Abstract
Balancing signals derived from the TGFβ family is crucial for regulating cell proliferation and differentiation, and in establishing the embryonic axis during development. TGFβ/BMP signaling leads to the activation and nuclear translocation of Smad proteins, which activate transcription of specific target genes by recruiting P/CAF and p300. The two members of the ZEB family of zinc finger factors (ZEB-1/δEF1 and ZEB-2/SIP1) regulate TGFβ/BMP signaling in opposite ways: ZEB-1/δEF1 synergizes with Smad-mediated transcriptional activation, while ZEB-2/SIP1 represses it. Here we report that these antagonistic effects by the ZEB proteins arise from the differential recruitment of transcriptional coactivators (p300 and P/CAF) and corepressors (CtBP) to the Smads. Thus, while ZEB-1/δEF1 binds to p300 and promotes the formation of a p300–Smad transcriptional complex, ZEB-2/SIP1 acts as a repressor by recruiting CtBP. This model of regulation by ZEB proteins also functions in vivo, where they have opposing effects on the regulation of TGFβ family-dependent genes during Xenopus development.
Keywords: CtBP/p300-P/CAF/Smad proteins/TGFβ/BMP/ZEB proteins
Introduction
Members of the TGFβ family of growth factors regulate cell growth and differentiation through their ability to regulate transcription of various target genes. Binding of TGFβ factors to specific transmembrane II receptors leads to the phosphorylation and activation of type I receptors, which in turn phosphorylates and activates the R-Smad proteins (reviewed in Massagué and Chen, 2000; Massagué et al., 2000; ten Dijke et al., 2000; Moustakas et al., 2001; Dennler et al., 2002). Broadly, while Smad1, Smad5 and Smad8 are activated in response to BMP and GDF factors, Smad2 and Smad3 respond to TGFβ, activin and nodal. Once activated, R-Smads bind to Co-Smad (Smad4 in mammals or Smad4α and Smad4β in Xenopus) and the resulting complexes translocate to the nucleus where they activate transcription of downstream target genes. In addition, Smad proteins bind to a number of coregulators that either repress or activate Smad-mediated transcription (e.g. Ski, TGIF, SNIP1, and FAST, TFE-III, OAZ, Mixer/Mix, respectively) (reviewed in Massagué and Wotton, 2000; Miyazono, 2000; Miyazono et al., 2001). Experiments with Xenopus have shown that regulation of Smad activity by these factors is crucial for a proper tissue specification and patterning during embryonic development (reviewed in Hill, 2001; von Bubnoff and Cho, 2001).
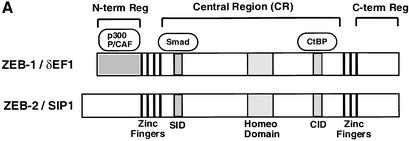
Proteins encoded by the Zfhx1a and Zfhx1b genes (ZEB-1/δEF1 and ZEB-2/SIP1, respectively) repress transcription of a wide number of key regulatory genes involved in different differentiation and developmental events (Genetta et al., 1994; Postigo and Dean, 1997; Postigo et al., 1997, 1999; Sekido et al., 1994; Takagi et al., 1998; Grooteclaes and Frisch, 2000; Murray et al., 2000; Comijn et al., 2001; Fontemaggi et al., 2001). ZEB repression is mediated through several repressor motifs within the central region of these proteins (Figure 1A; located in between the N- and C-terminal DNA-binding zinc finger clusters) through a mechanism that, at least in part, involves the NAD+-dependent corepressor CtBP (Postigo and Dean, 1999a, b, 2000; Zhang et al., 2002). CtBP also mediates the repressor activity of other key regulatory proteins including Snail, BKLF, TCF-3, Rb, BRCA-1 and Polycomb HPC2 (reviewed in Turner and Crossley, 2001; Chinnadurai, 2002). Interestingly, ZEB-1/δEF1 has also been shown to directly activate the transcription of the ovalbumin and vitamin D3 receptor genes, although the molecular mechanism behind these effects is unknown (Chamberlain and Sanders, 1999; Lazarova et al., 2001).
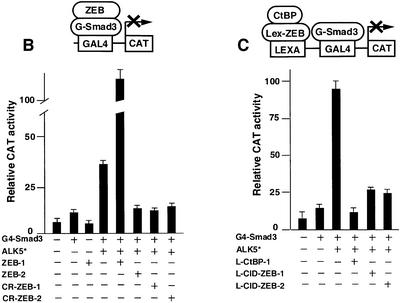
Fig. 1. (A) Schematic representation of the ZEB family (ZEB-1/δEF1 and ZEB-2/SIP1) of two-handed zinc finger factors. The central region (CR) in between both zinc finger clusters act as a repressor domain in part through the recruitment of the CtBP corepressor through a CtBP-interacting domain (CID). There is a region 3′ of the N-terminal zinc fingers that acts as a Smad-binding domain (SID). The N-terminal region of ZEB-1/δEF1 serves as a p300–p/CAF-binding domain (see Figure 2C). (B) The central repressor (CR) domain of both ZEB-1/δEF1 and ZEB-2/SIP1 inhibits the activity of Smad3 when brought directly to the promoter. 293T cells were transfected with 0.25 µg of a reporter construct (PG5-CAT) containing five Gal4 DNA-binding sites driving the CAT gene, 0.25 µg of Gal4–Smad3 and 0.1 µg of an expression vector for constitutively active ALK5 (T204D) (ALK5*) along with either 1 µg of expression vectors for either ZEB-1/δEF1 or ZEB-2/SIP1, 0.8 µg of either CR-ZEB-1 or CR-ZEB-2, or 0.7 µg of vector alone (CS2MT, not shown). (C) The CID of both ZEB proteins or CtBP-1 itself represses Smad3 transcriptional activation. 293T cells were cotransfected with 0.5 µg of a CAT reporter construct (PL6G5-CAT) containing both LexA and Gal4 DNA-binding sites along with 0.25 µg of Gal4–Smad3, 0.1 µg of an expression vector for constitutively active ALK5 (T204D) (ALK5*) and either 0.45 µg of LexA empty vector (PBXL3, not shown) or 0.5 µg of LexA-CtBP-1, LexA-CID-ZEB-1 or LexA-CID-ZEB-2 expression vectors. In all the above experiments, equal molar amounts of each empty vector were transfected as controls; 0.1 µg of SV40βgal was also cotransfected to control for transfection efficiency. Luciferase values were assessed as described in Material and methods.
We have recently found that ZEB proteins bind to Smad proteins and antagonistically regulate their transcriptional activity (Postigo, 2003). Thus, while ZEB-1/δEF1 activates Smad-mediated transcription, ZEB-2/SIP1 represses it. ZEB proteins also have an antagonistic role in several TGFβ and BMP functions, the molecular mechanism of which was previously unknown.
Here, we provide evidence that the two ZEB proteins regulate TGFβ/BMP signaling through differential recruitment of coactivators (p300 and P/CAF) and corepressors (CtBP) to the Smad complex. p300 and P/CAF are acetyltransferases that activate transcription by acetylating histones and loosening chromatin structure (Bannister and Kouzarides, 1996; Ogryzko et al., 1996; Yang et al., 1996). As with many other DNA-binding factors, Smad proteins recruit p300 and P/CAF to mediate their transcriptional effects, and this interaction is dependent on the phosphorylation/activation of R-Smad induced by TGFβ signaling (Feng et al., 1998; Janknecht et al., 1998; Pouponnot et al., 1998; Itoh et al., 2000).
We found that the N-terminal region of ZEB-1/δEF1 (but not ZEB-2/SIP1) binds both p300 and P/CAF. Moreover, the interaction between R-Smad and ZEB-1/δEF1 forms a complex that recruits p300 much more efficiently, thus accounting for their transcriptional synergy. In addition to acetylating histones, p300 and P/CAF acetylate a number of transcription factors (e.g. p53, E2F, Rb, etc.) to modulate their activity (Gu and Roeder, 1997; Martinez-Balbas et al., 2000; Chan et al., 2001; reviewed in Kouzarides, 2000). Binding of P/CAF to ZEB-1/δEF1 acetylates several lysine residues following the CtBP interaction domain (CID) of ZEB-1/δEF1, displacing CtBP and switching ZEB-1/δEF1 from a repressor to an activator. In contrast, ZEB-2/SIP1 is unable to interact with p300 or P/CAF and thus only serves as a transcriptional repressor by recruiting CtBP to the Smad complex. These opposing roles of ZEB-1/δEF1 and ZEB-2/SIP1 in cell culture systems were also found in vivo, where the ZEB proteins also show antagonistic roles in the regulation of TGFβ-dependent genes during Xenopus development.
