.9 | 6.0 | + | |
| P12 | RMQ11717 | Putative quinoprotein | 36.0 | 6.9 | + | ||
| 1 | RMQ02894 | GloA, lactoyglutathione lyase | pfam00903 | 16.5 | 5.7 | + | |
| n3 | RMQ09259 | Adenylate kinase | pfam00406 | 21.3 | 5.1 | + | |
| Transport | |||||||
| n4 | RMQ08930 | ABC-type Fe+ transport system, periplasmic component | 36.6 | 6.6 | + | ||
| n6 | RMQ06383 | ABC-type sulfate transport system, periplasmic component | 24.1 | 7.8 | + | ||
| 15 | RMQ02495 | Putative amino acid binding protein | pfam00497 | 28.5 | 5.2 | ∞ | |
| 31 | RMQ05493 | Putative oligopeptide binding protein | pfam00496 | 70.2 | 6.7 | + | |
| Stress proteins | |||||||
| P28 | RMQ01248 | DegP/HrA, Trypsin-like serine proteases | pfam00089 | 50.4 | 6.7 | ∞ | |
| 40 | RMQ04833 | DegP/HrA, Trypsin-like serine proteases | pfam00089 | 51.2 | 8.4 | ∞ | |
| n5 | RMQ08088 | ClpP, ATP-dependent Clp protease proteolytic subunit | pfam00574 | 23.1 | 5.8 | + | |
| 6 | RMQ05519 | Protease I (Serine protease) DJ-1/Pfpl family (GSP18) | pfam01965 | 20.7 | 4.9 | ∞ | |
| P22 | RMQ06501 | Hsp70, heat-shock protein 70 (DnaK) | pfam00012 | 51.9 | 4.9 | + | |
| P35 | RMQ06982 | Hsp70, heat-shock protein 70 (DnaK) | pfam00012 | 68.5 | 5.2 | + | |
| P4 | RMQ02206 | Hsp20, heat-shock protein 20 | pfam00011 | 18.4 | 5.2 | ∞ | |
| P15 | RMQ02531 | SodA, Superoxide dismutase | pfam02777 | 22.5 | 5.8 | + | |
| 37a | RMQ09549 | KatE, catalase (L48340) | pfam00199 | 63.5 | 7.1 | ∞ | |
| 37b | RMQ11789 | KatE, catalase | pfam00199 | 59.9 | 5.8 | ∞ | |
| 9 | RMQ05258 | Dps, DNA protection protein | pfam00210 | 19.9 | 5.0 | + | |
| Proteins of unknown function | |||||||
| 7, 12 | RMQ09016 | NfU-like/thioredoxin-like protein | pfam01106 | 20.3 | 4.8 | ∞ | |
| 43 n7 | RMQ10082 | Major royal jelly protein§ | pfam03022 | 41.4 | 6.3 | ∞ | + |
| P26 | RMQ06718 | Major royal jelly protein§ | pfam03022 | 40.2 | 5.4 | ∞ | |
| n12 | RMQ07439 | Protein of unknown function | 68.3 | 6.4 | + | ||
| 8 | RMQ10020 | Protein of unknown function | pfam05974 | 18.5 | 5.4 | ∞ | |
| 10 | RMQ09099 | Protein of unknown function | 21.1 | 5.9 | ∞ | ||
| 13 | RMQ08861 | Protein of unknown function | 20.3 | 5.3 | ∞ | ||
| 17 | RMQ03063 | Protein of unknown function | 31.7 | 4.9 | ∞ | ||
| 18 | RMQ00428 | Protein of unknown function§ | 31.9 | 5.3 | + | ||
| P30 | RMQ05730 | Protein of unknown function§ | 28.9 | 6.8 | + | ||
| 19 | RMQ01102 | Protein of unknown function | 43.5 | 5.0 | ∞ | ||
| P25 | RMQ00267 | Protein of unknown function | 39.1 | 5.2 | ∞ | ||
| 39 | RMQ03107 | Protein of unknown function | pfam00450 | 55.3 | 6.4 | ∞ | |
| 46 | RMQ03170 | Orf88, dioxygenase (AY034474) | pfam00903 | 36.4 | 6.2 | + | |
| 36 n8 | RMQ09688 | Putative nucleoside binding protein | 54.7 | 9.0 | ∞ | ∞ | |
| Regulator | |||||||
| P16 | RMQ08198 | Response regulator (PhyR, this work) | pfam06182 | 29.1 | 4.8 | ∞ | |
*Spot numbers that are preceded by “P” were identified on gels from independent experiments stained with silver nitrate rather than with SYPRO Ruby. Spot numbers preceded by “n” were detected to be induced in bacteria that were grown in the phyllosphere with respect to the rhizosphere. All other proteins were identified from the proteome of bacteria grown in the phyllosphere with respect to minimal medium supplemented with succinate.
