Abstract
Investigating how differential gene expression underlies caste determination in the social Hymenoptera is central to understanding how variation in gene expression underlies adaptive phenotypic diversity. We investigated for the first time the association between differential gene expression and queen–worker caste determination in the bumble-bee Bombus terrestris. Using suppression subtractive hybridization we isolated 12 genes that were differentially expressed in queen- and worker-destined larvae. We found that the sets of genes underlying caste differences in larvae and adults failed to overlap greatly. We also found that B. terrestris shares some of the genes whose differential expression is associated with caste determination in the honeybee, Apis mellifera, but their expression patterns were not identical. Instead, we found B. terrestris to exhibit a novel pattern, whereby most genes upregulated (i.e. showing relatively higher levels of expression) in queen-destined larvae early in development were upregulated in worker-destined larvae late in development. Overall, our results suggest that caste determination in B. terrestris involves a difference not so much in the identity of genes expressed by queen- and worker-destined larvae, but primarily in the relative timing of their expression. This conclusion is of potential importance in the further study of phenotypic diversification via differential gene expression.
Keywords: Bombus terrestris, caste determination, caste evolution, gene expression, social insects, suppression subtractive hybridization
1. Introduction
The evolution of sterile castes in social insects represents one of the major transitions in evolution (Maynard Smith & Szathmáry 1995). In common with other major transitions, such as the divergence of the germ line and somatic cells in multicellular organisms, it has involved units of biological organization at one level (individuals) cooperating to form a new unit of biological organization at a higher level (the colony), accompanied by the emergence of a reproductive division of labour among the lower-level units. In the advanced social Hymenoptera (ants, bees, wasps), this reproductive division of labour involves a polyphenism whereby adult females occur in two, adaptively interdependent castes. Females are morphologically, physiologically and behaviourally specialized as either reproductive queens or non-reproductive workers. With few exceptions (Julian et al. 2002; Volny & Gordon 2002; Helms Cahan & Keller 2003), whether female larvae develop into queens or workers depends not on genotypic differences, but on their environment at critical periods of caste determination (Wilson 1971; Wheeler 1986). Therefore, caste determination and differentiation must involve the differential expression of genes shared by queens and workers. Hence, the social Hymenoptera present a unique opportunity to investigate, in the fundamental context of one of evolution's major transitions, how variation in gene expression underlies adaptive morphological and behavioural diversity.
Comparative studies of caste-related gene expression provide essential insights into the nature, origin and evolution of caste polyphenism in social insects (Evans & Wheeler 1999, 2001; Robinson 2002a). One key question to be answered is whether those genes whose differential expression originally underlay reproductive differentiation in female adults of ‘primitively’ eusocial insects (those having queens and workers that do not differ in morphology) are also involved in larval caste determination in advanced social taxa (Evans & Wheeler 1999). Answering this question could reveal whether, at the proximate level, the evolution of a distinct morphological worker caste involves genes originally regulating reproductive differences among adult females. To date, however, the identity of differentially expressed genes associated with queen–worker caste determination in larvae has only been investigated in the advanced social honeybee Apis mellifera (Corona et al. 1999; Evans & Wheeler 1999, 2000; Hepperle & Hartfelder 2001). Related investigations have examined caste-associated gene expression in honeybee pupae and adults (Piulachs et al. 2003; Guidugli et al. 2004) and how variation in gene expression underlies behavioural differences among adult honeybee workers (Ben-Shahar et al. 2002; Kucharski & Maleszka 2002; Robinson 2002b; Whitfield et al. 2003), soldier caste differentiation (Miura et al. 1999; Miura 2001) and differences between worker and soldier castes in termites (Scharf et al. 2003), wing polymorphism in reproductive and sterile ant castes (Abouheif & Wray 2002), and physiological differences between alate and dealate queens in red fire ants (Tian et al. 2004).
Social insects exhibiting a less advanced level of sociality provide an excellent set of model systems to complement these studies. An appropriate species for investigation is the bumble-bee Bombus terrestris, which represents an intermediate stage between primitive and advanced sociality. Adult queens in B. terrestris are always larger than workers, but do not differ morphologically from them (e.g. both queens and workers have a sperm receptacle; M. J. Duchateau, personal communication). However, queens do differ physiologically and behaviourally from workers in that they accumulate glycogen and fat in their fat body, leave the nest to mate and are capable of overwintering (Michener 1974; Röseler 1991). Previous research has shown that caste membership in B. terrestris is determined at two stages in female larval development: if a colony queen is present and is producing a putative, caste-signalling pheromone, initially bipotential first- and second-instar larvae (i.e. larvae capable of developing into adults of either caste) are irreversibly determined towards development as workers; if such a queen is absent, larvae develop as normal queens provided that, in their fourth (last) instar, they also receive sufficient food (Röseler 1991; Cnaani & Hefetz 2001; Cnaani et al. 1997, 2000; Hartfelder et al. 2000; Bortolotti et al. 2001; Bourke & Ratnieks 2001; Pereboom et al. 2003). In addition, B. terrestris workers can be reproductive, since the end of the annual colony cycle (the so-called competition phase) is characterized by the emergence of dominant, reproductive (egg-laying) workers and subordinate, non-reproductive workers (Duchateau & Velthuis 1988; Bloch 1999; Bloch & Hefetz 1999). Therefore, by comparing the gene expression profiles underlying differences between adult queens and workers with those underlying differences between reproductive and non-reproductive adult workers, it is, in principle, possible to distinguish between differences owing to the presence or absence of reproduction and those owing to caste membership per se.