Results
The CID of both ZEB-1/δEF1 and ZEB-2/SIP1 represses Smad activity
ZEB proteins repress transcription of a wide number of genes involved in differentiation and development. The central region (between the N- and C-terminal zinc fingers) of both ZEB-1/δEF1 and ZEB-2/SIP1 serves as a transcriptional repressor domain in part as a result of the recruitment of CtBP through a specific CID (Figure 1A) (Postigo and Dean, 1999a, 2000; Grooteclaes and Frisch, 2000; Zhang et al., 2002). We have previously found that ZEB proteins have opposing effects on TGFβ signaling, with ZEB-1/δEF1 synergizing with Smad-mediated transcription and ZEB-2/SIP1 repressing it (Postigo, 2003).
Smad3 tethered to the DNA as a Gal4 fusion protein mediates transcription when activated by the cotransfection of a vector encoding for the constitutively active type I receptor ALK 5 (T204D) (Figure 1B). We found that, as when using natural TGFβ- and BMP-dependent promoters, the transcriptional activity of Gal4–Smad3 was enhanced by cotransfection of ZEB-1/δEF1 and repressed by ZEB-2/SIP1 (Figure 1B). Next, we investigated the effect of the central region of both ZEB proteins (CR-ZEB-1 and CR-ZEB-2) on the transcriptional activity mediated by TGFβ1. This central region includes the Smad-interaction domain (SID) and the CID region. As expected, the central repressor region of ZEB-2/SIP1 (CR-ZEB-2) repressed Gal4–Smad3 (Figure 1B). Surpris ingly, we found that the central repressor domain of ZEB-1/δEF1 (CR-ZEB-1) also inhibited Smad3 activity (Figure 1B).
Since both ZEB proteins repress transcription by mechanisms that involved, at least in part, the recruitment of CtBP, we tested whether CtBP is responsible for the repression of Smad3-mediated transcription by CR-ZEB-1 and CR-ZEB-2. To that effect, CtBP-1 itself or the CID regions of ZEB-1/δEF1 and ZEB-2/SIP1 were fused to the DNA-binding domain (DBD) of the bacterial protein LexA and tested for their ability to repress transcriptional activation mediated by Gal4–Smad3. We found that the CIDs of both ZEB proteins (as well as CtBP-1 itself) blocked transcriptional activation by Smad3 (Figure 1C).
These results indicated that in order for full-length ZEB-1/δEF1 to activate Smad-mediated transcription, ZEB-1/δEF1 has to have a domain outside the zinc finger clusters that is dominant over the repressor effect of the CID region. However, it remains unclear how full-length ZEB-1/δEF1 could augment Smad activity.
The N-terminal region of ZEB-1/δEF1 interacts with p300 and P/CAF to assemble a ZEB-1/δEF1–Smad activation complex
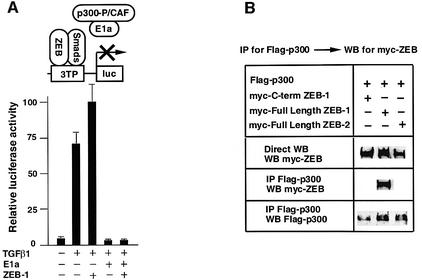
Smad proteins activate transcription of target genes through the recruitment of p300 and P/CAF coactivators (Feng et al., 1998; Janknecht et al., 1998; Pouponnot et al., 1998; Itoh et al., 2000). Therefore, we reasoned that ZEB-1/δEF1 might synergize with Smad proteins by promoting the recruitment of P/CAF–p300 to the complex. The adenovirus E1a protein, which binds and inactivates p300 (Chakravarti et al., 1999), has been shown to inhibit transcriptional activation of TGFβ-dependent reporters by TGFβ (Feng et al., 1998). As shown in Figure 2A, we found that E1a not only blocked the activation of the 3TP reporter by TGFβ but also blocked the synergy between ZEB-1/δEF1 and TGFβ.
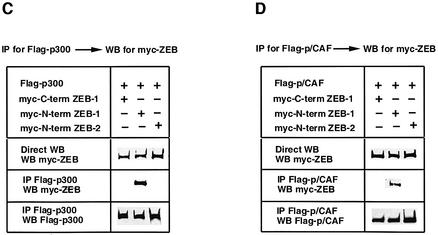
Fig. 2. The N-terminal domain of ZEB-1/δEF1 interacts with coactivators p300 and P/CAF. (A) ZEB-1/δEF1 synergy with TGFβ requires the p300 coactivator. 3TP-luc reporter (0.3 µg) was transfected in Mv1Lu along with 0.3 µg of an expression vector for E1A 12S and either 0.48 µg of CS2MT empty vector (not shown) or 0.7 µg of CS2MT-ZEB-1 either in the absence or presence of 25 pM of TGFβ1. SV40βgal (0.1 µg) was cotransfected in all points to control for transfection efficiency. (B) Full-length ZEB-1/δEF1, but not ZEB-2/SIP1, interacts with p300. 293T cells were cotransfected with 10 µg of a Flag-tagged p300 expression vector and 20 µg of either myc-tagged ZEB-1/δEF1 or myc-tagged ZEB-2/SIP1 expression vectors. After 48 h, cells lysates were immunoprecipitated (IP) for Flag-p300 and the binding to ZEB-1/δEF1 (but not to ZEB-2/SIP1) determined by western blotting (WB) with an anti-myc 9E10 mAb. (C) The N-terminal domain of ZEB-1/δEF1 interacts with p300. 293T cells were cotransfected with 10 µg of Flag-tagged p300 expression vector and 5 µg of the indicated myc-tagged ZEB expression vectors. Cell lysates were immunoprecipitated for Flag-p300 and binding to the different regions of ZEB-1/δEF1 and ZEB-2/SIP1 determined by western blotting with an anti-myc 9E10 mAb. (D) P/CAF interacts with the N-terminal region of ZEB-1/δEF1 but not ZEB-2/SIP1. Experiments were performed similarly to those in (C) except that P/CAF was expressed instead of p300.
The sensitivity of ZEB-1/δEF1–TGFβ synergy to E1a inhibition suggests that ZEB-1/δEF1 activity may depend on p300. Indeed, we found that full-length ZEB-1/δEF1, but not ZEB-2/SIP1, co-immunoprecipated with p300 (Figure 2B). From Figure 1B it appears that the region of ZEB-1/δEF1 responsible for synergy with Smads is outside the central region. Therefore, we investigated the N- and C-terminal regions of both ZEB proteins for binding to p300. As shown in Figure 2C, the N-terminal region of ZEB-1/δEF1 co-immunoprecipitated with p300, whereas the N-terminal region of ZEB-2/SIP1 or the C-terminal region of ZEB-1/δEF1 did not. Since Smad3 has also been shown to bind to P/CAF (Itoh et al., 2000), we investigated whether ZEB-1/δEF1 and ZEB-2/SIP1 could bind P/CAF. Indeed, we found that P/CAF also interacts with the N-terminal region of ZEB-1/δEF1 but not with ZEB-2/SIP1 (Figure 2D).