†Accession nos. are in parentheses.
‡Spots indicated as “∞” were only detectable in the proteome from bacteria grown in the phyllosphere and not in the references (mm and rhizosphere, respectively). Spots indicated as “+” were found to be at least 3-fold induced. Proteins were identified from 2D gels stained with SYPRO Ruby and found in two biological repetitions whereby the majority of the spots were in addition also found induced on gels that were stained with silver nitrate.
Among the proteins induced during bacterial growth in the phytosphere (leaf and root surface environments), key markers of methylotrophic metabolism (17) were found to be up-regulated (e.g., MxaF and Fae) with respect to growth on synthetic medium containing succinate as a carbon source (Table 1). The induction of these enzymes upon epiphytic growth is in agreement with a previous study in which an advantage of wild-type (WT) M. extorquens cells in competition with methylotrophy-minus mutants was demonstrated, suggesting methanol utilization by the methylotroph (14). Another protein induced during phyllosphere colonization was PhaA, which initiates synthesis of the reserve polyhydroxy butyrate (PHB) (ref. 18; Table 1). It has been shown that PHB formation is stimulated by a deficiency of nutrients such as NH4+, SO42−, Mg2+, Fe2+, or Mn2+ (19). The observed induction of PhaA might thus represent part of a general adaptation to nutrient-limiting conditions as would be expected for phyllospheric growth where the carbon source might not be a growth-limiting factor (14). The nature of this limiting factor is possibly suggested by the phyllosphere-specific induction of two putative periplasmic ABC transporter components predicted to be involved in iron and sulfate uptake (Table 1). Interestingly, iron and sulfate have been suggested to be critical for phyllosphere colonization in other organisms (9).
We also found several putative dehydrogenases/oxidoreductases to be induced during phytospheric growth (Table 1). Of these, RMQ03452 is phyllosphere-specific, sharing a high percentage of sequence identity with the AcoD of Ralstonia eutrophus, which is involved in the catabolism of acetoin and ethanol (20), and with AldB of Escherichia coli, which is thought to have a role in detoxifying alcohols and aldehydes (21). The RMQ03452 protein in M. extorquens might contribute to carbon dissimilation or detoxification of alcohols or aldehydes that are produced by plants (22). Methylobacterium spp. are specialists in dealing with toxic compounds, as is already clear when considering that formaldehyde is a central intermediate of methylotrophic metabolism (10, 15). This question of detoxification vs. catabolism also arises for a putative lactoylglutathione lyase (GloA) (Table 1). In E. coli, GloA is required for detoxification of methylglyoxal, which is known to cause DNA damage (23). Because it has been reported that methylglyoxal is formed during catabolism of certain amino acids and other compounds such as acetone (24), methylglyoxal might therefore also be an intermediate produced upon breakdown of nutrients of the facultative methylotroph.
The analysis of the in planta proteome of M. extorquens AM1 clearly reflects an adaptation to survival under stress conditions (Table 1). The identified stress proteins fall into two classes: chaperones/proteases and oxidative stress-related proteins. Among the former are two paralogues of the periplasmic DegP/HtrA family (25), which suggests a response to extracytoplasmic stress and/or the need for assistance in the maturation of components of the cell envelope required for epiphytic growth. In addition, there is a predicted protease of the DJ-1/Pfp-1 superfamily that is a homologue of the general stress protein 18 (GSP18) of Bacillus subtilis (26) and heat-shock proteins that are well known to be induced by various types of environmental stress (27). The oxidative stress response is suggested by the up-regulation of superoxide dismutase, catalases, and the Dps protein. Dps is a nonspecific DNA-binding protein and a key component of the protection strategy against H2O2 (28, 29), UV irradiation (30), and electrophiles such as methylglyoxal (23, 28) (see above). The formation of reactive oxygen species (ROS) is a normal event and a by-product of electron transport under aerobic conditions (31). Because the detoxification of ROS becomes particularly important under starvation conditions (32), the observed induction of ROS-removing enzymes in M. extorquens AM1 might thus be a reaction to endogenously formed ROS. On the other hand, it is well known that plant cells challenge bacteria by means of an oxidative burst (33) and superoxide dismutase (SOD), and Kat and Dps have been shown to counteract the toxic effects of ROS produced by plants (34, 35). All of the stress proteins that we identified appeared to be epiphytic-specific rather than phyllosphere-specific. However, another protein, ClpP, was found to be induced in the phyllophere rather than in the rhizosphere. Clp proteases of E. coli are known to play an important role in cytoplasmic quality control and participate in numerous regulatory mechanisms that are important in nongrowing or slow-growing cells. ClpP interacts with ClpX or ClpA, which exhibit different substrate specificities (36).
A Response Regulator Common to Alphaproteobacteria Essential for Plant Colonization.