In this study, we isolated and sequenced genes whose differential expression is associated with caste determination in female B. terrestris larvae. Capitalizing on the specific features of B. terrestris, we sought to test two previously untested hypotheses. First, by investigating whether these genes are also differentially expressed between adult queens and workers and between reproductive and non-reproductive adult workers in B. terrestris, we tested whether the same genes underlie caste differences in larvae and adults. Second, by comparing the genes isolated in B. terrestris with those known to be associated with caste determination in A. mellifera larvae (Corona et al. 1999; Evans & Wheeler 1999, 2000; Hepperle & Hartfelder 2001), we tested whether the identity of relevant genes is conserved across these related taxa and whether any conserved genes share patterns of differential expression. Overall, we therefore sought to investigate whether, in the evolution of advanced sociality, genes that originally underlay reproductive differences between adults have been co-opted for use in caste determination during larval development.
2. Material and methods
(a) Sample collection
All colonies used in the study were maintained in nest-boxes according to standard protocols (Duchateau & Velthuis 1988) in controlled-temperature rooms at the Institute of Zoology, London. We monitored colonies and the production of brood daily to evaluate colony development. Using suppression subtractive hybridization (SSH; Diatchenko et al. 1996) to enrich for differentially expressed genes, we created two paired subtracted cDNA libraries as follows: (i) from worker- and queen-destined larvae in the first and second instars (early-instar larvae); (ii) from worker- and queen-destined larvae in the fourth instar (last-instar larvae). Additionally, to investigate gene expression in adult phenotypes, we collected adult colony queens, and dominant, reproductive and subordinate, non-reproductive adult workers.
(i) Larval phenotypes
In June 2001, we raised colonies from Bombus terrestris dalmatinus queens obtained from stocks maintained at the Department of Behavioural Biology, University of Utrecht, and derived from a commercial supplier (Koppert Biological Systems, Berkel and Rodenrijs, the Netherlands; for stock maintenance methods see Duchateau 2000). We obtained early-instar (n=427) and last-instar (n=123) worker-destined larvae in July 2001 from 10 queenright colonies (i.e. colonies with a queen) producing worker brood only. Last-instar queen-destined larvae (n=161) were collected from the same 10 colonies in July and August 2001 after they had started producing queen brood. Larval instar, sex and caste were confirmed by determining body mass and by examining adults eclosing from a non-sampled brood from the same larval batches as the sampled larvae (Cnaani et al. 1997; Pereboom 1997). The number of non-sampled larvae (controls) remaining in the colony was always equal to or greater than the number of sampled larvae.
We removed the queen from five additional colonies on the eighth day after the start of the third brood (when a queen is still laying diploid, female eggs) in order to stimulate the development of new queens (Pereboom et al. 2003). We sampled early-instar queen-destined larvae (n=90) originating from the last batches of diploid eggs laid in the 7 days preceding the queens' removal. For the construction of library one (early instar), we only used larvae from batches that produced 100% queens in the control batch.
(ii) Adult phenotypes
Newly emerged queens produced by the first series of colonies in August 2001 were mated with B. t. dalmatinus males obtained from Koppert Biological Systems. After a period of seven months of artificial hibernation in boxes with peat dust at 0–4 °C and 80–100% relative humidity, 40 of these queens were allowed to found new colonies (see Duchateau 2000). From 15 of those colonies, we sampled the queen and the first 10 (non-reproductive) workers to eclose (n=150) in May 2002. We obtained dominant, laying workers (n=91) and subordinate, non-laying workers (n=92) in May and June 2002 from an additional 10 queenright colonies (9–10 workers per colony) that had entered their competition phase. Dominant workers were identified by being observed to behave aggressively (Duchateau & Velthuis 1988) within a period of 10 min, but were not removed until they were actually observed to lay eggs. In dominant workers, the rate of aggression is sufficiently high that a worker showing no aggressive behaviour in 10 min can be confidently classified as subordinate (J. J. M. Pereboom, personal observations; Lopez-Vaamonde et al. 2003). All removed individuals were immediately frozen at −80 °C.
(b) Selection of differentially expressed genes
(i) RNA extraction and mRNA purification
We extracted total RNA from homogenized pooled individuals of each of the phenotypes using TRIZOL reagent (Invitrogen) followed by treatment with DNase I to remove any residual genomic DNA. For library one, we pooled approximately 30 individuals and, for library two, we pooled four to six individuals to maximize our chances of detecting differential expression of genes that may show variation between individuals in the exact timing of expression. Sampled larvae originated from two to five different colonies, and individuals were selected to represent the full range of body sizes for early and last larval instars, respectively. Messenger RNA (mRNA) was purified from total RNA using a Nucleotrap kit (BD Clontech).
(ii) SSH and cDNA library construction
We synthesized complementary DNA (cDNA) from 2 μg mRNA and implemented SSH performing forward and reverse subtractions following the PCR-Select cDNA subtraction protocol (BD Clontech). We then ligated the PCR-amplified products into the pCR2.1 vector (T/A cloning kit, Invitrogen) according to the manufacturer's instructions. Single recombinant colonies of Escherichia coli were transferred to individual wells of standard 96-well microtitre plates each holding 150 μl LB medium, containing 20% glycerol and 50 μg ml−1 ampicillin. Colonies were allowed to grow in the medium at 37 °C overnight, and the libraries were then stored in this form at −80 °C.
