Abstract
The surveillance of the structural fidelity of the proteome is of utmost importance to all cells. The endoplasmic reticulum (ER) is the organelle responsible for proper folding and delivery of proteins to the secretory pathway. It contains a sophisticated protein proofreading and elimination mechanism. Failure of this machinery leads to disease and, finally, to cell death. Elimination of misfolded proteins requires retrograde transport across the ER membrane and depends on the central cytoplasmic proteolytic machinery involved in cellular regulation: the ubiquitin–proteasome system. The basics of this process as well as recent advances in the field are reviewed.
Keywords: chaperones/ERAD/proteasome/ubiquitin
Introduction
It is of great importance for the cell to regulate the individual entities of its proteome as well as to control the structural fidelity of each of its members. Proteins destined for secretion, the plasma membrane or the cell surface are translocated from the cytoplasm into the endoplasmic reticulum (ER), the central organelle for further delivery of these proteins to their site of action. Since proteins are translocated into the ER in an unfolded state, it is the primary function of this organelle to modify and fold the translocated proteins to acquire their biologically active conformation (Haigh and Johnson, 2002). In the ER, proteins undergo a quality control procedure that discriminates between properly folded proteins and terminally misfolded species as well as unassembled protein subunits (Ellgaard et al., 1999). The misfolded polypeptides and orphan subunits are subsequently subjected to ER-associated degradation (ERAD). The ERAD process requires retrotranslocation of the malfolded proteins across the ER membrane into the cytoplasm and subsequent degradation by the 26S proteasome (Sommer and Wolf, 1997; Brodsky and McCracken, 1999; Plemper and Wolf, 1999). ER degradation contributes to the molecular pathogenesis of many loss- and gain-of-toxic-function disorders (Aridor and Hannan, 2000; Kostova and Wolf, 2002; Rutishauser and Spiess, 2002). A detailed knowledge of this process is thus of great importance not only for our understanding of this basic cellular mechanism but also for the development of new strategies to treat a diverse set of diseases.
Protein folding and the unfolded protein response
Many proteins synthesized in the cytoplasm are transported via the secretory pathway to other compartments of the cell, the plasma membrane, or the extracellular space. Secretory proteins are first translocated into the ER in an unfolded state via an aqueous channel, the Sec61 translocon. In the ER lumen, proteins are properly folded, assembled into multisubunit complexes and covalently modified by a large array of ER-resident chaperones and enzymes, before they can enter the secretory pathway (Glick, 2002; Haigh and Johnson, 2002). The major components of this process in the ER are signal peptidase, which cleaves off the signal peptide from the newly translocated proteins; the oligosaccaryl-transferase complex (OST) which carries out N-glycosylation; and protein disulfide isomerase (PDI), which participates in disulfide bond formation. The two most studied examples of chaperones that assist proteins in their folding are the Hsp70 chaperone BiP (Kar2p in yeast), which recognizes hydrophobic patches on proteins, and calnexin, which binds carbohydrate moieties. Proteins are allowed to exit the ER and enter the secretory pathway only when they are properly folded and modified (Ellgaard et al., 1999). The importance of proper folding before ER exit is demonstrated by the existence of an unfolded protein response (UPR), a system that controls the level of the auxiliary folding proteins (Sidrauski et al., 2002). In Saccharomyces cerevisiae, the concentration of unfolded proteins in the ER is sensed by Ire1p, a transmembrane kinase localized to the ER/nuclear envelope, by virtue of a competition between the Ire1p lumenal domain and unfolded proteins for binding to Kar2p (BiP). Depletion of free Kar2p (BiP) molecules due to sequestration by increasing amounts of unfolded proteins leads to the dimerization of Ire1p, a conformational change that transmits a signal across the membrane and activates the cytoplasmic kinase activity. The kinase induces a non-canonical splicing of the HAC1 mRNA, allowing synthesis of the Hac1p transcription factor, which upregulates genes containing a UPR response element. This cascade of events leads to an increase in the levels of proteins required for folding and quality control (Travers et al., 2000; Sidrauski et al., 2002). The UPR and ER degradation are tightly interconnected. Overexpression or accumulation of unfolded proteins due to the absence of a component of the ER degradation machinery induces the UPR (Knop et al., 1996a) which, in turn, upregulates the overall level of components of the ER degradation machinery (Friedländer et al., 2000; Travers et al., 2000). Under normal conditions, cells are able to cope with the amount of naturally present unfolded proteins and do not require induction of the UPR. However, when the level of misfolded proteins rises above a certain level, the UPR becomes essential. In yeast, loss of function of components of both systems is lethal (Friedländer et al., 2000; Travers et al., 2000).