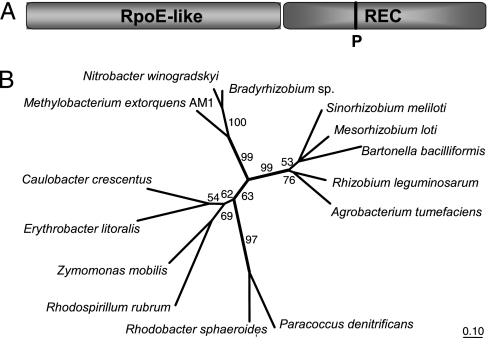
The analysis of the in planta proteome of M. extorquens AM1 revealed the induction of a putative two-component system response regulator, RMQ08198, that we named PhyR (for “phyllosphere-induced regulator”). This protein is interesting in several respects as follows. (i) A National Center for Biotechnology Information (NCBI) CD search revealed that PhyR carries a RpoE (σE)-like domain at the N terminus of the protein (Fig. 2A). A sigma-factor-like domain has not yet been described as part of a response regulator and might suggest that PhyR could possibly initiate transcription by itself. (ii) The sigma factor RpoE plays a major role in maintaining the integrity and function of the envelope and provides resistance to environmental stresses including desiccation and oxidative stress in Pseudomonas spp. (4, 37, 38). (iii) The domain structure of PhyR shows that the predicted phosphorable receiver domain is located at the C terminus (Fig. 2A), although this domain is usually located at the N terminus in described response regulators (39).
Fig. 2.
Structure and phylogeny of PhyR. (A) Predicted two-domain structure of PhyR. A strictly conserved aspartate residue (corresponding to position 190 of PhyR) is predicted to be the phosphorylation site (P) of the receiver domain according to Prosite (www.expasy.ch). (B) Phylogenetic tree of PhyR homologues in various Alphaproteobacteria using the treepuzzle algorithm and the Jones–Taylor–Thornton evolutionary model and based on the amino acid sequences aligned with ClustalW (see Fig. 4, which is published as supporting information on the PNAS web site). Branches that were recovered in <50% of 1,000 reconstructed treepuzzle trees are shown as multifurcations; percentage values for branches with ≥50% recovery are given in the tree. Original tree construction included all available PhyR homologue sequences currently available in the GenBank database. A selection was made of 13 representative sequences plus PhyR of M. extorquens AM1.
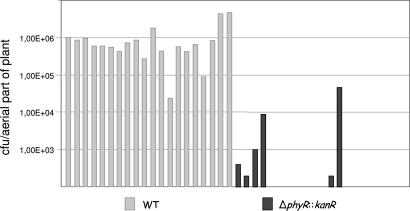
The identification of a previously undescribed type of response regulator in the proteome of in planta-grown M. extorquens AM1 prompted us to evaluate the importance of PhyR for plant colonization by constructing a deletion strain. Growth rates of the mutant were found to be unaltered with respect to WT when plant colonization was mimicked in vitro under mixed growth conditions (i.e., in the presence of succinate and methanol) (14). However, in planta colonization experiments revealed a severe growth defect of the PhyR deletion mutant (Fig. 3). Cell numbers of the mutant were below the detection limit for 65% of 3-week-old plants. When we then cloned the PhyR gene in trans, we were able to restore the colonization capacity to the WT level (data not shown).
Fig. 3.
Plant colonization of M. extorquens WT and the phyR deletion strain. The detection limit was at 102 cfu per plant. Three other independent experiments showed congruent results (data not shown).
Interestingly, a BLAST search revealed that PhyR homologues are present in essentially all free-living Alphaproteobacteria for which a genome sequence is available, but not in any other bacteria. This finding clearly indicates a more general function for PhyR homologues than adaptation to the phyllosphere and an ancient origin within this proteobacterial subgroup based on phylogenetic analysis (Fig. 2B).
Identification of Proteins That Are Positively Regulated by PhyR.
The phenotype of phyR mutants indicate that it is an important regulator and that one or more important physiological traits of phyllospheric growth are under PhyR control. To identify genes that are induced by PhyR, we performed proteome analysis with 2-DE so that we could readily recognize proteins that had been identified during the in planta proteome analysis. To this end, we cloned phyR in the expression vector pCM80 (40) and introduced the plasmid into M. extorquens AM1 ΔphyR. The proteome of this phyR-overexpressing strain was compared with the phyR-deficient strain containing the empty vector as control. The 42 proteins that we identified as PhyR-regulated are in Table 2.
Table 2.
