Abstract
Neutral mutations may hitchhike to high frequency when they are situated close to sites under positive selection, generating local reductions in genetic diversity. This process is thought to be an important determinant of levels of genomic variation in natural populations. The size of genome regions affected by genetic hitchhiking is expected to be dependent on the strength of selection, but there is little empirical data supporting this prediction. Here, we compare microsatellite variation around two drug resistance genes (chloroquine resistance transporter (pfcrt), chromosome 7, and dihydrofolate reductase (dhfr), chromosome 4) in malaria parasite populations exposed to strong (Thailand) or weak selection (Laos) by anti-malarial drugs. In each population, we examined the point mutations underlying resistance and length variation at 22 (chromosome 4) or 25 (chromosome 7) microsatellite markers across these chromosomes. All parasites from Thailand carried the K76T mutation in pfcrt conferring resistance to chloroquine (CQ) and 2–4 mutations in dhfr conferring resistance to pyrimethamine. By contrast, we found both wild-type and resistant alleles at both genes in Laos. There were dramatic differences in the extent of hitchhiking in the two countries. The size of genome regions affected was smaller in Laos than in Thailand. We observed significant reduction in variation relative to sensitive parasites for 34–64 kb (2–4 cM) in Laos on chromosome 4, compared with 98–137 kb (6–8 cM) in Thailand. Similarly, on chromosome 7, we observed reduced variation for 34–69 kb (2–4 cM) around pfcrt in Laos, but for 195–268 kb (11–16 cM) in Thailand. Reduction in genetic variation was also less extreme in Laos than in Thailand. Most loci were monomorphic in a 12 kb region surrounding both genes on resistant chromosomes from Thailand, whereas in Laos, even loci immediately proximal to selective sites showed some variation on resistant chromosomes. Finally, linkage disequilibrium (LD) decayed more rapidly around resistant pfcrt and dhfr alleles from Laos than from Thailand. These results demonstrate that different realizations of the same selective sweeps may vary considerably in size and shape, in a manner broadly consistent with selection history. From a practical perspective, genomic regions containing resistance genes may be most effectively located by genome-wide association in populations exposed to strong drug selection. However, the lower levels of LD surrounding resistance alleles in populations under weak selection may simplify identification of functional mutations.
Keywords: hitchhiking, selection, drug resistance, Plasmodium falciparum, chloroquine, pyrimethamine
1. Introduction
Selection intensity, rates of recombination and mutation, and population size are thought to be the critical parameters determining the impact of selection on neutral genetic variation surrounding a selected site. The most dramatic reductions in diversity around selected genes should result from strong selection acting on large populations of organisms with low rates of recombination and mutation (Wiehe 1998; Kim & Stephan 2002). However, even when all parameters are constant, simulations suggest that different realizations of selection events will show considerable variation in their impact on flanking genetic variability (Kim & Stephan 2002). Malaria parasites provide a useful system for exploring theoretical predictions about the genomic effects of selection. Dramatic selective sweeps have been described on both chromosomes 4 and 7 of Plasmodium falciparum (Wootton et al. 2002; Nair et al. 2003; Roper et al. 2003). On chromosome 7, variation has been removed for more than 200 kb around the chloroquine resistance transporter (pfcrt) as a consequence of selection by chloroquine (CQ; Wootton et al. 2002). Similarly, on chromosome 4, variation is reduced for approximately 100 kb (6 cM) around dihydrofolate reductase (dhfr), the gene underlying resistance to pyrimethamine in parasites from Thailand (Nair et al. 2003).
Resistance alleles at both pfcrt and dhfr have a single evolutionary origin in Southeast Asian countries (Wootton et al. 2002; Nair et al. 2003) and have subsequently invaded Africa resulting in selective sweeps on both chromosomes 4 and 7 spanning two continents (Roper et al. 2004). In this paper, we examine genomic variation resulting from different realizations of these two selection events in neighbouring countries adjacent to the Mekong River, Thailand and the Lao PDR (Laos). The strength of anti-malarial selection differs markedly in these two countries, allowing us to examine the impact of selection strength on hitchhiking. We cannot directly compare selection coefficients in the two locations. However, both clinical and molecular marker data suggest that selection is considerably stronger in Thailand than in Laos. This information is reviewed briefly below.
(a) Chloroquine
Resistance to CQ was first reported on the Thailand/Cambodia border in 1962 (Harinasuta et al. 1965) and in other countries on the in the Mekong region shortly afterwards (Cambodia, 1962; Vietnam, 1964; Burma, 1969). Resistance was suspected during this time in Laos, and clinical failures were reported in 1966 (Farid 1967) and soon after by Ebisawa et al. (1970). Hence, CQ resistance was first recorded in both countries only a few years apart. In Thailand, clinical resistance spread rapidly, rendering CQ ineffective by 1982. These clinical data suggest that CQ drug resistance genes reached fixation in Thailand in approximately 20 years, which is equivalent to approximately 120 parasite generations if we assume a generation time of two months in Plasmodium. Clinical resistance spread much more slowly in Laos, and CQ remains the first-line drug against malaria in this country. Recent clinical trials indicate unsatisfactory cure rates for CQ and only now are policy changes being considered (Guthmann et al. 2002; Mayxay et al. 2003; Schwobel et al. 2003). Molecular marker data are consistent with the clinical data. The CQ resistance transporter (pfcrt) is a major gene underlying CQ resistance. Southeast Asian resistance alleles have eight mutations that distinguish them from wild-type alleles, of which the K76T mutation is critical (Fidock et al. 2000). All parasites of more than 300 examined from four Thai locations carried the K76T mutation critical for CQ resistance (see Anderson et al. submitted). By comparison, the distribution of resistant alleles appears to be patchy in Laos, with some studies indicating high levels of the K76T mutations (Pillai et al. 2001; Berens et al. 2003), but other studies finding wild-type allele frequencies of 5–35% (Anderson et al. submitted). Taken together, the data demonstrate that K76T mutation has spread more rapidly in Thailand than in Laos.