ER quality control
Prion diseases like Scrapie (sheep), bovine spongiform encephalopathy (BSE, cattle), or Creutzfeldt–Jakob disease (CJD, human), and other neurodegenerative diseases such as Parkinson and Alzheimer, are the result of precipitated protein aggregates. On the other hand, human diseases such as cystic fibrosis and lung emphysema are caused by the rapid disappearance of crucial proteins, like the cystic fibrosis transmembrane conductance regulator (CFTR) and α-1-antitrypsin, respectively (Jensen et al., 1995; Ward et al., 1995; Qu et al., 1996; Kostova and Wolf, 2002; Rutishauser and Spiess, 2002). The discovery of a degradation process for mutated and misfolded ER proteins in yeast (Sommer and Wolf, 1997; Brodsky and McCracken, 1999; Plemper and Wolf, 1999; Kostova and Wolf, 2002) has shed light on the molecular mechanism underlying such seemingly different diseases.
Protein function depends greatly on a precise three-dimensional conformation, achieved following an uninterrupted, competent folding and assembly process. Mistakes are, however, a fact of life. Mutations leading to an incorrect final structure result in inactive proteins and, if not properly dealt with, in protein aggregates. To minimize folding mistakes, a complex chaperone system has evolved to assist the folding process and to prevent dead-end conformations (Kopito, 2000). This folding machinery is especially active in the ER, into which nearly all proteins enter in an unfolded state. Protein stretches that leave the Sec61 translocon are immediately occupied by chaperones, preventing hydrophobic surfaces from creating erroneous intra- and/or intermolecular contacts. How ever, even chaperone-assisted folding cannot guarantee perfection, especially when faced with a mutant protein. To overcome this problem, a quality control system has evolved in the ER to function as a checkpoint that detects improperly folded proteins and targets them for elimination (Ellgaard et al., 1999). The most influential ER folding components are the Hsp70 chaperone BiP (Kar2p), the lectin like chaperones calnexin and calreticulin, and enzymes involved in disulfide bond formation, such as PDI and oxidoreductase Erp57. All of these components also participate in ER quality control. Quite unexpectedly, the elimination of misfolded proteins from the ER is dependent on the cytoplasmic ubiquitin–proteasome system and, therefore, requires protein retrotranslocation (dislocation) across the ER membrane (Hiller et al., 1996; Werner et al., 1996; Wiertz et al., 1996; Plemper et al., 1997).