List of proteins found to be positively regulated by PhyR
| Spot no. | RMQ no. | Gene product* | CD search | Mr | pl | Ratio† phy R+ vs. phyR− |
|---|---|---|---|---|---|---|
| Found in phyllosphere proteome (see Table 1) | ||||||
| 20, 21 | RMQ08198 | (PhyR, response regulator) | pfam06182 | 29.1 | 4.8 | ∞ |
| 1, 2 | RMQ09549 | KatE, catalase (L48340) | pfam00199 | 63.5 | 7.1 | + |
| 12 | RMQ02531 | SodA, superoxide dismutase | pfam02777 | 22.5 | 5.8 | + |
| 26, 28 | RMQ02206 | Hsp20, heat-shock protein 20 | pfam00011 | 18.4 | 5.2 | ++ |
| 29 | RMQ05258 | Dps, DNA protection protein | pfam00210 | 19.9 | 5.0 | ∞ |
| 4 | RMQ11717 | Putative quinoprotein (glucose dehydrogenase) | 36.0 | 6.9 | + | |
| 38 | RMQ02894 | Putative lactoylglutathione lyase | pfam00903 | 16.5 | 5.7 | ∞ |
| 8 | RMQ03170 | Orf88, dioxygenase (glyoxalase family protein) (AY034474) | pfam00903 | 36.4 | 6.2 | ∞ |
| 43 | RMQ01365 | Gap20 (AF442748) | 19.0 | 5.8 | + | |
| 25 | RMQ09016 | NifU-like/thioredoxin-like protein | pfam01106 | 20.3 | 4.8 | ∞ |
| 24 | RMQ08861 | Protein of unknown function | 20.3 | 5.3 | ∞ | |
| 15 | RMQ00428 | Protein of unknown function | 31.9 | 5.3 | + | |
| Additional proteins under PhyR control | ||||||
| 3a | RMQ06018 | Glutathione-dependent formaldehyde dehydrogenase | pfam00107 | 42.2 | 5.7 | ∞ |
| 5 | RMQ01240 | ADH, alcohol dehydrogenase | pfam00107 | 39.1 | 6.5 | ∞ |
| 6 | RMQ00842 | ADH, alcohol dehydrogenase | pfam00107 | 35.2 | 6.3 | ∞ |
| 7a | RMQ07799 | Short-chain dehydrogenases/reductases (GSP39) | pfam00106 | 30.8 | 5.9 | ∞ |
| 23 | RMQ00500 | Short-chain dehydrogenases/reductases | pfam00106 | 27.6 | 5.1 | ∞ |
| 51 | RMQ11711 | PccA, propionyl-CoA carboxylase (AY181038) | pfam02786 | 75.6 | 4.9 | + |
| 14 | RMQ06958 | MclA, malyl-CoA lyase (U72662) | 38.0 | 5.6 | + | |
| 18 | RMQ06488 | MDH, malate dehydrogenase (L33465) | pfam02866 | 39.1 | 6.6 | ∞ |
| 30, 31 | RMQ02884 | Phosphoglycerate mutase | pfam00300 | 23.9 | 5.5 | + |
| 48 | RMQ06654 | NAD(P) transhydrogenase α-subunit | pfam01262 | 39.6 | 5.6 | ∞ |
| 19 | RMQ01528 | FixB, Electron transfer flavoprotein, α-subunit | pfam00766 | 32.5 | 4.9 | ∞ |
| 11 | RMQ02643 | WrbA, flavoprotein | pfam00258 | 21.1 | 6.2 | ∞ |
| 32 | RMQ00895 | Carbonic anhydrase | pfam00484 | 28.4 | 9.1 | + |
| 36 | RMQ06760 | Ohr, organic hydroperoxide resistance protein-like protein (GSP17o) | pfam02566 | 15.0 | 5.7 | ∞ |
| 40 | RMQ06032 | AhpC (alkyl hydroperoxide reductase)/TSA (thiol specific antioxidant) family protein | pfam00578 | 18.0 | 5.9 | + |
| 45 | RMQ07144 | Trx, thioredoxin | pfam00085 | 17.4 | 9.7 | + |
| 7b | RMQ06181 | Putative haloacetate dehalogenase/non-heme chloroperoxidase | pfam00561 | 30.6 | 5.8 | ∞ |
| 33 | RMQ07321 | Putative phospholipid-binding proteins | pfam01161 | 19.7 | 5.5 | ∞ |
| 10 | RMQ11238 | Putative 3-hydroxyisobutyrate dehydrogenase | 20.8 | 5.7 | + | |
| 22 | RMQ12169 | CinA-like protein | pfam00994 | 25.0 | 4.8 | + |
| 27 | RMQ09145 | Hbd, d-β-hydroxybutyrate dehydrogenase (AY391854) | pfam00106 | 31.9 | 8.7 | + |
| 42 | RMQ02665 | GreA, transcription elongation factor | pfam01272 | 22.1 | 6.9 | + |
| 13 | RMQ06933 | EF-Ts, elongation factor | pfam00889 | 35.1 | 5.3 | + |
| 50 | RMQ07718 | CzcB, Cobalt-zinc-cadmium resistance protein | pfam00529 | 46.3 | 6.2 | + |
| 3b | RMQ03224 | Hypothetical signaling protein | 42.4 | 8.2 | ∞ | |
| 34 | RMQ04652 | Protein of unknown function (GSP26) | pfam01243 | 19.0 | 5.7 | ∞ |
| 9 | RMQ05442 | Protein of unknown function | pfam01442 | 28.4 | 5.8 | + |
| 41 | RMQ01392 | Protein of unknown function | pfam03928 | 14.8 | 5.0 | + |
| 35 | RMQ12283 | Protein of unknown function | 14.9 | 5.6 | ∞ | |
| 37 | RMQ12368 | Protein of unknown function | 12.7 | 6.6 | ∞ | |
| 44 | RMQ07641 | Protein of unknown function | 16.8 | 5.4 | ∞ | |
*Accession nos. are in parentheses.