From Figure 2 C and Postigo (2003), it seems that ZEB-1/δEF1 contains independent binding sites for P/CAF–p300 and R-Smads. It was then tempting to speculate whether ZEB-1/δEF1 might serve as a scaffold to facilitate the assembly of a Smad–p300 transcriptional complex. To test this possibility we examined the effect of coexpressing ZEB-1/δEF1 on the interaction between Smad3 and p300. As reported (Feng et al., 1998; Janknecht et al., 1998; Pouponnot et al., 1998; Itoh et al., 2000), in the absence of TGFβ signaling we did not detect any interaction between Smad3 and p300, but we did when the constitutively active type I receptor ALK5 (T204D) was coexpressed (Figure 3A). Interestingly, coexpression of ZEB-1/δEF1 led to a much stronger co-immunoprecipitation of p300 by Smad3 (Figure 3A), suggesting that indeed ZEB-1/δEF1 enhances the assembly of a Smad3– p300 complex. We confirmed this result using a p300– VP16 fusion protein in a two-hybrid approach. As shown in Figure 3B, expression of ZEB-1/δEF1 further increased the recruitment of p300–VP16 to Smad3.
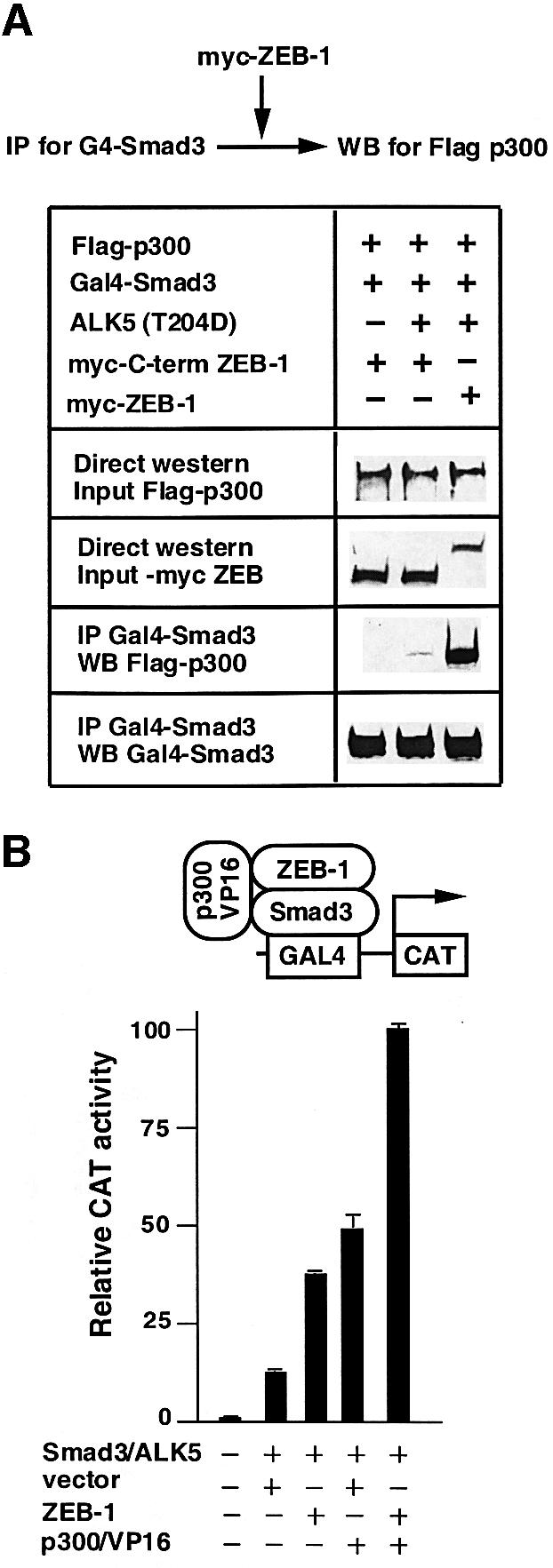
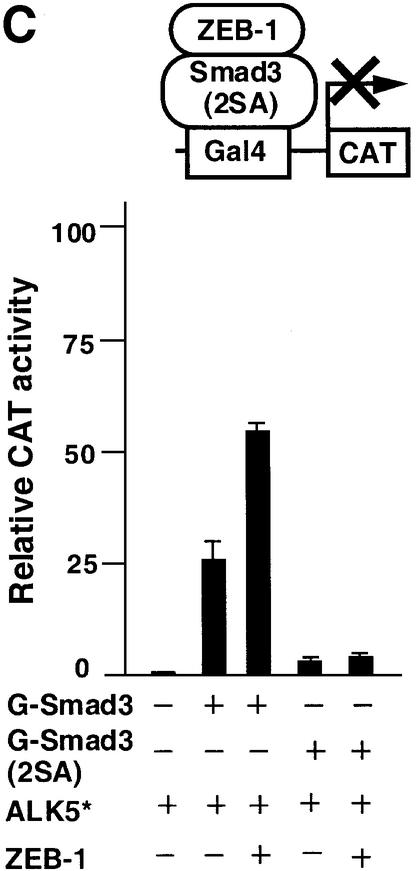
Fig. 3. ZEB-1/δEF1 promotes the assembly of a Smad3–p300 complex. (A) 293T cells were cotransfected with 5 µg of the indicated expression vectors and 0.8 µg of the constitutively active ALK5 (T204D). ZEB-1 contains the sequence of ZEB-1/δEF1 from the N-terminal region to the SID. After 48 h, cell lysates were immunoprecipitated (IP) for Smad3 and associated p300 was determined by western blotting (WB). The input (indicated as direct western) represents 15% of the lysate. (B) ZEB-1/δEF1 recruits p300 to Smad3. The PG5-CAT reporter containing five Gal4 sites fused to CAT (0.6 µg) was transfected in C33a cells along with the following expression vectors: 0.2 µg of Gal4–Smad3, 0.3 µg of constitutively active ALK5, either 0.48 µg of empty vector CS2MT or 0.7 µg of CS2MT-ZEB-1 and 0.25 µg of a p300–VP16 fusion protein. One-tenth of 1 µg of SV40βgal was cotransfected to control for transfection efficiency. CAT assays were performed as described in Materials and methods. (C) Formation of a p300–Smad3–ZEB-1/δEF1 complex requires the binding of p300 to Smad3. C33a cells were transfected with the same CAT reporter as in (B) along with either 0.2 µg of Gal4–Smad3 or Gal–Smad3 (2SA), 0.3 µg of constitutively active ALK5 (T204D) (ALK5*) and either 0.48 µg of empty vector CS2MT (not shown) or 0.7 µg of CS2MT-ZEB-1. One tenth of 1 µg of SV40βgal was cotransfected to control for transfection efficiency.
The assembly of a ZEB-1/δEF1–Smad3–p300 complex still requires interaction between Smad3 and p300, as a Smad3 mutant (2SA) deficient for binding to p300 not only did not activate transcription (as reported in Feng et al., 1998) but also failed to synergize with ZEB-1/δEF1 (Figure 3C).
Together, these results suggested that the N-terminal domain of ZEB-1/δEF1 allows the assembly of a Smad–ZEB-1/δEF1–p300 activation complex. In contrast, ZEB-2/SIP1, which lacks the p300-binding site, cannot assemble this coactivator complex and is thus a constitutive repressor by recruiting CtBP to the Smads.
P/CAF recruitment leads to the displacement of CtBP-1 from ZEB-1/δEF1
The interaction between ZEB-1/δEF1 and p300 explains the transcriptional synergy between ZEB-1/δEF1 and R-Smads. However, the above results did not clarify the interplay between coactivator (P/CAF–p300) and corepressor (CtBP-1), and how the binding of p300 and P/CAF to ZEB-1/δEF1 could be dominant over the repressor domain (the CID region) to switch ZEB-1/δEF1 from a repressor to an activator. One possibility was that the binding of P/CAF–p300 to ZEB-1/δEF1 leads to displacement of CtBP-1. It was recently noted by Goodman’s group that in many CtBP-binding proteins CID sites are followed by lysines and that P/CAF can acetylate these residues blocking CtBP binding (Zhang et al., 2000).
CtBP-binding sites in ZEB-1/δEF1 are also flanked by basic residues, and we found that ZEB-1/δEF1 was acetylated by P/CAF in vitro and that the mutation of the lysines following the CID sites to alanines inhibited acetylation (Figure 4A).