(iii) Differential screening
We used differential screening (Jin et al. 1997) to eliminate false positives (sequences falsely appearing to be differentially expressed by chance). Arrays of E. coli colonies derived from libraries were spotted onto duplicate nylon membranes (Hybond NX, Amersham) and cultured on LB agar medium (containing 50 μg ml−1 ampicillin) at 37 °C overnight. Colonies were denatured (0.5 M NaOH, 1.5 M NaCl) and neutralized (0.5 M Tris–HCl (pH 7.4), 1.5 M NaCl), and DNA was fixed to the membrane by baking at 80 °C.
Hybridization probes for differential screening of libraries were made from subtracted cDNA populations derived from SSH. Each part of a paired subtraction was radio-labelled separately by incorporation of (α-32P)-dATP using random priming (Random Primed DNA Labeling Kit, Roche). All prehybridizations, hybridizations and washes were carried out at 65 °C in glass bottles in a rotating incubator. Membranes were pre-hybridized for 30 min in Church buffer and, after addition of the denatured probe, hybridized overnight (Sambrook & Russell 2001). After hybridization, membranes were washed four times for 3 min in low-stringency wash buffer (2× SSC, 0.1% SDS), four times for 5 min in 1× SSC, 0.1% SDS wash buffer, and four times for 5 min in high-stringency wash buffer (0.1× SSC, 0.1% SDS). Membranes were wrapped in plastic and exposed to X-ray film (Kodak BioMax MS) with intensifying screens for 3–72 h, depending on the strength of the signal. Membranes were stripped of probe by washing in 0.1% SDS at 95 °C. Each duplicate membrane was hybridized in turn with each probe to control for possible differences in E. coli colony growth on the duplicates.
(iv) Sequencing and identification of selected sequences
We PCR-amplified candidate clones (selected by differential screening) using M13-forward and -reverse primers and sequenced both strands on an ABI-Prism 377 semi-automated sequencer. DNA sequence identification was achieved by comparing nucleotide and deduced amino acid sequences with published sequences of other organisms by means of homology searches of major databases (GenBank, EMBL, Swiss-Prot) using the Blast-X and Blast-N algorithms (Altschul et al. 1997). We set a cut-off for significant sequence homology at a probability score (expectation value or E-value, describing the probability of finding homologues by chance) of less than 10−3.
(c) Confirmation of differential expression
We isolated total RNA from pooled individuals of queen or worker phenotypes at different life-history stages (10 queen- and 10 worker-destined early-instar larvae; 3 queen- and 6 worker-destined last-instar larvae; 2 adult colony queens and 10 adult non-reproductive workers; 10 adult reproductive workers and 10 adult non-reproductive workers), with the larval phenotypes from the same set of colonies used for the construction of the corresponding subtracted libraries. For each phenotype we denatured and loaded 10 μg of total RNA on a 1.5% denaturing agarose gel (Sambrook & Russell 2001). After electrophoresis, samples were visualized by ethidium bromide staining, and gel images recorded using a CCD camera (Syngene GeneGenius system). RNA was transferred to nylon membrane by Northern blotting (Sambrook & Russell 2001) and fixed by baking at 80 °C.
We prepared Northern blots bearing RNA from each of the eight phenotypes. These were probed in turn with 17 unique sequences selected from the candidates isolated by differential screening that were successfully sequenced as described in the results. Hybridization probes were PCR-amplified from selected clones using M13-forward and -reverse primers. After agarose gel electrophoresis, probes were extracted from the gel using ‘QIAquick’ gel extraction (Qiagen) and radio-labelled with (α-32P)-dATP by random priming. Hybridization and washing conditions and exposure to X-ray film were as described above. Scanned images of the RNA gels and the autoradiographs were analysed on a Macintosh computer using the public domain software Nih Image (http://rsb.info.nih.gov/nih-image/). We calculated the relative optical density (OD) of the autoradiograph signals from pairs of phenotypes using gel plotting macros. These values were then normalized in proportion to the OD of the mRNA fluorescence on the RNA gel image to control for differences in RNA loading. We considered a minimum twofold difference (arbitrarily chosen) in OD between pairs of phenotypes as a criterion for differential expression.
(d) Data deposition
Differentially expressed sequences have been deposited in GenBank (dbEST) under accession numbers DN048368 (dbEST Id. 27769598) to DN048383 (dbEST Id. 27769613).
3. Results
(a) Confirmation of caste of sampled larvae
For the construction of the two cDNA libraries, the sex and caste of adults developing from non-sampled larvae confirmed the presumed sex and caste of the sampled larvae. The 10 queenright colonies providing early- (n=427) and last-instar (n=123) worker-destined larvae for RNA-extraction subsequently produced only workers (n≥427 and n≥123, respectively) from the control batches as expected. In the queen production stage, the same 10 colonies produced only queens (n≥161) from the control batches taken from the same groups of larvae from which last-instar queen-destined larvae (n=161) had been sampled. The body mass of a representative sample (including smallest and largest individuals) of the sampled last-instar worker- and queen-destined larvae was 273.3±61.4 mg (n=30) and 867.9±183.3 mg (n=30), respectively, which confirms their caste membership (in the last instar, worker-destined larvae weigh 70–380 mg and queen-destined larvae weigh 250–1300 mg; Cnaani et al. 1997; Pereboom 1997). In the five queenless colonies, all unsampled early-instar larvae (n=202) from the same batches as the sampled queen-destined larvae (n=90) developed as queens.