†Spots indicated as “∞” were only detectable in the proteome from M. extorquens AM1 ΔphyR pBG11 (phyR overexpression) and not in the strain containing pCM80 (phyR minus). Spots indicated as “+” were found to be at least 3-fold induced.
These results show that PhyR is partly responsible for the induction of some of the proteins that we had found to be induced during phyllospheric growth of the bacterium. However, we are unable to distinguish whether they are directly or indirectly regulated by PhyR. Among these proteins are KatE, SodA, Hsp20, Dps, GloA, and several uncharacterized proteins. As mentioned earlier, these proteins are known to be involved in coping with stress caused by electrophiles (GloA, Dps) and ROS (KatE, SodA, Dps). The latter group comprises proteins protective not only against superoxide anions and hydrogen peroxide but also alkyl hydroperoxides (see induction of Ohr and AhpC; Table 2) that are all important components of the plant defense response against microbial infection (33, 41) and by-products of aerobic metabolism (see above).
Several dehydrogenases (pfam00107 and -00106) were found to be induced by PhyR. The substrate spectrum and role of these dehydrogenases is unknown. They might be involved in substrate utilization during starvation to furnish the additional energy supply associated with processes such as repair of oxidized proteins and lipids. Nevertheless, a possible role for one of the putative dehydrogenases (RMQ06018) can be proposed. This protein represents a putative glutathione-dependent formaldehyde dehydrogenase (42) that might fulfill an auxiliary role coping with excess formaldehyde alongside the well described H4MPT- and H4F-dependent pathways for formaldehyde oxidation in Methylobacterium (15).
In several model bacteria, stress responses have been well studied. Whereas many regulators are specifically involved in one type of stress, other regulators control diverse functions. σS is a master regulator of the general stress response in bacteria that belong to Gammaproteobacteria (43). Mutants deficient in σS are less able to survive upon starvation and are more sensitive to oxidative and osmotic stress as well as UV and desiccation stress in both logarithmic- and stationary-phase cells in Enterobacteriaceae and pseudomonads (44–46). In Bacillus, σB has been postulated to be the functional homologue of σS. The general stress regulon of σB provides the cells with a nonspecific, multiple, and preventive stress resistance in which the protection against oxidative stress is an essential part of the response (47).
PhyR has a central role in the adaptation of Methylobacterium to the plant environment. Our proteome analysis points to a rather large PhyR-dependent regulon within which the oxidative stress response is an important part (Table 2), reminiscent of the role of σS/σB in adaptation for surviving stress and starvation in nature (43, 47). So far, the regulatory elements representing the functional homologues of σS/σB with their corresponding activation mechanisms in Alphaproteobacteria are unknown. It is therefore tempting to speculate that PhyR is involved in a σS/σB-like response. Biochemical analysis will be important to clarify whether PhyR represents a chimeric protein with a functional output domain that acts as a bona fide sigma factor, suggested by the σE-like domain.
Conclusions
Little is known about traits important for phyllosphere colonization, and even less is known about the regulatory mechanisms that determine the adaptation of plant epiphytes in general and Methylobacterium spp. in particular. Our proteome profiling approach for bacteria that have colonized the phyllosphere is clearly advantageous in detecting the up-regulation of proteins that might have partially overlapping functions, as suggested by the identification of protein paralogues. This work provides a list of candidate proteins that need to be analyzed in more detail with respect to their importance for bacterial fitness. In addition, we have identified a previously undescribed type of two-domain regulator termed PhyR, which plays a key role in the adaptation of bacteria for plant colonization. The PhyR regulon suggests a role in dealing with the various stresses that the bacteria are likely to encounter in the phyllosphere. We assume that this regulator is also of importance in other Alphaproteobacteria.
Experimental Procedures
Bacterial Growth Under in Vitro and in Planta Conditions.