Our current knowledge about the discovery of misfolded or unassembled proteins and their retention in the ER is still very limited. However, it is clear that the control mechanism works by structural rather than functional criteria. Mutations in CFTR and α-1-antitrypsin, for example, which do not perturb the biological activity of the proteins per se, lead to ER retention and elimination of the mutant molecules, thus causing disease (Jensen et al., 1995; Ward et al., 1995; Qu et al., 1996). To date, a chaperone-mediated retention mechanism has been described only for mutant glycoproteins in mammalian systems. As proteins become translocated across the ER membrane, core oligosaccharides of the Glc3Man9 GlcNAc2 structure are co-translationally attached to the side chains of asparagine residues within the Asn-X-Ser/Thr consensus sequence. As folding progresses, two of the outermost glucose residues of the N-linked glycan are trimmed by the glucosidases I and II. In mammalian cells, exposure of the innermost glucose leads to the binding of the monoglucosylated protein to calnexin and calreticulin. If the glycoproteins contain cysteine residues, mixed disulfide bonds are transiently formed by the thiol oxidoreductase ERp57, a member of the PDI family which specifically interacts with calnexin, calreticulin and monoglucosylated glycoproteins, and functions as a disulfide isomerase. When the remaining glucose residue is trimmed by glucosidase II, the complex dissociates, releasing a protein with the carbohydrate structure Man9GlcNAc2. If the glycoprotein is not properly folded, the N-glycan is reglucosylated at the same position by a UDP-glucose glucosyltransferase (UGGT). This induces a new round of calnexin/calreticulin binding, which prevents the escape of the glycoprotein from the ER. If this cycle persists due to the inability of the glycoprotein to reach its final native conformation, mannosidase I, a slow-acting enzyme, cleaves the α1,2-linked mannose of the middle branch, generating a glycan with the structure Man8GlcNAc2. Direct recognition of Man8GlcNAc2 by a specific lectin or attenuated release of the reglucosylated form (Glc1Man8GlcNAc2) from calnexin are events which determine the delivery of malfolded glycoproteins to the elimination machinery (Cabral et al., 2001).
In recent years, the yeast S.cerevisiae has set the pace in the field of protein quality control in the ER (Figure 1). One of the model proteins used to study ER quality control in this organism is a mutated vacuolar enzyme, carboxypeptidase yscY-S255R, commonly known as CPY*. This misfolded protein is translocated into the ER lumen normally; it is fully glycosylated but is never transported to the vacuole. Instead, it is retained in the ER and degraded by the proteasome (Hiller et al., 1996; Knop et al., 1996a). It is interesting that ER quality control leads to degradation of misfolded proteins by the cytoplasmic proteasome machinery rather than by the vacuole (lysosome). A carbohydrate-based retention mechanism, which appears very similar to the mechanism described for mammalian cells, has also been identified for CPY*. Glucosidases I and II (Gls1, Gls2), and α-1,2 mannosidase (Mns1) are necessary for the disposal of CPY* (Knop et al., 1996b; Jakob et al., 1998). As opposed to the mammalian system, there is no known glucosyltransferase in yeast that could reglucosylate the carbohydrate chains. Thus, trimming by glucosidases I and II, and later by α-1,2 mannosidase, is currently believed to be the timer for folding or, if unsuccessful, for degradation. The recently discovered lectin-like yeast protein Htm1/Mnl1p, and its mouse homolog EDEM, is believed to bind Man8GlcNAc2 and provides us with a link between recognition and targeting for degradation (Hosokawa et al., 2001; Jakob et al., 2001; Nakatsukasa et al., 2001) (Figure 1). Htm1p/Mnl1p is also necessary for the efficient degradation of other misfolded glycoproteins with differing topologies, such as Pdr5*, a mutated form of the ATP-binding cassette transporter Pdr5 (Plemper et al., 1998), and Stt3-7p, a mutant subunit of the OST complex (Jakob et al., 2001). On the other hand, degradation of Sec61-2p (Biederer et al., 1996), which is not N-glycosylated, is totally independent of Htm1p/Mnl1p, a finding that emphasizes the necessity of this lectin specifically for the recognition of misfolded glycoproteins (Jakob et al., 2001). At present, the overall impact of this carbohydrate-based recognition on the quality control and degradation process as a whole is not clear. Likewise, our knowledge of the mechanism underlying the discovery of misfolded or unassembled non-glycoproteins is minimal. An obvious possibility is recognition of exposed hydrophobic