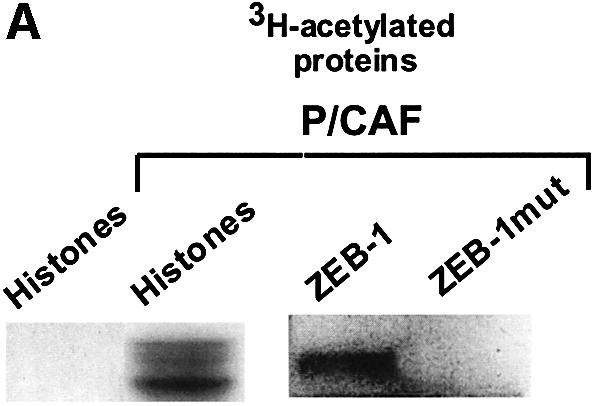
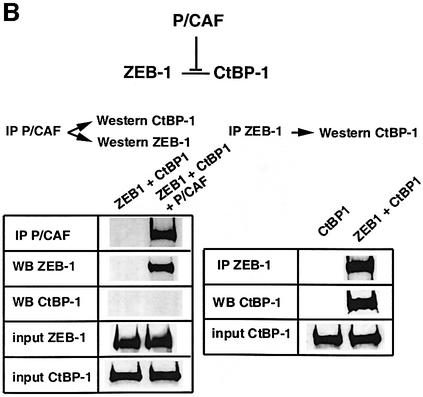
Fig. 4. P/CAF acetylates the lysine residues flanking the CID in ZEB-1/δEF1 and displaces CtBP. (A) Purified recombinant P/CAF was used in an in vitro assay to acetylate core histones or agarose beads coupled to either GST–CID-ZEB-1 (wild type) or GST–CID-ZEB-1 mutant (K741A, K774A, K775A). (B) Expression of P/CAF leads to displacement of CtBP-1 from ZEB-1/δEF1. The indicated tagged expression vectors (Flag-P/CAF, Gal4-ZEB-1/δEF1 and myc-CtBP) were transfected into 293T cells as in previous experiments and cell extracts were immunoprecipitated with antibodies to either Gal4-ZEB- 1/δEF1 or Flag-P/CAF. Note that the experiments were designed such that a similar amount of ZEB-1/δEF1 was immunoprecipitated directly (anti-Gal4-ZEB-1/δEF1) or co-immunoprecipitated by P/CAF (compare the left and right panels). No CtBP-1 was associated with ZEB-1/δEF1 when it was bound to P/CAF.
In immuno-coprecipitation assays we found that whereas ZEB-1/δEF1 interacts very efficiently with CtBP-1 (right panel in Figure 4B), the ZEB-1/δEF1 complex immunoprecipitated by P/CAF was completely devoid of CtBP-1 (left panel in Figure 4B). This experiment indicated that binding of P/CAF to ZEB-1/δEF1 displaces CtBP-1 from ZEB-1/δEF1. However, and as reported for the mutation of the lysines following the CID site in E1a (Zhang et al., 2000; Madison et al., 2002), the mutation of these residues in ZEB-1/δEF1 also had only a partial effect on the ability of ZEB-1/δEF1 to repress transcription through CtBP (data not shown). It is possible that in vivo acetylation of lysine residues by P/CAF is having a more dramatic effect on CtBP-1 binding than mutation. Alternatively, P/CAF may displace CtBP-1 from binding to CtBP-binding proteins by additional mechanisms such as a steric block.
Antagonistic roles of ZEB-1/δEF1 and ZEB-2/SIP1 in the regulation of activin- and BMP-dependent genes during Xenopus embryogenesis
The results described here and in Postigo (2003) indicate that ZEB proteins have opposing effects on TGFβ/BMP signaling. Regulation of activin and BMP signaling is crucial for establishing the dorso-ventral axis in vertebrate embryogenesis (Candia et al., 1997; reviewed in Dale and Wardle, 1999; McDowell and Gurdon, 1999; Hill, 2001; Muñoz-Sanjuan and Brinvalou, 2001; von Bubnoff and Cho, 2001). In view of our results in cell culture systems, we wondered whether ZEB proteins could also have opposing roles in the expression of genes targeted by activin and BMP signaling during Xenopus embryogenesis.
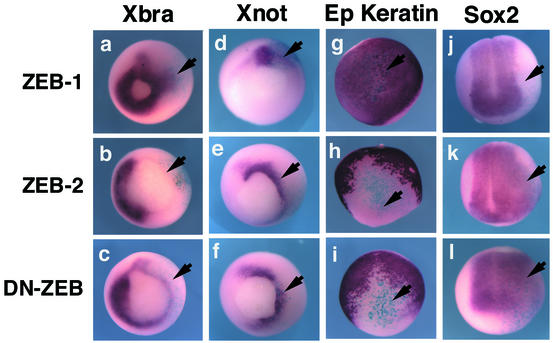
Injection of ZEB-2/SIP1 mRNA into Xenopus embryos has been examined previously (Remacle et al., 1999; Eisaki et al., 2000; Lerchner et al., 2000; Papin et al., 2002). Such ectopic expression of ZEB-2/SIP1 led to head abnormalities and thickening of the neural tube, induction of the neural marker NCAM and repression of the activin-dependent Brachyury gene (Eisaki et al., 2000; Lerchner et al., 2000). We found that injection of ZEB-2/SIP1 mRNA into two-cell stage Xenopus embryos, in addition to blocking Brachyury (arrow in Figure 6b), also inhibited expression of the BMP-regulated msx-1 gene (Figure 7) and induced a neuralization and dorsalization of the embryo (Figure 5). This was shown by ectopic expression of markers such as Sox2, Six3, Xnot and NCAM, and accompanying loss of epidermal keratin (Figures 6e, h and k and 7; data not shown).
Fig. 6. Regulation of activin/BMP-dependent genes and neural genes by ZEB-1/δEF1 and ZEB-2/SIP1. In situ hybridization for the indicated genes in embryos injected with ZEB-1/δEF1, ZEB-2/SIP1 or DN-ZEB RNAs along with mRNA encoding β-galactosidase (to mark the side of the injection) and allowed to develop to stage 11.5 (for expression of Xbrachyury, Xnot and epidermal keratin) or stage 13 (for Sox2). Whereas ZEB-1/δEF1 induced ectopic expression of Brachyury, ZEB-2/SIP1and DN-ZEB inhibited Brachyury expression. There was no effect on the control uninjected side (left side of each embryo).
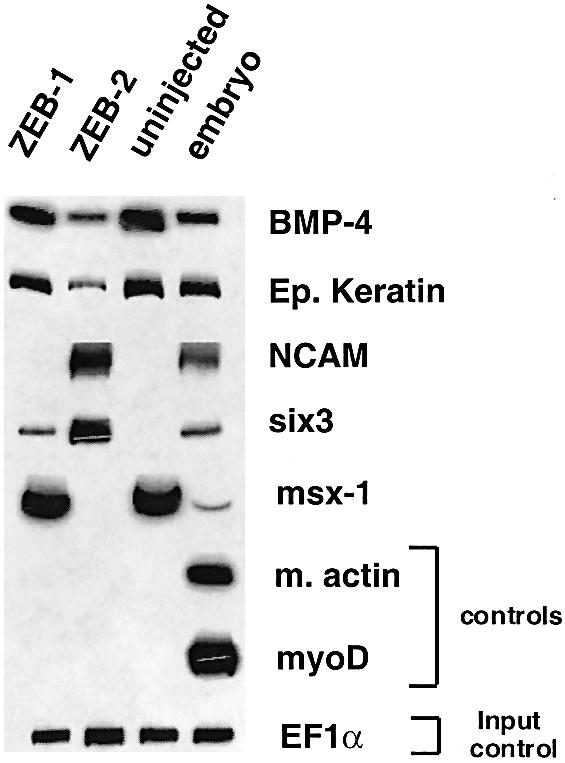
Fig. 7. Differential effect of ZEB-1/δEF1 versus ZEB-2/SIP1 on gene expression. mRNA isolated from animal caps of Xenopus embryos (either uninjected or injected with mRNA for ZEB-1/δEF1 or ZEB-2/SIP1) was used for RT–PCR analysis of the indicated genes (see Materials and methods for details).