For plant inoculation experiments, M. extorquens AM1 was grown in liquid MM (48) containing succinate (20 mM) to midexponential growth phase (OD600 = 1.2), centrifuged, washed, and resuspended in 10 mM MgCl2. The bacteria were adjusted to an OD600 of 1.0 (108 cfu per ml) and used for seed inoculation (4-h shaking at room temperature with slight moving) after sterilization of A. thaliana (ecotype Columbia) seeds. Sterilization of seeds was achieved through incubation in 2.4% hypochlorite for 5 min followed by eight washing steps. The plants were allowed to develop under controlled conditions on Murashige and Skoog medium in growth chambers under sterile conditions in Magenta boxes (1 week at 20°C, 16 h light/8 h darkness; 2 weeks at 22°C, 9 h light/15 h darkness). Preliminary experiments were performed to determine a suitable time point of harvest of M. extorquens AM1 from plants. It was chosen at 3 weeks to ensure that the overall bacterial population showed logarithmic development at this time point (average cell population: 106 cfu per aerial part of each plant). Sterility of uninoculated plants was verified by sonication of leaves in phosphate buffer and plating on KingB medium. For each experiment, ≈150 plants were grown. In addition, bacterial precultures (105 cfu/ml) were spread on the surface of agar-solidified MM complemented with 20 mM succinate for 5 days at 22°C, 9 h light/15 h darkness.
Harvest of Bacteria and Preparation of Cell Extracts.
The aerial parts of the plants were separated from the roots by cutting with a razor blade, and bacteria were harvested in aliquots of 15 plants in 50-ml plastic tubes filled with cooled TE buffer (10 mM Tris, pH 7.5/1 mM EDTA) supplemented with PMSF (0.3 mg/ml) and Percoll (GE Healthcare, Uppsala, Sweden; 20% final concentration) through alternating sonication and vortexing (45 s/30 s, 3 times). The suspension was centrifuged (12,000 × g, 4°C for 10 min) whereby the addition of Percoll facilitated sedimentation of the bacteria, leaving small plant debris in the supernatant. Cells from one experiment were washed, pooled, and frozen until further use. Bacteria from roots and in vitro conditions were treated in parallel in a similar way. Total proteins were extracted by using a French pressure cell at 108 Pa (two times, 4°C), and the cell extract was recovered after centrifugation (13,000 × g, 4°C for 30 min).
Proteome Analysis.
2-DE was performed with 18-cm immobilized pH gradient strips (4.0–7.0; GE Healthcare) as described (17). Five independent experiments were performed, whereby the material (aerial parts, roots, in vitro) from two experiments was subjected to SYPRO Ruby staining (Molecular Probes, Leiden, The Netherlands; using 350 μg of protein) and from three experiments to silver nitrate staining (using 120 μg of protein). To identify proteins associated with epiphytic growth of M. extorquens, images were analyzed by using the Delta 2D software package (Decodon, Greifswald, Germany). Only proteins that were at least 3-fold induced in the independent experiments were identified. Protein identification was performed by peptide mass fingerprinting as described (17) and liquid chromotography/tandem mass spectrometry (49). Identification of differentially expressed proteins was performed independently from the different gels that represent the different biological repetitions and that were stained with SYPRO Ruby and silver nitrate, respectively, and had to give congruent results.
Mutant Generation and Construction of Complementation Strains.
A phyR mutant was generated by using the suicide vector pCM184 (50). Complementation of the phyR deletion mutant was achieved through cloning of phyR with its presumed promoter region by using the forward primer Prom-Phy-f-BamHI tggatcctgccgcgactacgacaaacgag (located 454 nt upstream of the predicted start codon) and the reverse primer Phy-r-KpnI catcggccggtaccttttcacgg into the XbaI and HindIII sites of the broad host range cloning vector pCM62 (40) resulting in pBG17. The plasmid was subsequently introduced in the ΔphyR::kanR mutant. In addition, phyR was cloned downstream of the mxaF promoter into the PstI/KpnI site of the expression vector pCM80 (40) resulting in pBG11 by using the primer Phy-PstI-f: catggctgcagcagcaacg and Phy-CM80-r-KpnI mentioned above.
Phenotypic Analysis of M. extorquens AM1 Constructs and Analysis of the PhyR Regulon.
Colonization of the phyR deletion strain was compared with M. extorquens AM1 WT and the complemented strain ΔphyR::kanR pBG17. For this purpose, plant inoculation experiments were performed. For sampling, the aerial parts of the plants were placed individually in 1 ml of MM and sonicated for 5 min in an ultrasonication bath. Cell suspensions were then serially diluted and plated onto MM. To identify PhyR-regulated genes, we performed a differential proteome analysis by using M. extorquens AM1 ΔphyR::kanR containing pBG11 and pCM80, respectively, grown to midexponential growth phase in the presence of succinate. Cells were harvested, washed in ice-cold TE buffer supplemented with PMSF, and cell extracts were prepared, and proteome analysis was performed as described above.