patches by chaperone-like molecules. The major Hsp70 chaperone of the ER, BiP (Kar2p), is known to play a vital role in binding hydrophobic protein stretches to allow post-translational protein import into the ER and for protein folding and assembly within the ER (Johnson and van Waes, 1999; Haigh and Johnson, 2002). Its general function in protein quality control, therefore, seemed pretty straightforward. It is required for the degradation of soluble proteins such as mutant pro-α-factor and CPY* in yeast (Plemper et al., 1997; Brodsky et al., 1999). Surprisingly, Kar2p becomes dispensable when CPY* is anchored to the ER membrane. Basically, conversion of the same misfolded protein, CPY*, from a soluble form to a membrane-bound form abolishes the requirement for Kar2p (BiP) in ER degradation (C.Taxis and D.H.Wolf, manuscript submitted for publication). Degradation of other membrane proteins is also independent of Kar2p (Plemper et al., 1998). This indicates that Kar2p (BiP) and its DnaJ-like partners, Jem1p and Scj1p (Nishikawa et al., 2001), do not have a general function in the quality control process. Instead, Kar2p activity seems limited to soluble proteins (Figure 1). One function of Kar2p appears to reside in its ability to remain bound to misfolded proteins after their unsuccessful folding trials, essentially keeping them in soluble form. Scj1p and Jem1p may be necessary for triggering the release of Kar2p from such substrates in order for dislocation and degradation to take place. BiP and its partners may also play a role in the delivery of the soluble substrates to other, as yet unknown, components linking recognition to elimination (Nishikawa et al., 2001). BiP has also been implicated in acting as a seal for the protein translocation channel from the lumenal side (Johnson and van Waes, 1999; Haigh and Johnson, 2002). If BiP (Kar2p) has some function as a gatekeeper for the retrograde transport of malfolded proteins out of the ER, then this role would be limited to soluble proteins (C.Taxis and D.H.Wolf, manuscript submitted for publication). The requirement for Ca2+ ions in the ER for degradation of soluble CPY* may be partly explained by their regulatory role in Kar2p activity (Dürr et al., 1998). PDI, an oxidoreductase involved in disulfide bond formation in the ER (Regeimbal and Bardwell, 2002), is emerging as another component with multiple functions. While elimination of CPY* (Gillece et al., 1999) and unassembled Ig-µ chains (Fagioli et al., 2001) requires the enzymatic activity of PDI, elimination of the non-glycosylated and cysteine-free mutant pro-α-factor requires binding of PDI without enzymatic activity, suggesting a chaperone-like role for PDI under these circumstances (Gillece et al., 1999) (Figure 1). PDI has also been described to function as a redox-driven chaperone in the unfolding of the A1 chain of cholera toxin in the ER lumen, prior to its transport to the cytoplasm. In this scenario, PDI binds and unfolds the substrate in its reduced state. The complex is then targeted to the ER membrane, where it binds to a protein at the lumenal side of the membrane. Oxidation of PDI by Ero1p releases the substrate, possibly directly into the retrotranslocon (Tsai and Rapoport, 2002). However, this redox-dependent mechanism has been questioned by Lumb and Bulleid’s (2002) finding that PDI binding and release to two other substrates is not driven by the redox state of the protein. The necessity for Eps1p, a membrane-bound protein from the PDI family, in the elimination of a mutant form of the yeast membrane ATPase Pma1p (Wang and Chang, 1999), underlines the general necessity of this class of proteins as part of the ER protein quality control machinery. Calnexin represents yet another example of a multifunctional chaperone. In mammalian cells, it binds glycan moieties of proteins as expected from a lectin. In yeast, it is dispensable for CPY* degradation but it is involved in the elimination of the non-glycosylated mutant pro-α-factor (McCracken and Brodsky, 1996), suggesting additional non-lectin activity for this protein.
Fig. 1. Components of ER quality control and degradation in yeast.
The protein quality control machinery of the ER is also involved in the regulated degradation of ER-resident enzymes. The best-studied example in yeast is a key enzyme in the sterol biosynthetic pathway, HMGCoA-reductase 2. Signals from the mevalonate pathway are presumed to lead to conformational changes which channel the protein into the ER quality control pathway (Hampton, 2002). Elimination of this enzyme requires all of the major components of the ER degradation machinery (Hampton et al., 1996; Hampton and Bhakta, 1997; Gardner et al., 2000; Bays et al., 2001a, b).