(b) Sulfadoxine–pyrimethamine (SP)
In Thailand, SP was introduced as a first-line treatment in 1973. Even before this date, low levels of clinical failure were reported (Chin et al. 1973). On the Thailand–Burma (Myanmar) border, SP was widely used by 1976, and resistance spread to fixation in approximately 6 years (approximately 30 Plasmodium generations; White 1992; Nair et al. 2003). In Laos, SP was first used in the 1960s, and has been the secondly line anti-malarial drug in Laos since then. However, it is used infrequently in both the public and private sectors, in comparison with CQ. Clinical resistance was first reported in one out of 26 patients examined in 1970 (Ebisawa et al. 1970), suggesting that resistance mutations in dhfr were present at this time. However, unlike in Thailand, clinical resistance has spread slowly. Recent clinical trials show an 82% cure rate using this drug in both Phalanxay (Mayxay et al. 2003), one of the populations sampled for this study, and on the southern border of Laos (Schwobel et al. 2003). The current distribution of mutations underlying pyrimethamine resistance is consistent with the clinical data and also suggests weaker selection in Laos than in Thailand. All parasites examined from Mawker–Thai (Thailand) carry mutations conferring resistance in dhfr, while more than 30% of parasites carry wild-type alleles in the locations in Laos studied in this research (Nair et al. 2003). Furthermore, while most resistant alleles carry four mutations in Thailand, and more than 60% carry the critical I164L mutation that renders pyrimethamine treatment ineffective, in parasites from Laos, the predominant resistance alleles carry just two mutations and the I164L mutation is rare or absent (Berens et al. 2003; Nair et al. 2003). Furthermore, mutations in another gene, dihydropteroate synthase (dhps) that confer resistance to sulfaldoxine (the partner drug for pyrimethamine in SP) tell a similar story. All parasites examined in Mawker–Thai contain 2–3 mutations conferring resistance in dhps (Nair et al. 2002). By contrast, more than 80% of parasite from Laos contain wild-type dhps alleles (Anderson et al. submitted). The combination of clinical and molecular data suggests slower evolution of SP drug resistance in Laos than in Thailand.
We compare patterns of microsatellite length variation flanking pfcrt and dhfr in parasite populations collected from both Thailand and Laos. These comparisons reveal dramatic differences in the extent of hitchhiking in these two countries, with narrower and shallower valleys of reduced variation surrounding drug resistance mutations on parasite chromosomes from Laos.
2. Material and methods
(a) Collection of parasites
Collection locations in both Thailand and Laos have been described by Nair et al. (2003). In brief, we obtained venous blood from malaria patients visiting the malaria clinic in Mawker–Thai, while finger-prick blood samples (approximately 50 μl blood adsorbed onto 3 mm whatmann paper and air dried) were collected from patients in Sekong and Phalanxay in southern Laos (figure 1). DNA was prepared from venous blood by phenol/chloroform extraction and from filter-paper samples using the Gentra systems Card Capture kit.
Figure 1.
Map showing sample collection sites in Thailand and Laos.
(b) Removal of multiple clone infections
Infections containing multiple malaria clones were not included in the analysis since they complicate the construction of haplotypes. We genotyped all infections using a panel of six informative microsatellites to identify multiple clone infections and excluded samples showing more than one allele at any of the loci. The microsatellite loci amplified were ARA2 (chromosome 11), Polyα (chromosome 4), TA1 (chromosome 6), TA60 (chromosome 13), TA81 (chromosome 5) and G377 (chromosome 12). Oligo sequences and amplification conditions have been described previously (Anderson et al. 1999). Multiple clone infections were defined as those in which one or more of the six loci showed multiple alleles. Minor alleles were scored if they were more than 33% the height of the peaks corresponding to the predominant alleles.
(c) Genotyping of dhfr and pfcrt
We genotyped 4-point mutations in dhfr by primer extension using the SnaPshot kit (Applied Biosystems; Nair et al. 2002). These data have been reported previously for both Thai and Laos populations studied here (Nair et al. 2003). We genotyped the K76T mutation in pfcrt by digesting 6FAM end-labelled PCR products with ApoI and scoring the restriction fragments on a capillary sequencer (Anderson et al. 2003). Another seven amino acid mutations occur in resistant pfcrt in Southeast Asia (Fidock et al. 2000). These additional single nucleotide polymorphisms (SNPs) are in perfect linkage disequilibrium (LD) with the K76T mutation in Mawker–Thai (Anderson et al. submitted) and so were not genotyped here.
(d) Genotyping of flanking microsatellite markers
(i) Chromosome 4
Marker details and primer sequences, and patterns of variation in the Thai population have been described for markers flanking dhfr (Nair et al. 2003). To genotype these markers in parasites from Laos, we used a slightly different strategy, since finger-prick samples (rather than venous blood) were available. We designed an additional oligo and used a two round hemi-nested amplification strategy for each locus (Anderson et al. 1999). Twenty-two of the markers analysed previously could be amplified using this approach and were included in the study.
(ii) Chromosome 7
We searched chromosome 7 for perfect di-nucleotide microsatellites containing 10 or more (AT)n repeats. The arrangement of markers across chromosome 7 is shown in figure 2. The spacing is quite dense around pfcrt with five markers placed in a 22 kb region. Markers more than 10 kb from pfcrt were spaced at intervals of 13–80 kb (mean=36 kb). For samples from Laos, we designed an additional flanking oligo that was used to preamplify DNA as described above. The three oligos used for each microsatellite locus on chromosome 4 and 7 are listed in the Electronic Appendix.