Supplementary Material
Acknowledgments
We thank Claudia Knief (Laboratoire des Interactions Plantes Micro-Organismes) for tree calculation. This work was supported by a grant from the Centre National de la Recherche Scientifique “Program Protéomique et genie des protéines.” B.G. was supported by a fellowship of the French Ministry of Research. J.A.V. was supported by the Max-Planck-Gesellschaft.
Abbreviations
- MM
minimal medium
- ROS
reactive oxygen species
- 2-DE
2D gel electrophoresis.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ845291).
References
- 1.Lindow S. E., Brandl M. T. Appl. Environ. Microbiol. 2003;69:1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haefele D. M., Lindow S. E. Appl. Environ. Microbiol. 1987;53:2528–2533. doi: 10.1128/aem.53.10.2528-2533.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sundin G. W., Murillo J. Environ. Microbiol. 1999;1:75–87. doi: 10.1046/j.1462-2920.1999.00008.x. [DOI] [PubMed] [Google Scholar]
- 4.Yu J., Penaloza-Vazquez A., Chakrabarty A. M., Bender C. L. Mol. Microbiol. 1999;33:712–720. doi: 10.1046/j.1365-2958.1999.01516.x. [DOI] [PubMed] [Google Scholar]
- 5.Lindow S. E. Appl. Environ. Microbiol. 1993;59:1586–1592. doi: 10.1128/aem.59.5.1586-1592.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rainey P. B., Preston G. M. Curr. Opin. Biotechnol. 2000;11:440–444. doi: 10.1016/s0958-1669(00)00132-4. [DOI] [PubMed] [Google Scholar]
- 7.Boch J., Joardar V., Gao L., Robertson T. L., Lim M., Kunkel B. N. Mol. Microbiol. 2002;44:73–88. doi: 10.1046/j.1365-2958.2002.02877.x. [DOI] [PubMed] [Google Scholar]
- 8.Yang S., Perna N. T., Cooksey D. A., Okinaka Y., Lindow S. E., Ibekwe A. M., Keen N. T., Yang C. H. Mol. Plant Microb. Interact. 2004;17:999–1008. doi: 10.1094/MPMI.2004.17.9.999. [DOI] [PubMed] [Google Scholar]
- 9.Marco M. L., Legac J., Lindow S. E. Environ. Microbiol. 2005;7:1379–1391. doi: 10.1111/j.1462-2920.2005.00825.x. [DOI] [PubMed] [Google Scholar]
- 10.Chistoserdova L., Chen S. W., Lapidus A., Lidstrom M. E. J. Bacteriol. 2003;185:2980–2987. doi: 10.1128/JB.185.10.2980-2987.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirano S. S., Upper C. D. In: Microbial Ecology of Leaves. Andrews J. H., Hirano S. S., editors. New York: Springer; 1991. pp. 271–294. [Google Scholar]
- 12.Corpe W. A., Rheem S. FEMS Microbiol. Ecol. 1989;62:243–250. [Google Scholar]
- 13.Holland M. A. Recent Res. Dev. Plant Physiol. 1997;1:207–213. [Google Scholar]
- 14.Sy A., Timmers A. C., Knief C., Vorholt J. A. Appl. Environ. Microbiol. 2005;71:7245–7252. doi: 10.1128/AEM.71.11.7245-7252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vorholt J. A. Arch. Microbiol. 2002;178:239–249. doi: 10.1007/s00203-002-0450-2. [DOI] [PubMed] [Google Scholar]
- 16.Koenig R. L., Morris R. O., Polacco J. C. J. Bacteriol. 2002;184:1832–1842. doi: 10.1128/JB.184.7.1832-1842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laukel M., Rossignol M., Borderies G., Völker U., Vorholt J. A. Proteomics. 2004;4:1247–1264. doi: 10.1002/pmic.200300713. [DOI] [PubMed] [Google Scholar]
- 18.Korotkova N., Lidstrom M. E. J. Bacteriol. 2001;183:1038–1046. doi: 10.1128/JB.183.3.1038-1046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourque D., Pomerleau Y., Groleau D. Appl. Microbiol. Biotechnol. 1995;44:367–376. [Google Scholar]
- 20.Priefert H., Krüger N., Jendrossek D., Schmidt B., Steinbüchel A. J. Bacteriol. 1992;174:899–907. doi: 10.1128/jb.174.3.899-907.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J., Johnson R. C. J. Bacteriol. 1995;177:3166–3175. doi: 10.1128/jb.177.11.3166-3175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graus M., Schnitzler J. P., Hansel A., Cojocariu C., Rennenberg H., Wisthaler A., Kreuzwieser J. Plant Physiol. 2004;135:1967–1975. doi: 10.1104/pp.104.043240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Booth I. R., Ferguson G. P., Miller S., Li C., Gunasekera B., Kinghorn S. Biochem. Soc. Trans. 2003;31:1406–1408. doi: 10.1042/bst0311406. [DOI] [PubMed] [Google Scholar]