Retrotranslocation
It was initially believed that misfolded and orphan proteins of the ER were degraded by ER-resident proteinases and peptidases (Bonifacino and Klausner, 1994). However, the presence of unspecific proteinases in the ER was hard to reconcile with its primary function in folding and assembly. Rather, delivery of misfolded proteins to the lysosome/vacuole, the compartment of the cell associated with degradation, seemed more likely. The discovery of the involvement of the cytoplasmic ubiquitin–proteasome system in the degradation of mutant ER membrane proteins was an unexpected surprise (Sommer and Jentsch, 1993; Jensen et al., 1995; Ward et al., 1995). The findings, in S.cerevisiae, that the misfolded vacuolar peptidase CPY* (Hiller et al., 1996) and the mutant secretory protein pro-α-factor (Werner et al., 1996) were retained in the ER lumen and degraded in the cytoplasm by the ubiquitin–proteasome system, were crucial in establishing the concept of retrograde transport out of the ER. Genetic studies in yeast revealed a significantly retarded degradation of CPY* (Plemper et al., 1997), mutated pro-α-factor (Pilon et al., 1997) and a mutant polytopic membrane protein, Pdr5* (Plemper et al., 1998), in mutants defective in the Sec61 translocon. Co-immunoprecipitation studies indicated that Sec61β was associated with the major histocompatibility complex class I (MHC class I) heavy chains, with wild-type and mutant forms of the CFTR and with a mutant form of ribophorin I (RI332) during their dislocation from the ER (Kostova and Wolf, 2002). These studies support the notion that the Sec61 import channel may also be the export channel. It is very likely, though, that the retrotranslocation channel differs in its composition from the import channel. The Sec61 translocon is composed of three different subunits: Sec61α, Sec61β and Sec61γ (Sec61p, Sbh1p and Sss1p in yeast). Studies on the retrotranslocon have identified a genetic interaction between Hrd3p and Sec61p (Plemper et al., 1999a; P.Deak and D.H.Wolf, unpublished data) (Figure 1). The presence of two subsets of translocons can easily explain how this complicated two-way traffic across the membrane is regulated. Since CPY* leaves the translocon completely after import into the ER lumen (Plemper et al., 1999b), retrotranslocation also necessitates retargeting of the protein to the Sec61 channel. Possible components of this retargeting assembly are Der1p, an ER membrane protein of unknown function (Knop et al., 1996a), Kar2p (BiP), PDI, the lectin Htm1p/Mnl1p, and Hrd3p, an ER membrane protein that functions together with the ubiquitin–protein ligase Der3p/Hrd1p (Plemper et al., 1999a; Gardner et al., 2000; Deak and Wolf, 2001). Given the cooperativity between targeting, dislocation and degradation, it is very likely that these components do not act independently of each other. A recent report suggests that a fusion protein with an enhanced GFP and an MHC class I heavy chain (EGFP-HCI) domain may be retrotranslocated without unfolding of the EGFP domain (Fiebiger et al., 2002). The inner diameter of the translocon pore is estimated to fluctuate between 15 Å, in its inactive state, and 40–60 Å during protein translocation (Johnson and van Waes, 1999; Haigh and Johnson, 2002). Therefore, it is possible that in its active state the translocon could accommodate a folded GFP molecule with a length of 24 Å and a diameter of 42 Å. The retrotranslocon could even accommodate larger molecules through rearrangements of translocon subunits. However, additional biochemical proof is essential for the characterization of the dislocation channel.