Figure 2.
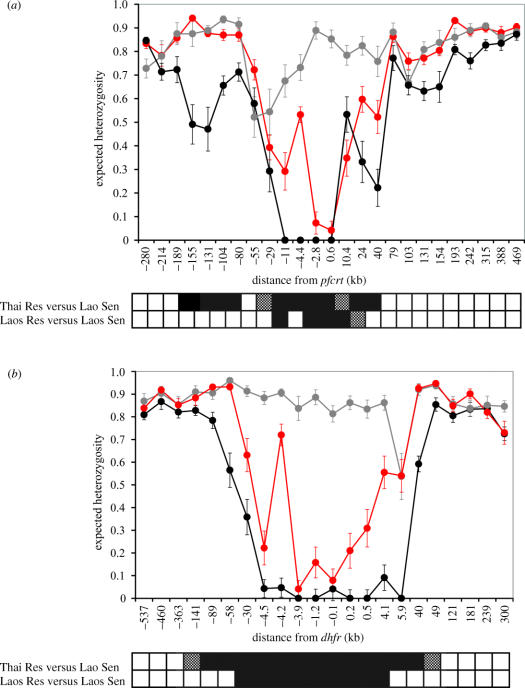
Patterns of genetic variation around pfcrt and dhfr. (a) and (b) show He (±1 s.d.) plotted against position on the chromosomes relative to pfcrt (chromosome 7) and dhfr (chromosome 4). The markers are ordered along the x-axis, which is not linear. Chromosomes bearing wild-type alleles are shown in grey, those bearing resistant alleles are shown in red (Laos) or black (Thailand). The distance from pfcrt and dhfr at which He differs significantly (p<0.01) between wild-type and resistant chromosomes is shown in the bars underneath the graph. Significance values were derived by permutation: black, p<0.001; stippled, p<0.01.
(e) Analysis
We measured expected heterozygosity (He) at each locus using the formula , where n is the number of infections sampled and pi is the frequency of the ith allele. For haploid blood stage malaria parasites, this statistic measures the probability of drawing two different alleles from a population. The sampling variance of He was calculated as (a slight modification of the standard diploid variance; Nei 1987). We then plotted He (±1 s.d.) across both sensitive and resistant chromosomes from the two countries. We measured the significance of differences in diversity in sensitive and resistant chromosomes by permutation. We measured the diversity ratio () in both observed data and in 10 000 datasets in which alleles at each locus were reshuffled among parasites. To evaluate the significance of differences in He at each marker location, we counted the number of the permuted diversity ratio statistics that were greater than the observed value.
To examine patterns of LD surrounding sensitive and resistant alleles at dhfr and pfcrt, we measured extended haplotype homozygosity (EHH; Sabeti et al. 2002), where EHH at a distance x from the gene of interest is defined as the probability that two randomly chosen haplotypes are homozygous for all microsatellites in this region. These statistics (±1 s.d.) were measured using the EHH calculator (Mueller & Andreoli 2004). Measurement of EHH requires complete haplotype data. We retained haplotypes containing missing data at two or fewer microsatellite loci. Missing data at each locus were replaced with alleles randomly drawn from the allele distribution at that locus. This introduces recombination into the data and can reduce EHH, particularly in chromosomes showing high levels of LD. Since our aim was to compare relative, rather than absolute, amounts of LD on wild-type and resistant chromosomes, this procedure is conservative.
3. Results
Out of infections from Mawker, 28% contained multiple infections, as did 40% from Phalanxay and 42% from Sekong. Following exclusion of multiple infections, we were left with 83 single clone infections from Laos and 49 from Thailand. In the samples from Thailand, we found the K76T mutation that is critical for CQ resistance in all parasites examined, while in Laos the wild-type amino acid (K) was found in 5% of samples from Sekong and 36% of samples from Phalanxay. Thai parasites also contained 2–4 mutations in dhfr conferring resistance to pyrimethamine (Nair et al. 2003). By contrast, in Laos, wild-type alleles were present in 33% of parasites analysed from Sekong and 40% of parasites from Phalanxay (table 1). The samples from both locations in Laos were combined for these analyses, since patterns of variation flanking pfcrt and dhfr were very similar in these two locations.
Table 1.
Allele frequencies at dhfr and pfcrt in single clone infections from Laos and Thailand. (The four-letter codes for dhfr alleles describe amino acids present at codons 51, 59, 108 and 164. Alleles are listed in order of increasing resistance, with bold indicating changes from the wild-type allele.)