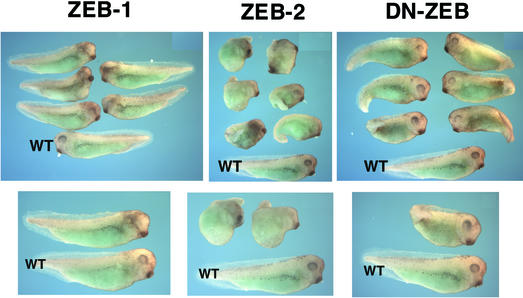
Fig. 5. Regulation of Xenopus development by ZEB-1/δEF1 and ZEB-2/SIP1 in vivo. Phenotype of Xenopus embryos injected at the two-cell stage with RNA for ZEB-1/δEF1, ZEB-2/SIP1 or DN-ZEB. Embryos were allowed to develop until tailbud (stage 29–30). Embryos injected with ZEB-2/SIP1 or DN-ZEB mRNA were significantly dorsalized. In contrast, ZEB-1/δEF1 showed no dorsalizing effect, and there was disruption of eye development in the injected side. In each panel, a wild-type embryo is shown at the bottom (indicated as WT).
BMP-4 signaling is known to inhibit neural induction in Xenopus (Jones et al., 1992; reviewed in Dale and Wardle, 1999). Interestingly, the BMP-4 promoter contains ZEB sites and is itself BMP dependent (Vainio et al., 1993; Ebara et al., 1997; Kawasaki et al., 1999). As shown in Figure 7, we found that BMP-4 expression was inhibited by ZEB-2/SIP1, which could explain the neuralization/dorsalization effect of ZEB-2/SIP1. This in vivo repression of Smad-dependent genes by ZEB-2/SIP1 is also consistent with the inhibition of Smad signaling by ZEB-2/SIP1 that we observed in cell line systems, as described in this manuscript and in Postigo (2003).
Next, we examined the effect of ZEB-1/δEF1 RNA injection into Xenopus embryos. Consistent with our results in cultured cell lines showing a synergy between TGFβ family members and ZEB-1/δEF1, we found that microinjection of ZEB-1/δEF1 also had an opposite effect to ZEB-2/SIP1 in vivo, as it induced the ectopic expression of the activin-dependent Brachyury gene (arrow in Figure 6a).
From these results, it seems that ZEB-1/δEF1 and ZEB-2/SIP1 can regulate activin/BMP signaling in vivo. However, the above studies relied on ectopic overexpression of ZEB-1/δEF1 and ZEB-2/SIP1 in Xenopus embryos. Therefore, it was still unclear whether endogenous ZEB proteins do play a role. To address this we decided to use a dominant-negative approach to displace ZEB proteins from binding to their DNA-binding sites in activin/BMP-dependent promoters. ZEB-1/δEF1 and ZEB-2/SIP1 share high homology in their zinc finger clusters that serve as DBDs (Genetta et al., 1994; Sekido et al., 1997). We and others have shown that the C-terminal zinc finger region has higher affinity for DNA than the full-length proteins, displaces endogenous ZEB proteins from DNA-binding sites and serves as an efficient dominant negative for both ZEB proteins (Postigo and Dean, 1997; Remacle et al., 1999; Grooteclaes and Frisch, 2000; Fontemaggi et al., 2001). Therefore, we reasoned that this dominant negative (DN-ZEB) could be used to test the role of endogenous ZEB proteins in activin and BMP signaling in Xenopus embryogenesis.
As in other ZEB-dependent genes, the particular configuration and arrangement of the ZEB DNA-binding sites in the promoter of the Xenopus Brachyury gene is important to determine the specificity of these sites for ZEB regulation (Remacle et al., 1999; Lerchner et al., 2000). We examined the effect of injecting DN-ZEB mRNA on the expression of Brachyury. Interestingly, we found that DN-ZEB had the same effect as ZEB-2/SIP1, namely it inhibited the expression of the activin-dependent Brachyury gene (arrow in Figure 6c). Likewise, DN-ZEB also induced dorsalization of the embryo, ectopic neural differentiation and expression of neural markers (Figures 5 and 6f, i and l).
Because ZEB-2/SIP1 and DN-ZEB led to a similar phenotype, we suggest that DN-ZEB and ZEB-2/SIP1 are displacing an activator from the Brachyury promoter. This activator is likely to be ZEB-1/δEF1 (i) because of the already mentioned specificity of the ZEB sites in the Brachyury gene (Remacle et al., 1999; Lerchner et al., 2000) and (ii) because if DN-ZEB and ZEB-2/SIP1 were displacing an activator other than ZEB-1/δEF1, injection of ZEB-1/δEF1 should have also displaced this activator and had an effect similar to ZEB-2/SIP1 and DN-ZEB (namely the repression of Brachyury); instead, injection of ZEB-1/δEF1 induced ectopic expression of Brachyury (arrow in Figure 6a).
The effect of injecting ZEB-2/SIP1 RNA appears to be somewhat more dramatic than that caused by DN-ZEB injection, perhaps reflecting the importance of active repression versus simple displacement. Both ZEB-2/SIP1 and DN-ZEB would displace ZEB-1/δEF1 from the Brachyury promoter, but ZEB-2/SIP1 has an active transcriptional repressor domain that DN-ZEB does not have. Thus, expression of ZEB-2/SIP1 is likely to replace ZEB-1/δEF (associated with Smads and p300) with an active repressor complex containing ZEB-2/SIP1 and CtBP. Interestingly, ZEB-2/SIP1 expression is induced by TGFβ (Comijn et al., 2001; our own data not shown), suggesting the possibility of a feedback mechanism between both ZEB proteins (see Discussion in Postigo, 2003).
Taken altogether, the above results indicate that ZEB-1/δEF1 and ZEB-2/SIP1 not only antagonize in their regulation of Smad-mediated functions in tissue culture systems, but they also have opposing roles in modulating TGFβ family signaling in vivo.
Discussion
ZEB-1/δEF1 and ZEB-2/SIP1 are structurally quite similar. Both contain N- and C-terminal zinc fingers, which serve as DBDs, and their central region interacts with the corepressor CtBP (Genetta et al., 1994; Sekido et al., 1994, 1997; Postigo and Dean, 1999a, 2000; Verschueren et al., 1999). Although ZEB proteins have been classically described as transcriptional repressors, they function antagonistically in regulating TGFβ/BMP signaling (Postigo, 2003). In this paper, we present evidence indicating that this antagonism results from the differential recruitment of coactivators (p300 and P/CAF) and corepressors (CtBP) by ZEB-1/δEF1 and ZEB-2/SIP1. TGIF also recruits CtBP to repress Smads but it does not require direct binding to DNA to mediate its repressor effects (Melhuish and Wotton, 2000). In contrast, regulation of TGFβ signaling by ZEB proteins is unique not only because of the specificity dictated by their DNA-binding sites in TGFβ-dependent promoters, but also because of their mechanism of action. Whereas ZEB-2/SIP1 only binds CtBP and acts as a Smad repressor, ZEB-1/δEF1 recruits p300 and P/CAF to the Smads to form a complex that not only activates transcription of TGFβ-dependent genes more efficiently but also displaces CtBP from ZEB-1/δEF1. p300 has also been shown to act as a bridge between Smad1 and Stat3 in the developing brain (Nakashima et al., 1999). ZEB-1/δEF1 activates the vitamin D3 receptor and the estrogen-responsive ovalbumin gene in a cell-specific manner (Chamberlain and Sanders, 1999; Lazarova et al., 2001; Dillner and Sanders, 2002). Although the molecular mechanism remains to be determined, it is tempting to speculate that the activation of these two genes by ZEB-1/δEF1 also involves recruitment of p300 and P/CAF and displacement of CtBP.
In the last few years, there have been a number of reports indicating that p300 and P/CAF acetylate proteins other than histones, including many transcription factors (reviewed in Kouzarides, 2000). The effect of acetylation on transcription factors varies from protein to protein (Gu and Roeder, 1997; Martinez-Balbas et al., 2000; reviewed in Kouzarides, 2000). However, and even for p53 (where the effect of acetylation has been extensively studied), there is still controversy in the literature about the consequences of its acetylation that could reflect specific functions for the acetylation of individual lysines or different experimental conditions (Nakamura et al., 2000; Barlev et al., 2001; Espinosa and Emerson, 2001; Prives and Manley, 2001). In general, acetylation of transcription factors has been associated with stimulation of transcription. In this line, our results indicated that P/CAF switches ZEB-1/δEF1 to an activator. When the N-terminal p300–P/CAF binding site in ZEB-1/δEF1 was deleted (as in the case of CR-ZEB-1 in Figure 1A), the truncated ZEB-1/δEF1 behaved in the same fashion as ZEB-2/SIP1: it became a repressor of Smad activity.