Ubiquitylation and targeting to the proteasome
Following retrotranslocation from the ER, nearly all misfolded proteins are polyubiquitylated prior to degradation (Thrower et al., 2000; Kostova and Wolf, 2002). One exception is the mutated pro-α-factor (Werner et al., 1996), which, although not ubiquitylated, is still targeted to the proteasome. Genetic and biochemical studies in yeast characterized the ubiquitylation machinery and localized it to the cytoplasmic side of the ER membrane. Ubc6p, a tail-anchored E2 (Sommer and Jentsch, 1993) and Ubc7p, a soluble cytoplasmic E2 recruited to the ER membrane via Cue1p (Biederer et al., 1997), are the two best-studied components of the ubiquitylation machinery (Biederer et al., 1996; Hiller et al., 1996; Plemper et al., 1998). Recently, Ubc1p has also been implicated as an E2 involved in CPY* ubiquitylation (Friedländer et al., 2000). The ubiquitin–protein ligase (E3) cooperating with Ubc1p and Ubc7p is the RING-H2 finger domain ER membrane protein Der3/Hrd1p (Bays et al., 2001a; Deak and Wolf, 2001). Der3/Hrd1p activity is dependent on the presence of an interacting ER membrane protein, Hrd3p (Hampton et al., 1996). Hrd3p is believed to play a role in signaling events between the ER lumen and the cytoplasmic ubiquitylation machinery (Plemper et al., 1999a; Gardner et al., 2000; Deak and Wolf, 2001) and is necessary for the degradation of both soluble and polytopic membrane proteins (Figure 1). Doa10/Ssm4p is the second E3 described in yeast as involved in ER-associated degradation. In addition to its involvement in the ubiquitylation of the Deg1 domain of the transcriptional repressor MATα2, Doa10/Ssm4p is also involved in the degradation of the tail-anchored ER membrane protein Ubc6p (Swanson et al., 2001). Interestingly, neither the ubiquitin–protein ligase Der3/Hrd1p, its interaction partner Hrd3p, nor the Sec61 channel participate in the elimination of Ubc6p (Walter et al., 2001). The elimination of the vacuolar membrane ATPase subunit Vph1p (Hill and Cooper, 2000; Wilhovsky et al., 2000) is also independent of the function of the Der3/Hrd1p–Hrd3p complex. It is still an open question whether Doa10/Ssm4p is involved. The ubiquitin conjugating enzymes MmUbc6 and MmUbc7 (Tiwari and Weissman, 2001), and the RING-finger E3, gp78 (Fang et al., 2001), are the mammalian orthologs of the yeast ER-ubiquitylation machinery. Interestingly, gp78 recruits MmUbc7 to the ER membrane through a region that exhibits homology to yeast Cue1p, representing a case of convergent evolution of two functions into a single polypeptide. These proteins have been shown to participate in proteasomal degradation of mammalian ERAD substrates (Fang et al., 2001).
ER degradation requires directionality from the ER lumen or membrane to the proteasome. Once the protein is threaded into the retrotranslocon, there needs to be a pulling or pushing force to direct the substrate to the proteasome. It is clear now that lack of polyubiquitylation not only prevents proteasomal degradation (Hiller et al., 1996; Biederer et al., 1997) but also leads to failure in transport out to the cytoplasm. The substrate seems to remain or slips back into the ER (Biederer et al., 1997; Bordallo et al., 1998; Shamu et al., 2001), which suggests that polyubiquitylation is necessary for retrotranslocation. Modification of the protein may occur when the N-terminus or the first lysine residue becomes accessible to the ubiquitylation machinery (Hershko and Ciechanover, 1998). Progressive polyubiquitylation may serve as a ratcheting mechanism moving the polypeptide from the retrotranslocation channel into the cytoplasm, where the long and bulky polyubiquitin chains prevent the polypeptide from slipping back into the ER. Consistent with this idea are the findings that a hypo-ubiquitylated CPY* fails to be completely transported into the cytosol (Jarosch et al., 2002). Recent data indicate that Cdc48 (p97), a member of the AAA family of ATPases, together with two partner proteins, Ufd1p and Npl4p, forms a complex essential for the extraction and degradation of a number of substrate proteins (Bays et al., 2001a; Ye et al., 2001; Braun et al., 2002; Jarosch et al., 2002; Rabinovich et al., 2002). In the absence of the Cdc48 complex, polyubiquitylated substrates accumulate at the ER membrane, suggesting a role downstream of polyubiquitylation. At the moment there is controversy concerning the temporal organization and association of this complex with