| Laos | Thailand | ||
|---|---|---|---|
| Sekong | Phalanxay | Mawker–Thai | |
| dhfr | n=36 | n=45 | n=49 |
| NCSI | 0.33 | 0.40 | — |
| NCNI | 0.03 | 0.07 | — |
| NRNI | 0.47 | 0.44 | 0.10 |
| IRNI | 0.17 | 0.09 | 0.12 |
| NRNL | — | — | 0.14 |
| IRNL | — | — | 0.63 |
| pfcrt | n=39 | n=47 | n=44 |
| 76K | 0.05 | 0.36 | — |
| 76T | 0.95 | 0.64 | 1 |
(a) Diversity
Microsatellite heterozygosity around both dhfr and pfcrt is shown in figure 2. We compared He on resistant chromosomes from both Laos and Thailand with He on sensitive chromosomes from Laos. Chromosomes bearing wild-type dhfr or pfcrt alleles were highly variable. Mean He on chromosomes 4 and 7 were 0.86 and 0.81, respectively. We investigated the significance of reductions in variation by permutation. On chromosome 4, we observed significant reduction (p<0.01) in resistant relative to sensitive parasites for 98–137 kb (6–8 cM) on chromosome 4 in Thailand, compared with 34–64 kb (2–4 cM) in Laos. Similarly, on chromosome 7, we observed reduced variation in markers spanning a 195–268 kb region around pfcrt in Thailand, while significant reduction was only observed 34–69 kb (2–4 cM) in Laos. Valleys of reduced variation were also deeper in Thailand. On chromosome 4, seven out of nine markers genotyped within 6 kb of dhfr were monomorphic on resistant chromosomes. By comparison, all nine markers showed variation on resistant chromosomes from Laos, with a minimal He of approximately 0.1 in markers close to dhfr. The pattern is similar on chromosome 7. Four out of five markers genotyped within 11 kb of pfcrt were monomorphic on resistant chromosomes from Thailand, while He only dipped to less than 0.1 in two markers immediately adjacent to pfcrt in resistant chromosomes from Laos.
(b) Linkage disequilibrium
To investigate LD, we removed samples containing missing data for two or more samples. This resulted in reduction of the dataset to 116 samples (48 from Thailand and 68 from Laos) for chromosome 4, and to 100 (34 from Thailand and 66 from Laos) for chromosomes 7. Figure 3 shows the decay in LD on either side of the two resistance genes. EHH decays to zero within 5 kb (figure 3) of wild-type alleles on both chromosomes 4 and 7. By contrast, EHH decays more slowly on chromosomes bearing resistance alleles. There are dramatic and consistent differences in the rates of decay around resistant alleles in the two countries. EHH is lower around resistant alleles in Laos than in Thailand at all distances from pfcrt except 10.4 kb. Similarly, on chromosome 4, EHH decays more rapidly around resistant alleles from Laos than those from Thailand, and EHH drops to zero within 5 kb on either side of sensitive alleles.
Figure 3.
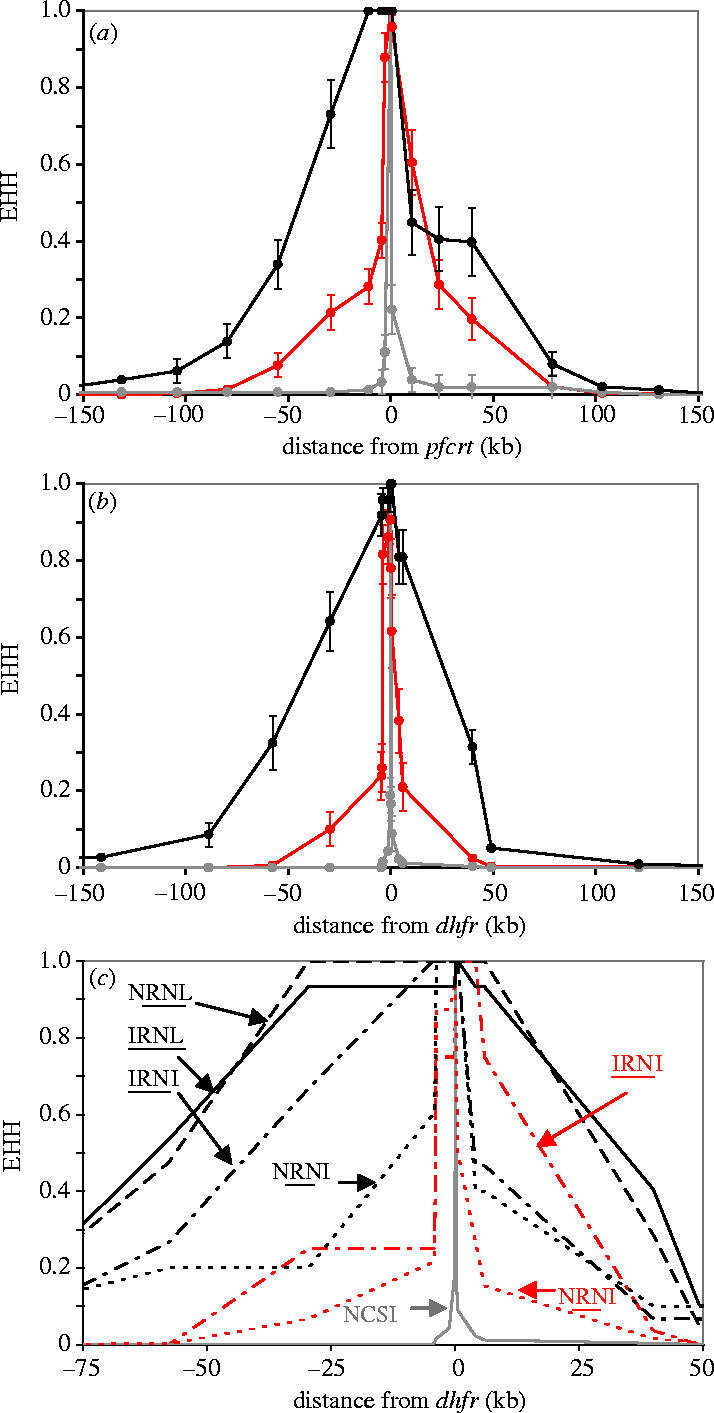
Decline in linkage disequilibrium around pfcrt and dhfr. (a) and (b) show the decline in LD around wild-type and resistant alleles at pfcrt and dhfr. The x-axes are shown to scale. In (b), resistant alleles bearing between one and four mutations are grouped. These graphs are shown with the x-axis drawn to scale. LD is measured using EHH (±1 s.d.). (c) EHH plotted around dhfr alleles bearing different resistance mutations. EHH surrounding resistant alleles from Thailand is shown in black and those from Laos in red, while EHH decay around sensitive alleles is shown in grey. Four-letter codes indicate amino acids present at positions 51, 59, 108 and 164, while underlined letters are those underlying resistance. These codes are marked on the graph for clarity. Data describing EHH decline are not shown for the NCNI allele, as there were only three parasites carrying this allele.