How is the switch of ZEB-1/δEF1 from a transcriptional repressor to a transcriptional activator regulated? P/CAF and p300 are crucial for the activity of differentiation-promoting transcription factors such as the retinoic acid receptor and the myogenic factor myoD (Puri et al., 1997; Kawasaki et al., 1998; reviewed in Goodman and Smolik, 2000). At least in the case of myogenesis, it has been demonstrated that P/CAF–p300 is unable to associate with myoD until myoblasts receive a signal to differentiate into myotubes. It is then possible that endogenous ZEB-1/δEF1 does not associate with endogenous P/CAF–p300 at significant levels until such differentiation signals occur. This would allow the ZEB-1/δEF1–CtBP complex (which would be predominant in the absence of P/CAF–p300 binding) to actively repress transcription of target genes prior to the onset of differentiation and TGFβ/BMP signals. This model of regulation would be somewhat analogous to cell cycle control, where other transcription factors, E2F proteins, oscillate between activation and repression of target genes through differential interaction with P/CAF–p300 and the retinoblastoma protein Rb (along with other corepressors), respectively (reviewed in Harbour and Dean, 2000).
Altogether, the results presented here indicate that ZEB proteins play an important role in the regulation of the TGFβ family activities during cell differentiation and embryonic development. Moreover, their intricate mechanism of action also offers a unique model to study the transcriptional regulation of Smad-mediated signaling.
Materials and methods
Cells and cell culture
C33a and Mv1Lu cells were obtained from the American Type Culture Collection (Manassas, VA). 293T cells were obtained from Dr S.J.Korsmeyer (Dana Farber Cancer Institute, Boston, MA). Cell lines were maintained in Dulbecco’s modified Eagle’s medium containing 12% fetal calf serum (Gibco-Life Technologies, Grand Island, NY).
Plasmid construction
The 3TP-lux reporter was obtained from Dr J.Massagué (Sloan Kettering Cancer Center, New York, NY). A constitutively active form for activin-like kinases ALK5 (T204D) was obtained from Dr K.Miyazono (Cancer Institute, Japanese Foundation for Cancer Research, Tokyo, Japan).
The different myc-tagged versions of ZEB proteins have been described (Postigo, 2003). Mutations of individual amino acids in the context of either full-length ZEB-1/δEF1 or the CID region were generated using the QuikChange XL Directed-Mutagenesis kit (Stratagene, La Jolla, CA). PG5-CAT containing five Gal4 DNA-binding sites upstream of the fibronectin TATA box and fused to the chloramphenicol acetyl transferase (CAT) gene and PL6G5-CAT (as PG5-CAT but containing also six Lex DNA-binding sites) reporters have been described previously (Postigo et al., 1997). Gal4–Smad3 was constructed by cloning the Smad3 cDNA in-frame with the DBD of the Gal4 protein in the PM1 vector. Gal4–Smad3 (2SA) was obtained from Dr X.H.Feng (Baylor College, Houston, TX) (Feng et al., 1998). LexA-CtBP-1, LexA-CID-ZEB-1 and LexA-CID-ZEB-2 were described previously (Postigo and Dean, 2000). pCI-neo-Flag-p300 and pCI-neo-Flag P/CAF were obtained from Dr R.L.Schiltz (NIH, Bethesda). CMV-p300/VP16 was obtained from Dr D.Livingston (Dana Farber Cancer Institute, Boston, MA).
Transfections
Cells were transfected by the calcium phosphate method or with Superfectin reagent (Qiagen, Hilden, Germany) as described previously (Postigo and Dean, 2000). For luciferase assays, 5–12 h after transfection, cells were washed and activated with the indicated amounts of TGFβ-1. Twenty-four hours later, luciferase activity was assessed. As an internal control for transfection, cells were cotransfected with SV40 β-gal (Promega Corp., Madison, WI), the values of which are determined using a luminescent β-galactosidase detection Kit II® (Clontech Laboratories, Palo Alto, CA). Transcriptional assays using a CAT reporter gene were performed as described previously (Postigo and Dean, 2000).
In vitro acetylation assays
Purified core chicken histones (Upstate Biotechnologies, UBI) or either GST–CID-ZEB-1 and GST–CID-ZEB-1 (K745A; KK774/775AA) bound to beads were incubated with recombinant P/CAF (UBI) in a 30 µl reaction containing acetylation buffer (10 mM Tris–HCl pH 8.0, 10% glycerol, 1 mM EDTA, 10 mM sodium butyrate, 1 mM DTT) and [3H]acetyl coA (Amersham Pharmacia Biotech) and incubated for 30 min at 30°C. Reactions were loaded in a 10% polyacrylamide gel and analyzed by fluorography.
Protein co-immunoprecipitations and western blot assays
Immunoprecipitation and western blots were performed as described previously (Postigo and Dean, 2000). Where indicated, lysates were immunoprecipitated with agarose-conjugated anti-Flag M2 (Sigma Chemicals, St Louis, MO), anti-Gal4-DBD, anti-ZEB-1 (ZEB C-20) and anti-myc 9E10 (Santa Cruz Biotechnologies, Santa Cruz, CA) antibodies. Also where indicated, 15% of the lysate was directly loaded into the gel as input control for direct western analysis. The following HRP-conjugated antibodies were used for western blotting: Gal4-DBD polyclonal antibody (Santa Cruz Biotechnologies), anti-LexA polyclonal antibody (Santa Cruz Biotechnologies), anti-Flag M2 mAb (Sigma Chemicals) or anti-myc 9E10 mAb (Santa Cruz Biotechnologies).
Xenopus embryo culture and in situ hybridization analysis
Xenopus laevis embryos were obtained and cultured as described previously (Peng, 1991). CS2MT, CS2MT-ZEB-1, CS2MT-ZEB-2 and CS2MT-DN-ZEB plasmids were linearized with NotI and used to produced capped RNA with a SP6-Message Machine Kit® (Ambion). A total of 250–500 pg of each RNA was dissolved in water and injected into one blastomere at the two-cell stage along with β-galactoside mRNA to trace the site of injection. Embryos were cultured in 0.2× Marc’s Modified Ringer’s solution (MMR) containing 4% Ficoll and 50 µg/ml gentamycin. In situ hybridization assays were carried out as described previously (Kroll et al., 1998). Embryos were injected in both cells at the two-cell stage for animal cap isolations. Animal cap explants were made at stage 8 and raised to stage 18 in 0.7× MMR supplemented with 50 µg/ml gentamycin and 1 mg/ml bovine serum albumin.
RT–PCR analysis of animal caps
To analyze gene expression in animal caps, we extracted RNA at stage 18 (late neurula) from 10 animal caps and synthesized cDNA using an Omni-Script kit (Qiagen). The resulting cDNA was amplified by RT–PCR as described previously (Kroll et al., 1998). Primers for BMP4, six3 and EF-1α have been reported previously (Rao, 1994; Wilson and Melton, 1994; Kroll et al., 1998). Other primers for PCR included: epidermal keratin (5′-CACCAGAACACAGAGTAC-3′ and 5′-CAACCTTCCCATCAACCA-3′), NCAM (5-CACAGTTCCACCAAATGC-3′ and 5′-GGAATCAAGCGGTACAGA-3′), muscle actin (5′-GCTGACAGAATGCAGAAG-3′ and 5′-TTGCTTGGAGGAGTGTGT-3′), myoD (5′-GTGTTG TTGTCCATCCATGAC-3′ and 5′-AGGTCCAACTGCTCCGACGGCATGAA-3′) and msx.1 (5′-GGCAGTCTGCAACTACTA-3′ and 5′-CTATGTCATGTCACCCTC-3′). For each primer set, we determined whether amplification was within a linear range by quantification with a PhosphorImager (Molecular Dynamics) using serial dilutions of cDNA from embryonic samples.