(b) Differential screening and sequence identification
We successfully constructed both parts of library one, but differential screening and sequencing suggested SSH was unsuccessful for the worker part of library two, despite our repeating the SSH procedure twice. Library one thus consisted of 960 clones and library two (queen part) of 480 clones. We screened all clones from both libraries using differential screening (figure 1) and selected 100 candidate clones (at random from those clones showing the strongest differential hybridization) from library one and 50 candidate clones (on the same basis) from library two (queen part) to be sequenced. Of 129 successfully sequenced clones, 37 (28.7%) showed no significant homology to any known DNA or deduced amino acid sequences. Some of the remaining 92 sequences were abundant, with multiple clones present in the subtracted libraries (e.g. hexamerin, 10/92 clones; CG-5520 protein, 9/92 clones; chymotrypsin, 11/92 clones).
Figure 1.
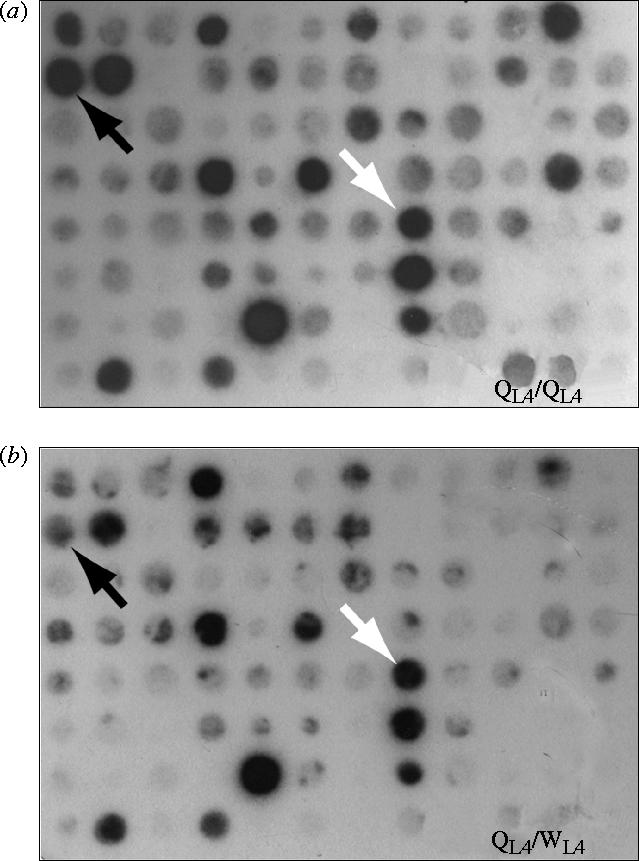
Array of 96 potentially differentially expressed clones from the last-instar queen library (library two, QL4), first probed with cDNA from last-instar queen-destined larvae (a; QL4), and stripped and re-probed with cDNA from last-instar worker-destined larvae (b; WL4). The black arrow indicates a clone expressed at a higher level in last-instar queen-destined larvae compared with worker-destined larvae (i.e. stronger signal after hybridization to queen cDNA). A white arrow indicate clones expressed equally in worker- and queen-destined larvae (i.e. false positives).
(c) Northern blots: patterns of expression
Sixteen sequences of the 129 successfully sequenced clones were selected for probing the Northern blots (table 1). The selected sequences were chosen to include: (i) all sequences (n=4) found to show significant homology to genes known to be differentially expressed across castes in A. mellifera, (ii) a set (n=9) of sequences with significant homology to non-Apis genes, and (iii) a set (n=3) of sequences with no known homologues. From the Northern blots, we found that four sequences showed no detectable expression and 12 sequences exhibited differences in expression between queen- and worker-destined larvae, including five sequences exhibiting differences in expression of molecular size variants (table 1; figure 2). Such variants may represent multiple loci encoding similar protein products, or they may be the result of alternative splicing of the mRNA from a single locus (Boue et al. 2003). Results of the Northern blots were not always consistent with results from differential screening. The 16 candidate sequences had been selected based on displaying relatively higher levels of expression in early-instar queen-destined larvae (n=6), early-instar worker-destined larvae (n=6) or last-instar queen-destined larvae (n=4). However, in the Northern blots, 9 sequences showed a higher level of expression in early-instar queen-destined larvae, and 12 showed a higher level of expression in last-instar worker-destined larvae. The reasons for these discrepancies are unknown, but similar discrepancies have been recorded previously (Cao et al. 2004). They do not affect our conclusions because results from Northern blots provided clear evidence of differential expression.
Table 1.
Relative expression levels (A versus B, i.e. phenotype A versus phenotype B) measured as the optical density of autoradiograph signals of one phenotype (XA), divided by the sum of the optical densities of signals of both phenotypes (XA+XB).