The dhfr alleles in Thailand contain between two and four mutations, whereas those from Laos contain one to three mutations. We observed dramatic differences in LD surrounding different dhfr alleles (figure 3c). In particular, we found a relationship between the number of mutations within dhfr and hitchhiking, with greater LD around alleles bearing three or more mutations than those bearing two mutations. Dhfr alleles IRNI and NRNI occur in both Thailand and Laos. We observe a slower decline in EHH around these alleles in Thailand than in Laos.
4. Discussion
The genome regions affected by hitchhiking around both dhfr and pfcrt in Laos are quite modest in size relative to previous reports (Wootton et al. 2002; Nair et al. 2003). Around dhfr, hitchhiking is detectable for less than half the distance observed in neighbouring Thailand, extending just 34–64 kb (2–4 cM) in Laos, compared with 98–137 kb (6–8 cM) in Thailand. Similarly, around pfcrt, hitchhiking has detectable effects on genetic variation for 34–69 kb in Laos. This is less than a quarter the distance observed in a heterogeneous collection of parasites from different Southeast Asian locations (Wootton et al. 2002) and also less than we observe in parasites from a single clinic on the Thailand/Myanmar border. We have estimated the extent of hitchhiking effects by comparing heterozygosity in sensitive and resistant chromosomes. In Laos, we have compared resistant and wild-type chromosomes from the same location so the results are geographically controlled. In Thailand, we found no sensitive chromosomes, so we have compared diversity in Thai parasites with that observed in sensitive chromosomes from Laos. As such, both geographical variation and selection may contribute to the differences observed. We do not believe that the geographical factor results in important bias in our conclusions for two reasons. First, in chromosomal regions distant from pfcrt and dhfr, we observe no significant difference in He between the two countries. Second, synonymous SNPs show minimal differentiation between populations from Thailand and Laos, with FST values of less than 0.04 for 11 loci examined (Anderson et al. submitted). Furthermore, we observe two other features that suggest greater hitchhiking in Thailand. First, LD declines more steeply with genetic distance in Laos than in Thailand. Second, the valleys of reduced variation surrounding resistant alleles at both genes are considerably shallower in Laos than in Thailand.
These data demonstrate that selection for the same gene in neighbouring countries may have very different impacts on flanking neutral variation. We discuss the reasons for the observed differences, and practical implications for association mapping of genes underlying adaptive traits.
(a) Why do sweeps differ in size and shape?
(i) Transit time of selected alleles
The most obvious difference between parasite populations in Thailand and Laos is the time taken for resistant alleles to spread to fixation (transit time). In Thailand, CQ resistance reached fixation in the 1980s, and even 30 years after CQ was abandoned, we have not detected sensitive alleles in five populations examined (Anderson et al. submitted). By comparison, in Laos, sensitive alleles are still present, with frequencies of between 5 and 36% in Sekong and Phalanxay. Similarly, pyrimethamine resistance spread extremely rapidly in Thailand and, today, four-fifths of the populations examined contain no sensitive alleles, while wild-type alleles constitute more than 35% of the parasite population in Laos (Nair et al. 2003). Hence, while transit times were rapid in Thailand, resistance alleles have still not reached fixation in Laos. Consequently, diversity can be restored in Laos rapidly owing to recombination between sensitive and resistant chromosomes. In Thailand, where the selective sweeps on both chromosomes have effectively purged all standing variation in the vicinity of around resistance loci, mutation is the only force restoring variation.
The underlying reasons for the differences in selection pressure in the two countries are less easy to quantify. At least three factors probably play a role. First, Thailand is a rich country relative to Laos, with GDPs of US$6400 and US$1620, respectively. Therefore, health care is more developed in Thailand and treatment coverage is better. Second, malaria in Thailand is localized to a few border provinces and is largely associated with migrant populations that have been the focus of intensive anti-malarial treatment campaigns. By comparison, malaria is found in all 17 provinces in Laos (Pholsena 1992), and malaria-control campaigns have had poor coverage and have frequently been interrupted by political upheaval or war (Watson 1999). Third, asymptomatic cases may be more common in Laos than in Thailand (Toma et al. 2001; Vythilingam et al. 2003), resulting in a reservoir of parasites in Laos that are not exposed to malaria treatment.
(ii) Fitness differences between dhfr alleles
Two additional factors may contribute to the differences in the transit times of resistant dhfr alleles and hitchhiking in Thailand and Laos. First, dhfr resistance alleles containing the I164L mutation conferring high levels of pyrimethamine resistance (Plowe et al. 1995) are common in Thailand but absent in the populations studied from Laos (Nair et al. 2003). Hence, dhfr alleles from Thailand will have higher fitness than those from Laos in the presence of drug selection and will therefore spread more rapidly, giving rise to more extensive hitchhiking. We can examine allelic effects by comparing hitchhiking around different dhfr alleles (figure 3c). Three alleles bearing 3–4 mutations conferring resistance (IRNI, NRNL, IRNL) show greater hitchhiking than the allele bearing two mutations (NRNI). This is consistent with clinical data indicating more rapid clearance and lower levels of gametocyte production in double mutant parasites relative to triple and quadruple mutants (Mendez et al. 2002). These data suggest that differences in the relative fitness of dhfr alleles in the two countries contribute to the differences in allele transit time and hitchhiking observed. Two dhfr alleles occur in both Thailand and Laos (NRNI and IRNI). The decline in LD around these two alleles is more rapid in Laos than in Thailand, consistent with weaker selection in Laos. Therefore, both fitness differences between dhfr alleles and differences in selection intensity play a role in explaining the differences observed.