Acknowledgments
Acknowledgements
We are grateful to Drs X.H.Feng, E.Harlow, S.J.Korsmeyer, D.Livingston, J.Massagué, K.Miyazono and R.L.Schiltz for providing us with valuable constructs and cell lines. A.A.P. is supported by a Career Development Award (Special Fellow) from the Leukemia and Lymphoma Society (formerly Leukemia Society of America). This work was partially funded by grants from the March of Dimes and Whitehall Foundations to K.L.K. J.J.T. is supported by a predoctoral fellowship award from the HHMI.
References
- Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- Barlev N.A., Liu,L., Chehab,N.H., Mansfield,K., Harris,K.G., Halazonetis,T.D. and Berger,S.L. (2001) Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell, 8, 1243–1254. [DOI] [PubMed] [Google Scholar]
- Candia A.F., Watabe,T., Hawley,S.H., Onichtchouk,D., Zhang,Y., Derynck,R., Niehrs,C. and Cho,K.W. (1997) Cellular interpretation of multiple TGF-β signals: intracellular antagonism between activin/BVg1 and BMP-2/4 signaling mediated by Smads. Development, 124, 4467–4480. [DOI] [PubMed] [Google Scholar]
- Chakravarti D., Ogryzko,V., Kao,H.Y., Nash,A., Chen,H., Nakatani,Y. and Evans,R,M. (1999) A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell, 96, 393–403. [DOI] [PubMed] [Google Scholar]
- Chamberlain E.M. and Sanders,M.M. (1999) Identification of the novel player δEF1 in estrogen transcriptional cascades. Mol. Cell. Biol., 19, 3600–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H.M., Krstic-Demonacos,M., Smith,L., Demonacos,C. and La Thangue,N.B. (2001) Acetylation control of the retinoblastoma tumour-suppressor protein. Nat. Cell Biol., 3, 667–674. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G. (2002) CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell, 9, 213–224. [DOI] [PubMed] [Google Scholar]
- Comijn J., Berx,G., Vermassen,P., Verschueren,K., van Grunsven,L., Bruyneel,E., Mareel,M., Huylebroeck,D. and van Roy,F. (2001) The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell, 7, 1267–1278. [DOI] [PubMed] [Google Scholar]
- Dale L. and Wardle,F.C. (1999) A gradient of BMP activity specifies dorsal-ventral fates in early Xenopus embryos. Semin. Cell. Dev. Biol., 10, 319–326. [DOI] [PubMed] [Google Scholar]
- Dennler S., Goumans,M.J. and ten Dijke,P. (2002) Transforming growth factor β signal transduction. J. Leuk. Biol., 1, 731–740. [PubMed] [Google Scholar]
- Dillner N.B. and Sanders,M.M. (2002) The zinc finger/homeodomain protein δEF1 mediates estrogen-specific induction of the ovalbumin gene. Mol. Cell. Endocrinol., 192, 85–91. [DOI] [PubMed] [Google Scholar]
- Ebara S., Kawasaki,S., Nakamura,I., Tsutsumimoto,T., Nakayama,K., Nikaido,T. and Takaoka,K. (1997) Transcriptional regulation of the mBMP-4 gene through an E-box in the 5′-flanking promoter region involving USF. Biochem. Biophys. Res. Commun., 240, 136–141. [DOI] [PubMed] [Google Scholar]
- Eisaki A., Kurosda,H., Fukui,A. and Asashima,M. (2000) XSIP1, a member of the two handed zinc finger proteins induced anterior neural markers in Xenopus laevis animal cap. Biochem. Biophys. Res. Commun., 271, 151–157. [DOI] [PubMed] [Google Scholar]
- Espinosa J.M. and Emerson,B.M. (2001) Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell, 8, 57–69. [DOI] [PubMed] [Google Scholar]
- Feng X.H., Zhang,Y., Wu,R.Y. and Derynck,R. (1998) The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for Smad3 in TGFβ-induced transcriptional activation. Genes Dev., 12, 2153–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontemaggi G., Gurtner,A., Strano,S., Higashi,Y., Sacchi,A., Piaggio,G. and Blandino,G. (2001) The transcriptional repressor ZEB regulates p73 expression at the crossroad between proliferation and differentiation. Mol. Cell. Biol., 21, 8461–8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genetta T., Ruezinsky,D. and Kadesch,T. (1994) Displacement of an E-box-binding repressor by basic helix–loop–helix proteins: implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol. Cell. Biol., 14, 6153–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman R.H. and Smolik,S. (2000) CBP/p300 in cell growth, transformation and development. Genes Dev., 14, 1553–1577. [PubMed] [Google Scholar]
- Grooteclaes M.L. and Frisch,S.M. (2000) Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene, 19, 3823–3828. [DOI] [PubMed] [Google Scholar]
- Gu W. and Roeder,R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell, 90, 595–606. [DOI] [PubMed] [Google Scholar]
- Harbour J.W. and Dean,D.C. (2000) The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev., 14, 2393–2409. [DOI] [PubMed] [Google Scholar]
- Hill C.S. (2001) TGF-β signalling pathways in early Xenopus development. Curr. Opin. Genet. Dev., 11, 533–540. [DOI] [PubMed] [Google Scholar]
- Itoh S., Ericsson,J., Nishikawa,J.-i., Heldin,C.-H. and ten Dijke,P. (2000) The transcriptional co-activator P/CAF potentiates TGFβ/Smad signaling. Nucleic Acids Res., 28, 4291–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janknecht R., Wells,N.J. and Hunter,T. (1998) TGFβ-stimulated cooperation of Smad proteins with the coactivators CBP/p300. Genes Dev., 12, 2114–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C.M., Lyons,K.M., Lapan,P.M., Wright,C.M.E. and Hogan,B.L.M. (1992) DVR-4 (Bone morphogenetic protein-4) as a posterior-ventralizing factor in Xenopus mesoderm induction. Development, 115, 639–647. [DOI] [PubMed] [Google Scholar]
- Kawasaki H., Eckner,R., Yao,T.P., Taira,K., Chiu,R., Livingston,D.M. and Yokoyama,K.K. (1998) Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature, 393, 284–289. [DOI] [PubMed] [Google Scholar]
- Kawasaki S., Ebara,S., Nakayama,K. and Takaoka,K. (1999) The E-Box motif, recognized by tissue-specific nuclear factor(s), is important for BMP-4 gene expression in osteogenic cells. Biochem. Biophys. Res. Commun., 263, 560–565. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2000) Acetylation: a regulatory modification to rival phosphorylation. EMBO J., 19, 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll K.L., Salic,A.N., Evans,L.M. and Kirschner,M.W. (1998) Geminin, a neuralizing molecule that demarcates the future neural plate at the onset of gastrulation. Development, 125, 3247–3258. [DOI] [PubMed] [Google Scholar]
- Lazarova D.L., Bordonaro,M. and Sartorelli,A.C. (2001) Transcriptional regulation of the vitamin D3 receptor gene by ZEB. Cell Growth Differ., 12, 319–326. [PubMed] [Google Scholar]
- Lerchner W., Latinkic,B.V., Remacle,J.E., Huylebroeck,D. and Smith,J.C. (2000) Region-specific activation of the Xenopus Brachyury promoter involves active repression in ectoderm and endoderm: a study using transgenic frog embryos. Development, 127, 2729–2739. [DOI] [PubMed] [Google Scholar]
- Madison D.L., Yaciuk,P., Kwok,R.P. and Lundblad,J.R. (2002) Acetylation of the adenovirus-transforming protein E1A determines nuclear localization by disrupting association with importin-α. J. Biol. Chem., 277, 38755–38763. [DOI] [PubMed] [Google Scholar]
- Martinez-Balbas M.A., Bauer,U.M., Nielsen,S.J., Brehm,A. and Kouzarides,T. (2000) Regulation of E2F1 activity by acetylation. EMBO J., 19, 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. and Chen,Y.G. (2000) Controlling TGFβ signaling. Genes Dev., 14, 627–644. [PubMed] [Google Scholar]