| clone number | GenBank acc. no. (dbEST Id.) | sequence similarity (GenBank acc. no.; E-value) | functional group | larval phenotypes | adult phenotypes | |||
|---|---|---|---|---|---|---|---|---|
| QL1 versus WL1 | QL4 versus WL4 | Qadult versus Wadult | Wrep versus Wnon-rep | |||||
| WL1 79 | DN048368 (27769598) | larval cuticle protein‡ (O02387; E=3×10−11) | structural protein | 0.68 | 0.09 | * | * | |
| QL4 20 | DN048376 (27769606) | hexamerin‡ (AY601637; E=5×10−53) | storage protein | band 1 | * | 0.02 | * | * |
| band 2 | * | 0.24 | * | * | ||||
| QL1 39 | DN048371 (27769601) | unknown sequence | band 1 | * | 0.21 | * | * | |
| band 2 | * | 0.86 | * | * | ||||
| QL1 93 | DN048374 (27769604) | unknown sequence | band 1 | 0.92 | 0.02 | * | * | |
| band 2 | * | 0.00 | * | * | ||||
| WL1 21 | DN048379 (27769609) | 60-S ribosomal protein (AY072286; E=5×10−14) | protein synthesis | band 1 | 0.97 | 0.00 | * | * |
| band 2 | 0.80 | 0.11 | 0.35 | 0.66 | ||||
| WL1 28 | DN048380 (27769610) | chymotrypsin‡ (P00768 E=4×10−31) | enzyme | 0.97 | 0.01 | 0.46 | 0.27 | |
| WL1 67 | DN048381 (27769611) | cytochrome oxidase I‡ (AY181169; E=2×10−32) | electron transport | 0.94 | 0.11 | 0.33 | 0.14 | |
| WL1 23 | DN048378 (27769608) | hypothetical protein (AABL0106743; E=7×10−11) | unknown | 0.89 | 0.00 | 0.00 | 0.38 | |
| WL1 68 | DN048382 (27769612) | peroxiredoxin (AY438331; E=3×10−34) | enzyme | 0.92 | 0.25 | 0.69 | 0.11 | |
| QL4 34 | DN048377 (27769607) | fatty acyl CoA-desaturase (AF417841; E=5×10−44) | enzyme | 0.87 | 0.00 | 0.45 | 0.55 | |
| QL4 18 | DN048375 (27769605) | ATP synthase beta subunit (AY580209; E=2×10−93) | electron transport | 0.92 | 0.00 | 0.29 | 0.53 | |
| QL4 14 | DN048383 (27769613) | unknown sequence | band 1 | 0.65 | 0.20 | 0.14 | 0.46 | |
| band 2 | * | * | 0.00 | * | ||||
| band 3 | * | * | 0.00 | * | ||||
| QL1 42 | DN048370 (27769600) | imaginal disk growth factor (Q9W303; E=3×10−30) | enzyme | * | * | * | * | |
| QL1 32 | DN048369 (27769599) | super oxide dismutase 1 (AY4624198; E=4×10−20) | enzyme | * | * | * | * | |
| QL1 40 | DN048372 (27769602) | CG 5520 protein (AE003763; E=3×10−4) | signal transduction | * | * | * | * | |
| QL1 43 | DN048373 (27769603) | CG31605-PA (AE003619; E=1×10−13) | immunity | * | * | * | * | |
1.00, Expressed in phenotype A only; 0.67 or >0.67, expressed two or more times more highly in phenotype A than in phenotype B; 0.5, no differential expression; 0.33 or <0.33, expressed two or more times more highly in phenotype B than in phenotype A; 0.00, expressed in phenotype B only (twofold or greater differences are printed in bold type). *, no signal detected in either phenotype. ‡, Northern analysis was performed in duplicate, using separate blots. Expression was assayed in early-instar queen- and worker-destined larvae (QL1 and WL1), in last-instar queen- and worker-destined larvae (QL4 and WL4), in adult colony queens and non-reproductive adult workers (Qadult and Wadult) and in reproductive and non-reproductive adult workers (Wrep and Wnon-rep. First column: clone numbers refer to the subtracted library from which the clone was retrieved. Italics indicate clones for which Northern results were inconsistent with the subtracted library from which they were captured. Second column: GenBank accession number and dbEST Id (between brackets) of the submitted sequences. Third column: sequences are named according to results of BLAST homology searches; the GenBank accession number and E-values of the closest hit are provided between brackets (E-values express the probability of finding homologues by chance; significance at E<10−3). Sequences with no significant homologues are referred to as ‘unknown sequence’. Gene functional group classification is based on the gene ontology classification used in Adams et al. (2000). A number of phenotypes produced multi-banded patterns as indicated.
Figure 2.

Autoradiographs of three representative Northern blot results. (a) Larval cuticle protein was upregulated in early-instar queen-destined larvae (QL1) and in last-instar worker-destined larvae (WL4), but showed no expression in adult phenotypes (Qad (Qadult in table 1) versus Wnon-rep). (b) Chymotrypsin was exclusively expressed in early-instar queen-destined larvae and last-instar worker-destined larva, and showed expression at approximately similar levels in all adult phenotypes. (c) 60S Ribosomal protein showed multiple bands in early-instar queen-destined larvae (QL1), early-instar worker-destined larvae (WL1) and last-instar worker-destined larvae (WL4). Relative levels of expression are given in table 1, where all additional abbreviations are defined.
In the early instars, most sequences (9/12) displayed relatively higher levels (≥twofold difference) of expression in queen-destined larvae than in worker-destined larvae, but, in the last instar, all 12 sequences were expressed at higher levels in worker-destined larvae than in queen-destined larvae (with the exception of the second band in clone no. QL1 39; table 1). In those cases where sequences were expressed in both larvae and adults (8/12), four displayed higher levels of expression in adult workers compared with queens (Cytochrome Oxidase I, Hypothetical Protein, ATP-synthase beta chain, and the unknown sequence from clone no. QL4 14; table 1) and one sequence (Peroxiredoxin) displayed higher levels of expression in adult queens compared with workers. Three sequences (Chymotrypsin, Cytochrome Oxidase I and Peroxiredoxin) were expressed at higher levels in non-reproductive workers compared with reproductive workers.
The genes associated with caste membership and/or being reproductive (adult queens versus non-reproductive workers) and being reproductive alone (reproductive versus non-reproductive adult workers) tended not to share patterns of differential expression (table 1). Those genes showing higher levels of expression in adult workers versus queens, but no differences in expression between reproductive and non-reproductive workers (Hypothetical Protein, ATP-synthase beta subunit, and unknown sequence QL4 14) are presumably associated with membership of the worker caste only. The only gene suggesting an association with the absence of reproduction is Cytochrome Oxidase I, being upregulated in adult workers (versus queens) and in non-reproductive workers (versus reproductive workers). No genes appeared to be associated with the presence of reproduction alone.