In addition, genetic background may play an important role in determining the fitness of dhfr alleles. Dhfr alleles are selected by treatment with the drug combination, SP. Mutations in another gene in the folate pathway, dhps, result in resistance to sulfadoxine. All parasites examined from Mawker–Thai contained 2–3 mutations in dhps in addition to 2–4 mutations in dhfr. By contrast, more than 80% of parasites from Laos still carry wild-type dhps alleles (Anderson et al. submitted). Parasites carrying mutations that confer resistance to both compounds survive better than parasites with mutations in dhfr only (Kublin et al. 2002). Hence, dhfr alleles will spread more rapidly, with greater hitchhiking of flanking neutral mutations, when accompanied by sulfadoxine-resistant dhps alleles.
(iii) Inbreeding and recombination rates
Transmission intensity, inbreeding and recombination are intimately linked in P. falciparum, with low transmission associated with inbreeding and reduced recombination rates (Conway et al. 1999; Anderson et al. 2000; Anderson 2004). Two lines of evidence suggest that levels of transmission are higher in Laos than in Thailand. First, prevalence of P. falciparum is higher in Laos. In 1998, the countrywide prevalence of P. falciparum was 14% (Pillai et al. 2001). In southern Laos, where we collected the samples analysed here, a prevalence of P. falciparum of more than 30% has been reported (Toma et al. 2001; Vythilingam et al. 2003). By comparison, cross-sectional surveys along the Thailand/Burma border revealed prevalences of 1–4% (Luxemburger et al. 1996). Similarly, the number of infective mosquito bites has been estimated at between 1.9 and 7.5 per person per month in Sekong (Vythilingam et al. 2003). By comparison, people on the Thailand/Myanmar border are exposed to more than one infective bite per year (Nosten et al. 2000). However, transmission in Thailand was higher in the past, and has been reduced by treatment with mefloquine–artemisinin combinations since 1994 (Nosten et al. 2000). Hence, the extent to which differences in transmission and parasite recombination rate contributed to hitchhiking effects in early stages of these selective sweeps is difficult to assess.
(b) Asymmetry of selective valleys
The reductions in variation on each side of dhfr and pfcrt are asymmetric. For example, on chromosome 4 in Thailand, variation is restored to background levels 30 kb (approximately 2 cM) to the 3′ of dhfr, while variation is still significantly reduced for 58 kb on the 5′ side. In Laos, the asymmetry is even more dramatic. Variation is restored to background levels within 6 kb (more than 0.5 cM) of the 3′ of dhfr, while reduced variation of 30–58 kb is seen to the 5′ of dhfr. Similarly, on chromosome 7, variation is restored to background 80 kb to the 3′ of pfcrt, while reduced variation is still apparent at −155 kb. Simulation models of hitchhiking indicate that valleys of reduced variation generated following a selective event may be asymmetric even when rates of recombination and mutation are constant (Kim & Stephan 2002). However, given that the direction of asymmetry on chromosome 4 is the same in two Southeast Asian locations and also in African sites (R. Pearce, personal communication), local heterogeneity in recombination rates is the more probable explanation in this case. We investigated local recombination rates by comparing LD between microsatellite loci on either side of dhfr in the 30 chromosomes from Laos containing wild-type dhfr alleles. Out of 10 pairwise comparisons between loci within 6 kb to the 5′ of dhfr, five showed significant LD (p<0.05). By comparison, only one of 10 pairwise comparisons among loci within 6 kb of dhfr to the 3′ showed significant LD. These data are consistent with there being lower recombination rates and more extensive hitchhiking on the 5′ than on the 3′ of dhfr.
(c) Implications for genetic mapping
The dramatic impact of drug selection on flanking genetic variation observed around pfcrt and dhfr provided demonstrations that genome-wide association could be used to locate selected genes in Plasmodium using a relatively low density of genetic markers (Wootton et al. 2002; Nair et al. 2003; Anderson 2004). The data presented in this study reveal that drug-associated sweeps have considerably less impact on flanking genetic variation in Laos, the regions affected spanning just 34–64 kb on chromosome 4 and 34–69 kb on chromosome 7. Furthermore, variation is restored on resistant chromosomes as close as 6 kb from dhfr in Laos. These data clearly demonstrate the importance of choosing parasite populations exposed to strong recent selection for mapping studies of genomic regions containing resistance genes (Anderson 2004). Parasite populations exposed to strong recent selection are ideal for mapping genomic regions containing resistance genes, since such regions can be detected with a relatively low density of microsatellite markers. However, the more localized reduction in variability and reduced LD around drug resistance genes in weakly selected populations may be more useful for fine-scale mapping of functional variation associated with resistance. Similar suggestions have been made by Kohn et al. (2000), who observed similar patterns of LD around genes underlying resistance to warfarin in rat populations exposed to strong and weak selection. The differences in hitchhiking between Thailand and Laos are most simply explained by differences in selection pressure. However, we note that differences in inbreeding may have similar effects on genetic hitchhiking, with highly inbred parasites being optimal for genome-wide mapping and outbred parasite optimal for fine-mapping (Nair et al. 2003; Anderson 2004).