- Massagué J. and Wotton,D. (2000) Transcriptional control by the TGFβ/Smad signaling system. EMBO J., 19, 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J., Blain,S.W. and Lo,R.S. (2000) TGFβ signaling in growth control, cancer and heritable disorders. Cell, 103, 295–309. [DOI] [PubMed] [Google Scholar]
- McDowell N. and Gurdon,J.B. (1999) Activin as a morphogen in Xenopus mesoderm induction. Semin. Cell. Dev. Biol., 10, 311–317. [DOI] [PubMed] [Google Scholar]
- Melhuish T.A. and Wotton,D. (2000) The interaction of the carboxyl terminus-binding protein with the Smad corepressor TGIF is disrupted by a holoprosencephaly mutation in TGIF. J. Biol. Chem., 275, 39762–39766. [DOI] [PubMed] [Google Scholar]
- Miyazono K. (2000) Positive and negative regulation of TGFβ signaling. J. Cell Sci., 113, 101–109. [DOI] [PubMed] [Google Scholar]
- Miyazono K., Kusanagi,K. and Inoue,H. (2001) Divergence and convergence of TGF-β/BMP signaling. J. Cell Physiol., 187, 265–276. [DOI] [PubMed] [Google Scholar]
- Moustakas A., Souchelnytskyi,S. and Heldin,C.H. (2001) Smad regulation in TGF-β signal transduction. J. Cell Sci., 114, 4359–4369. [DOI] [PubMed] [Google Scholar]
- Muñoz-Sanjuan I. and Brivanlou,A.H. (2001) Early posterior/ventral fate specification in the vertebrate embryo. Dev. Biol., 237, 1–17. [DOI] [PubMed] [Google Scholar]
- Murray D., Precht,P., Balakir,R. and Horton,W.E.,Jr (2000) The transcription factor δEF1 is inversely expressed with type II collagen mRNA and can repress Col2a1 promoter activity in transfected chondrocytes. J. Biol. Chem., 275, 3610–3618. [DOI] [PubMed] [Google Scholar]
- Nakamura S., Roth,J.A. and Mukhopadhyay,T. (2000) Multiple lysine mutations in the C-terminal domain of p53 interfere with MDM2-dependent protein degradation and ubiquitination. Mol. Cell. Biol., 20, 9391–9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Yanagisawa,M., Arakawa,H., Kimura,N., Hisatsune,T., Kawabata,M., Miyazono,K. and Taga,T. (1999) Synergistic signaling in fetal brain by STAT3–Smad1 complex bridged by p300. Science, 284, 479–482. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Shiltz,R.L., Russanova,V., Howard,B.H. and Nakatani,Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- Papin C., van Grunsven,L.A., Verschueren,K., Huylebroeck,D. and Smith,J.C. (2002) Dynamic regulation of Brachyury expression in the amphibian embryo by XSIP1. Mech. Dev., 111, 37–46. [DOI] [PubMed] [Google Scholar]
- Peng H.B. (1991) Solutions and protocols. In Kay,B.K. and Peng,H.B. (eds), Xenopus laevis: Practical Uses in Cell and Molecular Biology. Academic Press, San Diego, CA, p. 657. [PubMed]
- Postigo A.A. (2003) Opposing functions of ZEB proteins in the regulation of the TGFβ/BMP signaling pathway. EMBO J., 22, 2443–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo A.A and Dean,D.C. (1997) ZEB, a vertebrate homolog of Drosophila Zfh-1, is a negative regulator of muscle differentiation. EMBO J., 16, 3935–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo A.A. and Dean,D.C. (1999a) ZEB represses transcription through interaction with the corepressor CtBP. Proc. Natl Acad. Sci. USA, 96, 6683–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo A.A. and Dean,D.C. (1999b) Independent repressor domains in ZEB regulate muscle and T-cell differentiation. Mol. Cell. Biol., 19, 7961–7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo A.A. and Dean,D.C. (2000) Differential expression and function of members of the zfh-1 family of zinc finger/homeodomain repressors. Proc. Natl Acad. Sci. USA, 97, 6391–6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo A.A., Sheppard,A.M., Mucenski,M.L. and Dean,D.C. (1997) c-myb and ets factors synergize to overcome transcriptional repression by ZEB. EMBO J., 16, 3924–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo A.A., Ward,E., Skeath,J.B. and Dean,D.C. (1999) zfh-1, the Drosophila homologue of ZEB, is a transcriptional repressor that regulates somatic myogenesis. Mol. Cell. Biol., 19, 7255–7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouponnot C., Jayaraman,L. and Massague,J. (1998) Physical and functional interaction of SMADs and p300/CBP. J. Biol. Chem., 273, 22865–22868. [DOI] [PubMed] [Google Scholar]
- Prives C. and Manley,J.L. (2001) Why is p53 acetylated? Cell, 107, 815–818. [DOI] [PubMed] [Google Scholar]
- Puri P.L. et al. (1997) Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell, 1, 35–45. [DOI] [PubMed] [Google Scholar]
- Rao Y. (1994) Conversion of a mesodermalizing molecule, the Xenopus Brachyury gene, into a neuralizing factor. Genes Dev., 8, 939–947. [DOI] [PubMed] [Google Scholar]
- Remacle J.E., Kraft,H., Lerchner,W., Wuytens,G., Collart,C., Verschueren,K., Smith,J.C. and Huylebroeck,D. (1999) New mode of DNA binding of multi-zinc finger transcription factors: δEF1 family members bind with two hands to two target sites. EMBO J., 18, 5073–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekido R., Murai,K., Funahashi,J., Kamachi,Y., Fujisawa-Sehara,A., Nabeshima,Y. and Kondoh,H. (1994) The δ-crystallin enhancer-binding protein δEF1 is a repressor of E2-box-mediated gene activation. Mol. Cell. Biol., 14, 5692–5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekido R., Murai,K., Kamachi,Y. and Kondoh,H. (1997) Two mechanisms in the action of repressor δEF1: binding site competition with an activator and active repression. Genes Cells, 2, 771–783. [DOI] [PubMed] [Google Scholar]
- Takagi T., Moribe,H., Kondoh,H. and Higashi,Y. (1998) δEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development, 125, 21–31. [DOI] [PubMed] [Google Scholar]
- ten Dijke P., Miyazono,K. and Heldin,C.H. (2000) Signaling inputs converge on nuclear effectors in TGFβ signaling. Trends Biochem. Sci., 25, 64–70. [DOI] [PubMed] [Google Scholar]
- Turner J. and Crossley,M. (2001) The CtBP family: enigmatic and enzymatic transcriptional co-repressors. BioEssays, 23, 683–690. [DOI] [PubMed] [Google Scholar]
- Vainio S., Karavanova,I., Jowett,A. and Thesleff,I. (1993) Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell, 75, 45–58. [PubMed] [Google Scholar]
- Verschueren K. et al. (1999) SIP1, a novel zinc finger/homeodomain repressor interacts with Smad proteins and bind to 5′-CACCT sequences in candidate target genes. J. Biol. Chem., 274, 20489–20498. [DOI] [PubMed] [Google Scholar]
- von Bubnoff A. and Cho,KW. (2001) Intracellular BMP signaling regulation in vertebrates: pathway or network? Dev. Biol., 239, 1–14. [DOI] [PubMed] [Google Scholar]
- Wilson P.A. and Melton,D.A. (1994) Mesodermal patterning by an inducer gradient depends on secondary cell–cell communication. Curr. Biol., 4, 676–686. [DOI] [PubMed] [Google Scholar]
- Yang X.J., Ogryzko,V.V., Nishikawa,J., Howard,B.H. and Nakatani,Y. (1996) A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature, 382, 319–324. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Yao,H., Vo,N. and Goodman,R.H. (2000) Acetylation of adenovirus E1A regulates binding of the transcriptional corepressor CtBP. Proc. Natl Acad. Sci. USA, 97, 14323–14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Piston,D.W. and Goodman,R.H. (2002) Regulation of corepressor function by nuclear NADH. Science, 295, 1895–1897. [DOI] [PubMed] [Google Scholar]