All four genes (Hexamerin, Larval Cuticle protein, ATP-synthase beta subunit and Cytochrome Oxidase I) previously known to be differentially expressed between queen- and worker-destined larvae of the honeybee, A. mellifera (Corona et al. 1999; Evans & Wheeler 1999, 2000), and identified as candidates for differential expression in B. terrestris by differential screening, were confirmed to be differentially expressed in B. terrestris larvae using Northern blots. Hexamerin shared the same expression pattern in A. mellifera and B. terrestris, being upregulated in last-instar worker-destined larvae in both species. However, the remaining three sequences were expressed at higher levels in last-instar queen-destined A. mellifera larvae, but in B. terrestris they were expressed at higher levels in last-instar worker-destined larvae. In B. terrestris, these genes were also expressed at higher levels in early-instar queen-destined larvae.
4. Discussion
Using SSH, we isolated 12 genes whose differential expression is associated with caste determination and differentiation of queen- and worker-destined bumble-bee (B. terrestris) larvae. Differential expression was confirmed in both the early instars and the last instar, consistent with previous research that demonstrates these instars to be the sensitive periods for queen–worker caste determination in B. terrestris (see §1). The sets of genes associated with caste-related expression in larvae tended not to be differentially expressed between adult queens and workers (7/12), or between reproductive and non-reproductive workers (9/12). By comparing both adult queens with workers, and reproductive with non-reproductive adult workers, we were able to discriminate one gene associated with the absence of reproduction, and three genes associated with membership of the worker caste. Sequence analysis showed that four sequences differentially expressed in queen- and worker-destined Bombus larvae are differentially expressed in queen- and worker-destined honeybee (A. mellifera) larvae, although the expression pattern of these genes was not always the same. An unexpected finding in B. terrestris was that, in queen-destined larvae, genes expressed in early instars were downregulated in the last instar, whereas, in worker-destined larvae, the same genes were not expressed in early instars and were upregulated in the last instar (e.g. Chymotrypsin; figure 2b). Overall, the differentially expressed genes isolated in B. terrestris did not appear to fall preferentially into any one functional group (table 1). In addition, the underlying reasons for the observed expression differences in specific genes were not obvious, and therefore they remain to be determined. For example, although early-instar queen-destined larvae exhibited higher expression of the Larval Cuticle protein gene than early-instar worker-destined larvae (table 1), this could not be attributed to differences in the growth rates of these instars, which are absent (Cnaani et al. 1997).
In this study, the first hypothesis we sought to test was whether genes differentially expressed between queen- and worker-destined larvae are also differentially expressed between adult queens and workers, and between reproductive and non-reproductive adult workers. Half (6/12) of the genes we identified as associated with caste-determining processes in larvae were not expressed differentially or not expressed at detectable levels in adult queen and worker phenotypes. This suggests that, in B. terrestris, different genes often underlie caste differences in larvae and adults. Although larval and adult phenotypes would be expected not to share the differential expression of some genes (e.g. genes associated with ovary activation in adults), it is conceivable that some genes (e.g. regulatory genes associated with the switch to development as one phenotype or another) do share patterns of differential expression across larvae and adults. Our results suggest that the latter class of genes do not represent the majority.
The second hypothesis we sought to test was whether genes associated with caste determination in B. terrestris larvae are identical with those known to be associated with caste determination in A. mellifera larvae (Corona et al. 1999; Evans & Wheeler 1999, 2000; Hepperle & Hartfelder 2001) and whether such genes share patterns of differential expression. We found four genes to be differentially expressed in queen- and worker-destined larvae of both B. terrestris and A. mellifera. This occurred even though recent molecular evidence suggests independent origins of eusociality in Apis and Bombus (Cameron & Mardulyn 2001). However, the precise pattern of differential gene expression was not identical. First, three of the four genes upregulated in last-instar worker-destined B. terrestris larvae were upregulated in last-instar queen-destined A. mellifera larvae. Second, in A. mellifera, last-instar queen-destined larvae downregulate many of the genes expressed by young bipotential larvae, but last-instar worker-destined larvae show similar expression profiles to those of young bipotential larvae (Evans & Wheeler 2000). In B. terrestris, however, genes expressed in early-instar queen-destined larvae are downregulated in early-instar worker-destined larvae. Third, in contrast to B. terrestris larvae, A. mellifera larvae express a distinct set of worker- or queen-related genes late in development (Evans & Wheeler 2000).
An unanticipated result of this study was our finding that genes differentially expressed in larvae were upregulated in queen-destined larvae early in development and upregulated in worker-destined larvae late in development. Explaining this novel pattern of gene expression remains a challenge for future studies. Together with the contrasting pattern of differential gene expression in B. terrestris and A. mellifera, it suggests that caste determination in B. terrestris involves a difference not so much in the type of genes expressed by queen- and worker-destined larvae, but primarily in the relative timing of their expression during development. This conclusion is of potential importance in the further study of caste evolution and, hence, of the evolution of other forms of phenotypic diversification via differential gene expression. In Bombus, comparative studies encompassing both species that lack a sensitive period of caste determination in the early larval instars (e.g. Bombus hypnorum; Röseler 1991), and the obligate social parasites (formerly Psithyrus species) that lack a worker caste altogether (Alford 1975), as well as experimental manipulations of gene expression using RNAi techniques (e.g. Beye et al. 2002), should prove fruitful for advancing still further our understanding of how differential gene expression underpins caste determination.