- 24.Cooper R. A. Annu. Rev. Microbiol. 1984;38:49–68. doi: 10.1146/annurev.mi.38.100184.000405. [DOI] [PubMed] [Google Scholar]
- 25.Spiess C., Beil A., Ehrmann M. Cell. 1999;97:339–347. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- 26.Antelmann H., Bernhardt J., Schmid R., Mach H., Volker U., Hecker M. Electrophoresis. 1997;18:1451–1463. doi: 10.1002/elps.1150180820. [DOI] [PubMed] [Google Scholar]
- 27.Weiner L., Model P. Proc. Natl. Acad. Sci. USA. 1994;91:2191–2195. doi: 10.1073/pnas.91.6.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez A., Kolter R. J. Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L., Helmann J. D. Mol. Microbiol. 1995;18:295–300. doi: 10.1111/j.1365-2958.1995.mmi_18020295.x. [DOI] [PubMed] [Google Scholar]
- 30.Nair S., Finkel S. E. J. Bacteriol. 2004;186:4192–4198. doi: 10.1128/JB.186.13.4192-4198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez-Flecha B., Dimple B. J. Bacteriol. 1997;179:382–388. doi: 10.1128/jb.179.2.382-388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nystrom T. Annu. Rev. Microbiol. 2004;58:161–181. doi: 10.1146/annurev.micro.58.030603.123818. [DOI] [PubMed] [Google Scholar]
- 33.Levine A., Tenhaken R., Dixon R., Lamb C. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 34.Ceci P., Ilari A., Falvo E., Chiancone E. J. Biol. Chem. 2003;278:20319–20326. doi: 10.1074/jbc.M302114200. [DOI] [PubMed] [Google Scholar]
- 35.Kim Y. C., Miller C. D., Anderson A. J. Appl. Environ. Microbiol. 2000;66:1460–1467. doi: 10.1128/aem.66.4.1460-1467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weichart D., Querfurth N., Dreger M., Hengge-Aronis R. J. Bacteriol. 2003;185:115–125. doi: 10.1128/JB.185.1.115-125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schnider-Keel U., Lejbolle K. B., Baehler E., Haas D., Keel C. Appl. Environ. Microbiol. 2001;67:5683–5693. doi: 10.1128/AEM.67.12.5683-5693.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keith L. M., Bender C. L. J. Bacteriol. 1999;181:7176–7184. doi: 10.1128/jb.181.23.7176-7184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West A. H., Stock A. M. Trends Biochem. Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 40.Marx C. J., Lidstrom M. E. Microbiology. 2001;147:2065–2075. doi: 10.1099/00221287-147-8-2065. [DOI] [PubMed] [Google Scholar]
- 41.Jalloul A., Montillet J. L., Assigbetse K., Agnel J. P., Delannoy E., Triantaphylides C., Daniel J. F., Marmey P., Geiger J. P., Nicole M. Plant J. 2002;32:1–12. doi: 10.1046/j.1365-313x.2002.01393.x. [DOI] [PubMed] [Google Scholar]
- 42.Gutheil W. G., Kasimoglu E., Nicholson P. C. Biochem. Biophys. Res. Commun. 1997;238:693–696. doi: 10.1016/s0006-291x(00)90000-7. [DOI] [PubMed] [Google Scholar]
- 43.Hengge-Aronis R. Microbiol. Mol. Biol. Rev. 2002;66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarniguet A., Kraus J., Henkels M. D., Muehlchen A. M., Loper J. E. Proc. Natl. Acad. Sci. USA. 1995;92:12255–12259. doi: 10.1073/pnas.92.26.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller C. D., Kim Y. C., Anderson A. J. Can. J. Microbiol. 2001;47:41–48. doi: 10.1139/w00-123. [DOI] [PubMed] [Google Scholar]
- 46.Stockwell V. O., Loper J. E. Microbiol. 2005;151:3001–3009. doi: 10.1099/mic.0.28077-0. [DOI] [PubMed] [Google Scholar]
- 47.Hecker M., Völker U. Adv. Microb. Physiol. 2001;44:35–91. doi: 10.1016/s0065-2911(01)44011-2. [DOI] [PubMed] [Google Scholar]
- 48.Harder W., Attwood M., Quayle J. R. J. Gen. Microbiol. 1973;78:155–163. [Google Scholar]
- 49.Boudart G., Jamet E., Rossignol M., Lafitte C., Borderies G., Jauneau A., Esquerre-Tugaye M. T., Pont-Lezica R. Proteomics. 2005;5:212–221. doi: 10.1002/pmic.200400882. [DOI] [PubMed] [Google Scholar]
- 50.Marx C. J., Lidstrom M. E. BioTechniques. 2002;33:1062–1067. doi: 10.2144/02335rr01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.