Acknowledgments
This research was supported by an NIH grant AI48071 (to T.J.C.A.). We thank Jeff Williams for statistical advice. The SMRU is part of the Wellcome Trust Mahidol University Oxford Tropical Medicine Research Programme supported by the Wellcome Trust of Great Britain. F. N. is a Wellcome Trust Senior Clinical Fellow. The following people assisted in sample collection: the staff of the SMRU; Samlane Phompida, Rattanaxay Phetsouvanh, Maniphone Khanthavone, Tiengkham Pongvongsa and the staff of the Centre for Malariology, Parasitology and Entomology in Vientiane and Savannakhet, Laos.
Supplementary Material
References
- Anderson T.J. Mapping drug resistance genes in Plasmodium falciparum by genome-wide association. Curr. Drug Targets Infect. Disord. 2004;4:65–78. doi: 10.2174/1568005043480943. [DOI] [PubMed] [Google Scholar]
- Anderson T.J, Su X.Z, Bockarie M, Lagog M, Day K.P. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology. 1999;119:113–125. doi: 10.1017/s0031182099004552. [DOI] [PubMed] [Google Scholar]
- Anderson T.J, et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol. Biol. Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- Anderson T.J, Nair S, Jacobzone C, Zavai A, Balkan S. Molecular assessment of drug resistance in Plasmodium falciparum from Bahr El Gazal province, Sudan. Trop. Med. Int. Health. 2003;8:1068–1073. doi: 10.1046/j.1360-2276.2003.01144.x. [DOI] [PubMed] [Google Scholar]
- Anderson T.J, Nair S, Qin H, Singlam S, Brockman A, Paiphun L, Nosten F. Are transporter genes other than the chloroquine resistance locus (pfcrt) and multi drug resistance gene (pfmdr) associated with antimalarial drug resistance? Antimicrob. Agents Chemother. In press doi: 10.1128/AAC.49.6.2180-2188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson T.J.C, Nair S, Williams J.T, Mayxay M, Newton P.N, Guthmann J.-P, Smithuis F.M, Tran Tinh Hien, White N.J, Nosten F. Geographical distribution of drug resistance mutations and putatively neutral SNPs in SE Asian malaria parasites. Mol. Biol. Evol. Submitted doi: 10.1093/molbev/msi235. [DOI] [PubMed] [Google Scholar]
- Berens N, Schwoebel B, Jordan S, Vanisaveth V, Phetsouvanh R, Christophel E.M, Phompida S, Jelinek T. Plasmodium falciparum: correlation of in vivo resistance to chloroquine and antifolates with genetic polymorphisms in isolates from the south of Lao PDR. Trop. Med. Int. Health. 2003;8:775–782. doi: 10.1046/j.1365-3156.2003.01099.x. [DOI] [PubMed] [Google Scholar]
- Chin W, Bear D.M, Colwell E.J, Kosakal S. A comparative evaluation of sulfalene–trimethoprim and sulphormethoxine–pyrimethamine against falciparum malaria in Thailand. Am. J. Trop. Med. Hyg. 1973;22:308–312. doi: 10.4269/ajtmh.1973.22.308. [DOI] [PubMed] [Google Scholar]
- Conway D.J, Roper C, Oduola A.M, Arnot D.E, Kremsner P.G, Grobusch M.P, Curtis C.F, Greenwood B.M. High recombination rate in natural populations of Plasmodium falciparum. Proc. Natl Acad. Sci. USA. 1999;96:4506–4511. doi: 10.1073/pnas.96.8.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisawa I, Muto T, Kameko S, Mitsui G. Response of Laotian malaria strains to chemotherapy. Jpn. J. Exp. Med. 1970;40:151–157. [PubMed] [Google Scholar]
- Farid, M. A. 1967 Report on a field visit to Laos, 11–16 March 1967 WHO (WP/M2/76/2).
- Fidock D.A, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthmann J.P, Kasparian S, Phetsouvanh R, Nathan N, Garcia M, Phompida S, Brockman A, Gastellu M, Legros D. The efficacy of chloroquine for the treatment of acute, uncomplicated, Plasmodium falciparum malaria in Laos. Ann. Trop. Med. Parasitol. 2002;96:553–557. doi: 10.1179/000349802125001654. [DOI] [PubMed] [Google Scholar]
- Harinasuta T, Suntharasamai P, Viravan C. Chloroquine-resistant falciparum malaria in Thailand. Lancet. 1965;2:657–660. doi: 10.1016/s0140-6736(65)90395-8. [DOI] [PubMed] [Google Scholar]
- Kim Y, Stephan W. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics. 2002;160:765–777. doi: 10.1093/genetics/160.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn M.H, Pelz H.J, Wayne R.K. Natural selection mapping of the warfarin-resistance gene. Proc. Natl Acad. Sci. USA. 2000;97:7911–7915. doi: 10.1073/pnas.97.14.7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kublin J.G, et al. Molecular markers for failure of sulfadoxine–pyrimethamine and chlorproguanil–dapsone treatment of Plasmodium falciparum malaria. J. Infect. Dis. 2002;185:380–388. doi: 10.1086/338566. [DOI] [PubMed] [Google Scholar]
- Luxemburger C, Thwai K.L, White N.J, Webster H.K, Kyle D.E, Maelankirri L, Chongsuphajaisiddhi T, Nosten F. The epidemiology of malaria in a Karen population on the western border of Thailand. Trans. R. Soc. Trop. Med. Hyg. 1996;90:105–111. doi: 10.1016/s0035-9203(96)90102-9. [DOI] [PubMed] [Google Scholar]