Acknowledgments
We thank Marie José Duchateau for providing bumble-bee stocks, Jay Evans for helpful advice and encouragement at the start of this project, Sam Martin for advice on SSH, and two anonymous referees for commenting on an earlier version of this paper. The Leverhulme Trust provided financial support for this work through a research grant to A.F.G.B., W.C.J. and R.L.H.
References
- Abouheif E, Wray G.A. Evolution of the gene network underlying wing polyphenism in ants. Science. 2002;297:249–252. doi: 10.1126/science.1071468. [DOI] [PubMed] [Google Scholar]
- Adams M.D, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Alford D.V. Davis-Poynter; London: 1975. Bumblebees. [Google Scholar]
- Altschul S.F, Madden T.L, Schaffer A.A, Zhang J, Zhang Z, Miller W, Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shahar Y, Robichon A, Sokolowski M.B, Robinson G.E. Influence of gene action across different time scales on behavior. Science. 2002;296:741–744. doi: 10.1126/science.1069911. [DOI] [PubMed] [Google Scholar]
- Beye M, Härtel S, Hagen A, Hasselmann M, Omholt S.W. Specific developmental gene silencing in the honey bee using a homeobox motif. Insect Mol. Biol. 2002;11:527–532. doi: 10.1046/j.1365-2583.2002.00361.x. [DOI] [PubMed] [Google Scholar]
- Bloch G. Regulation of queen–worker conflicts in bumble-bee (Bombus terrestris) colonies. Proc. R. Soc. B. 1999;266:2465–2469. doi: 10.1098/rspb.1999.0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch G, Hefetz A. Regulation of reproduction by workers in bumble-bee Bombus terrestris queenright colonies. Behav. Ecol. Sociobiol. 1999;45:125–135. [Google Scholar]
- Bortolotti L, Duchateau M.J, Sbrenna G. Effect of juvenile hormone on caste determination and colony processes in the bumblebee Bombus terrestris. Entomol. Exp. Appl. 2001;101:143–158. [Google Scholar]
- Boue S, Letunic I, Bork P. Alternative splicing and evolution. Bioessays. 2003;25:1031–1034. doi: 10.1002/bies.10371. [DOI] [PubMed] [Google Scholar]
- Bourke A.F.G, Ratnieks F.L.W. Kin-selected conflict in the bumble-bee Bombus terrestris (Hymenoptera: Apidae) Proc. R. Soc. B. 2001;268:347–355. doi: 10.1098/rspb.2000.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S.A, Mardulyn P. Multiple molecular data sets suggest independent origins of highly eusocial behavior in bees (Hymenoptera: Apinae) Syst. Biol. 2001;50:194–214. [PubMed] [Google Scholar]
- Cao W, et al. Comparing gene discovery from Affymetrix GeneChip microarrays and Clontech PCR-select cDNA subtraction: a case study. BMC Genomics. 2004 doi: 10.1186/1471-2164-5-26. (See http://www.biomedcentral.com/1471-2164/5/26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnaani J, Hefetz A. Are queen Bombus terrestris giant workers or are workers dwarf queens? Solving the ‘chicken and egg’ problem in bumblebee species. Naturwissenschaften. 2001;88:85–87. doi: 10.1007/s001140000202. [DOI] [PubMed] [Google Scholar]
- Cnaani J, Borst D.W, Huang Z.Y, Robinson G.E, Hefetz A. Caste determination in Bombus terrestris: differences in development and rates of JH biosynthesis between queen and worker larvae. J. Insect Physiol. 1997;43:373–381. doi: 10.1016/s0022-1910(96)00106-0. [DOI] [PubMed] [Google Scholar]
- Cnaani J, Robinson G.E, Hefetz A. The critical period for caste determination in Bombus terrestris and its juvenile hormone correlates. J. Comp. Physiol. A. 2000;186:1089–1094. doi: 10.1007/s003590000163. [DOI] [PubMed] [Google Scholar]
- Corona M, Estrada E, Zurita M. Differential expression of mitochondrial genes between queens and workers during caste determination in the honeybee Apis mellifera. J. Exp. Biol. 1999;202:929–938. doi: 10.1242/jeb.202.8.929. [DOI] [PubMed] [Google Scholar]
- Diatchenko L, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl Acad. Sci. USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchateau M.J. Biological aspects of rearing bumblebees for pollination. In: Sommeijer M.J, de Ruijter A, editors. Insect pollination in greenhouses. Utrecht University; Utrecht, The Netherlands: 2000. pp. 25–29. [Google Scholar]
- Duchateau M.J, Velthuis H.H.W. Development and reproductive strategies in Bombus terrestris colonies. Behaviour. 1988;197:186–207. [Google Scholar]
- Evans J.D, Wheeler D.E. Differential gene expression between developing queens and workers in the honey bee, Apis mellifera. Proc. Natl Acad. Sci. USA. 1999;96:5575–5580. doi: 10.1073/pnas.96.10.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J.D, Wheeler D.E. Expression profiles during honeybee caste determination. Genome Biol. 2000;2:1–6. doi: 10.1186/gb-2000-2-1-research0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J.D, Wheeler D.E. Gene expression and the evolution of insect polyphenisms. Bioessays. 2001;23:62–68. doi: 10.1002/1521-1878(200101)23:1<62::AID-BIES1008>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Guidugli K.R, Hepperle C, Hartfelder K. A member of the short-chain dehydrogenase/reductase (SDR) superfamily is a target of the ecdysone response in honey bee (Apis mellifera) caste development. Apidologie. 2004;35:37–47. [Google Scholar]
- Hartfelder K, Cnaani J, Hefetz A. Caste-specific differences in ecdysteroid titers in early larval stages of the bumblebee Bombus terrestris. J. Insect Physiol. 2000;46:1433–1439. doi: 10.1016/s0022-1910(00)00067-6. [DOI] [PubMed] [Google Scholar]
- Helms Cahan S, Keller L. Complex hybrid origin of genetic caste determination in harvester ants. Nature. 2003;424:306–309. doi: 10.1038/nature01744. [DOI] [PubMed] [Google Scholar]
- Hepperle C, Hartfelder K. Differentially expressed regulatory genes in honey bee caste development. Naturwissenschaften. 2001;88:113–116. doi: 10.1007/s001140000196. [DOI] [PubMed] [Google Scholar]
- Jin H, Cheng X, Diatchenko L, Siebert P.D, Huang C.C. Differential screening of a subtracted cDNA library: a method to search for genes preferentially expressed in multiple tissues. Biotechniques. 1997;23:1084–1086. doi: 10.2144/97236st05. [DOI] [PubMed] [Google Scholar]
- Julian G.E, Fewell J.H, Gadau J, Johnson R.A, Larrabee D. Genetic determination of the queen caste in an ant hybrid zone. Proc. Natl Acad. Sci. USA. 2002;99:8157–8160. doi: 10.1073/pnas.112222099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski R, Maleszka R. Evaluation of differential gene expression during behavioral development in the honeybee using microarrays and northern blots. Genome Biol. 2002;3:1–9. doi: 10.1186/gb-2002-3-2-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Vaamonde C, Koning J.W, Jordan W.C, Bourke A.F.G. No evidence that reproductive bumblebee workers reduce the production of new queens. Anim. Behav. 2003;66:577–584. [Google Scholar]
- Maynard Smith J, Szathmáry E. W.H. Freeman; Oxford: 1995. The major transitions in evolution. [Google Scholar]
- Michener C.D. Harvard University Press; Cambridge, MA: 1974. The social behaviour of the bees. [Google Scholar]
- Miura T. Morphogenesis and gene expression in the soldier-caste differentiation of termites. Insectes Soc. 2001;48:216–223. [Google Scholar]
- Miura T, Kamikouchi A, Sawata M, Takeuchi H, Natori S, Kubo T, Matsumoto T. Soldier caste-specific gene expression in the mandibular glands of Hodotermopsis japonica (Isoptera: Termopsidae) Proc. Natl Acad. Sci. USA. 1999;96:13 874–13 879. doi: 10.1073/pnas.96.24.13874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereboom, J. J. M. 1997 The significance of trophogenic and social factors on caste determination and differentiation in the bumblebee Bombus terrestris Ph.D. thesis, University of Utrecht, The Netherlands.
- Pereboom J.J.M, Velthuis H.H.W, Duchateau M.J. The organisation of larval feeding in bumblebees (Hymenoptera, Apidae) and its significance to caste differentiation. Insectes Soc. 2003;50:127–133. [Google Scholar]
- Piulachs M.D, Guidugli K.R, Barchuk A.R, Cruz J, Simões Z.L.P, Bellés X. The vitellogenin of the honey bee, Apis mellifera: structural analysis of the cDNA and expression studies. Insect Biochem. Mol. Biol. 2003;33:459–465. doi: 10.1016/s0965-1748(03)00021-3. [DOI] [PubMed] [Google Scholar]
- Robinson G.E. Sociogenomics takes flight. Science. 2002a;297:204–205. doi: 10.1126/science.1074493. [DOI] [PubMed] [Google Scholar]
- Robinson G.E. Genomics and integrative analyses of division of labor in honeybee colonies. Am. Nat. 2002b;160:S160–S172. doi: 10.1086/342901. [DOI] [PubMed] [Google Scholar]
- Röseler P.-F. Roles of morphogenetic hormones in caste polymorphism in bumblebees. In: Gupta A.P, editor. Morphogenetic hormones of arthropods. Roles in histogenesis, organogenesis, and morphogenesis. Rutgers University Press; New Brunswick: 1991. pp. 384–399. [Google Scholar]
- Sambrook J, Russell D.W. 3rd edn. Cold Spring Harbor Laboratory Press; New York: 2001. Molecular cloning: a laboratory manual. [Google Scholar]
- Scharf M.E, Wu-Scharf D, Pittendrigh B.R, Bennet G.W. Caste- and development associated gene expression in a lower termite. Genome Biol. 2003;4:R62. doi: 10.1186/gb-2003-4-10-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H.S, Vinson B, Coates C.J. Differential gene expression between alate and dealate queens in the red imported fire ant, Solenopsis invicta Buren (Hymenoptera: Formicidae) Insect Biochem. Mol. Biol. 2004;34:937–949. doi: 10.1016/j.ibmb.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Volny V.P, Gordon D.M. Genetic basis for queen-worker dimorphism in a social insect. Proc. Natl Acad. Sci. USA. 2002;99:6108–6111. doi: 10.1073/pnas.092066699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D.E. Developmental and physiological determinants of caste in social Hymenoptera: evolutionary implications. Am. Nat. 1986;128:13–34. [Google Scholar]
- Whitfield C.W, Cziko A.-M, Robinson G.E. Gene expression profiles in the brain predict behavior in individual honey bees. Science. 2003;302:296–299. doi: 10.1126/science.1086807. [DOI] [PubMed] [Google Scholar]
- Wilson E.O. Belknap Press; Cambridge, MA: 1971. The insect societies. [Google Scholar]