- Mayxay M, Newton P.N, Khanthavong M, Tiengkham P, Phetsouvanh R, Phompida S, Brockman A, White N.J. Chloroquine versus sulfadoxine–pyrimethamine for treatment of Plasmodium falciparum malaria in Savannakhet province, Lao people's democratic republic: an assessment of national antimalarial drug recommendations. Clin. Infect. Dis. 2003;37:1021–1028. doi: 10.1086/377734. [DOI] [PubMed] [Google Scholar]
- Mendez F, Munoz A, Carrasquilla G, Jurado D, Arevalo-Herrera M, Cortese J.F, Plowe C.V. Determinants of treatment response to sulfadoxine–pyrimethamine and subsequent transmission potential in falciparum malaria. Am. J. Epidemiol. 2002;156:230–238. doi: 10.1093/aje/kwf030. [DOI] [PubMed] [Google Scholar]
- Mueller J.C, Andreoli C. Plotting haplotype-specific linkage disequilibrium patterns by extended haplotype homozygosity. Bioinformatics. 2004;20:786–787. doi: 10.1093/bioinformatics/btg481. [DOI] [PubMed] [Google Scholar]
- Nair S, Brockman A, Paiphun L, Nosten F, Anderson T.J. Rapid genotyping of loci involved in antifolate drug resistance in Plasmodium falciparum by primer extension. Int. J. Parasitol. 2002;32:852–858. doi: 10.1016/s0020-7519(02)00033-4. [DOI] [PubMed] [Google Scholar]
- Nair S, et al. A selective sweep driven by pyrimethamine treatment in SE Asian malaria parasites. Mol. Biol. Evol. 2003;20:1526–1536. doi: 10.1093/molbev/msg162. [DOI] [PubMed] [Google Scholar]
- Nei M. Columbia University Press; New York: 1987. Molecular evolutionary genetics. [Google Scholar]
- Nosten F, van Vugt M, Price R, Luxemburger C, Thway K.L, Brockman A, McGready R, ter Kuile F, Looareesuwan S, White N.J. Effects of artesunate–mefloquine combination on incidence of Plasmodium falciparum malaria and mefloquine resistance in western Thailand: a prospective study. Lancet. 2000;356:297–302. doi: 10.1016/s0140-6736(00)02505-8. [DOI] [PubMed] [Google Scholar]
- Pholsena K. The malaria situation and antimalaria program in Laos. Southeast Asian J. Trop. Med. Public Health. 1992;23(Suppl. 4):39–42. [PubMed] [Google Scholar]
- Pillai D.R, Labbe A.C, Vanisaveth V, Hongvangthong B, Pomphida S, Inkathone S, Zhong K, Kain K.C. Plasmodium falciparum malaria in Laos: chloroquine treatment outcome and predictive value of molecular markers. J. Infect. Dis. 2001;183:789–795. doi: 10.1086/318836. [DOI] [PubMed] [Google Scholar]
- Plowe C.V, Djimde A, Bouare M, Doumbo O, Wellems T.E. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am. J. Trop. Med. Hyg. 1995;52:565–568. doi: 10.4269/ajtmh.1995.52.565. [DOI] [PubMed] [Google Scholar]
- Roper C, Pearce R, Bredenkamp B, Gumede J, Drakeley C, Mosha F, Chandramohan D, Sharp B. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet. 2003;361:1174–1181. doi: 10.1016/S0140-6736(03)12951-0. [DOI] [PubMed] [Google Scholar]
- Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T.J. Intercontinental spread of pyrimethamine resistant malaria. Science. 2004;305:1124. doi: 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- Sabeti P.C, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Schwobel B, Jordan S, Vanisaveth V, Phetsouvanh R, Christophel E.M, Phompida S, von Sonnenburg F, Jelinek T. Therapeutic efficacy of chloroquine plus sulphadoxine/pyrimethamine compared with monotherapy with either chloroquine or sulphadoxine/pyrimethamine in uncomplicated Plasmodium falciparum malaria in Laos. Trop. Med. Int. Health. 2003;8:19–24. doi: 10.1046/j.1365-3156.2003.00977.x. [DOI] [PubMed] [Google Scholar]
- Toma H, Kobayashi J, Vannachone B, Arakawa T, Sato Y, Nambanya S, Manivong K, Inthakone S. A field study on malaria prevalence in southeastern Laos by polymerase chain reaction assay. Am. J. Trop. Med. Hyg. 2001;64:257–261. doi: 10.4269/ajtmh.2001.64.257. [DOI] [PubMed] [Google Scholar]
- Vythilingam I, Phetsouvanh R, Keokenchanh K, Yengmala V, Vanisaveth V, Phompida S, Hakim S.L. The prevalence of Anopheles (Diptera: Culicidae) mosquitoes in Sekong province, Lao PDR in relation to malaria transmission. Trop. Med. Int. Health. 2003;8:525–535. doi: 10.1046/j.1365-3156.2003.01052.x. [DOI] [PubMed] [Google Scholar]
- Watson L. Lao Malaria Review. Unpublished European Union malaria project report. 1999 [Google Scholar]
- White N.J. Antimalarial drug resistance: the pace quickens. J. Antimicrob. Chemother. 1992;30:571–585. doi: 10.1093/jac/30.5.571. [DOI] [PubMed] [Google Scholar]
- Wiehe T. The effect of selective sweeps on the variance of the allele distribution of a linked multiallele locus: hitchhiking of microsatellites. Theor. Popul. Biol. 1998;53:272–283. doi: 10.1006/tpbi.1997.1346. [DOI] [PubMed] [Google Scholar]
- Wootton J.C, Feng X, Ferdig M.T, Cooper R.A, Mu J, Baruch D.I, Magill A.J, Su X.Z. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.