the ER membrane and the subunit responsible for ubiquitin binding (Dai and Li, 2001; Meyer et al., 2002). Cdc48 exists as homo-hexameric rings, which undergo strong conformational changes upon ATP hydrolysis (Rouiller et al., 2000; Zhang et al., 2000), critical for substrate dislocation from the ER (Ye et al., 2001). There is convincing evidence that the Cdc48–Ufd1–Npl4 complex can separate a tightly associated protein at the ER membrane, a processed dimer of the Spt23 transcription factor, and, by this, release active transcription factors into the cytosol (Rape et al., 2001). The general picture that emerges is the following: after polyubiquitylation and partial dislocation of the substrate from the retrotranslocon, the ER-associated Cdc48–Ufd1p–Npl4 complex binds the ubiquitylated protein. Rounds of ATP hydrolysis then induce conformational changes in the Cdc48 complex, which progressively pulls the polyubiquitylated substrate away from the ER membrane and hands it over to the proteasome for degradation (Figure 1). In mutants defective either in proteasome activity or in one of the 19S cap ATPases, CPY* accumulates to a large extent in the cytoplasm (Jarosch et al., 2002). Similarly, MHC class I molecules are found as soluble ubiquitylated intermediates in HCMV US11 expressing cells when the proteasome is inhibited (Wiertz et al., 1996; Shamu et al., 2001). These data confirm that the proteasome acts after release of the ubiquitylated substrate from the ER membrane. In a few cases, however, the proteasome itself seems to have an active role in the dislocation of membrane-bound proteins (Mayer et al., 1998; Walter et al., 2001). Since the role of Cdc48 in the extraction of these membrane proteins has yet to be tested, it is not clear whether there are different modes for ER-membrane extraction. It is certainly possible that other as yet unknown components exist as mediators between the Cdc48 complex and the proteasome.
Glycosylated proteins of the ER are retrotranslocated into the cytoplasm in a glycosylated state (Hiller et al., 1996; Wiertz et al., 1996). The discovery of de-N-glycosylated intermediates in the cytosol upon inhibition of the proteasome alludes to the activity of a cytoplasmic N-glycanase prior to proteasomal degradation. Recently, a highly conserved cytoplasmic N-glycanase (Png1p) has been identified in yeast. PNG1 is not an essential gene but its product is required for efficient degradation of CPY* (Suzuki et al., 2000). Rad23, a protein that interacts with the proteasome via its ubiquitin-like (Ubl) domain escorts Png1p to the proteasome where it is thought to act in a complex to efficiently deglycosylate the substrate prior to degradation (Suzuki et al., 2001).
The involvement of cytoplasmic chaperones in the degradation of misfolded soluble ER lumenal proteins is an open question. In contrast, misfolded ER membrane proteins with tightly folded cytoplasmic domains seem to require additional chaperone activity for efficient degradation. Elimination of the CFTR expressed in yeast is dependent on the presence of cytoplasmic Hsp70 chaperones (Zhang et al., 2001). The use of modular substrates containing the misfolded CPY* domain fused to a membrane anchor, with (CTG*), or without (CT*), an added cytosolic GFP domain, has given clues about the possible requirements for cytosolic chaperones. While soluble CPY* and membrane-anchored CT* do not require additional cytoplasmic components other than the Cdc48 complex, degradation of membrane-bound CTG* is dependent on Hsp70, Hsp40, and, to a certain extent, Hsp104 chaperone activity (Figure 1) (C.Taxis and D.H.Wolf, manuscript submitted for publication). One may, therefore, speculate that the tighter the folding of the cytoplasmic domain of a malfolded ER membrane protein, the stronger is the requirement for cytoplasmic chaperone activity for degradation.
It has recently been shown that efficient ER degradation of soluble malfolded proteins requires an efficient ER–Golgi traffic (Caldwell et al., 2001; Vashist et al., 2001; Taxis et al., 2002). Possible explanations include the necessity of transport of the malfolded proteins from ER to Golgi for further modification, or the ER–Golgi cycling of an as yet unknown factor as a prerequisite for efficient degradation (Caldwell et al., 2001; Vashist et al., 2001). A recent study reports that overexpression of CPY* and other malfolded proteins leads to dependence on the activity of the Golgi-associated E3 Rsp5, rather than the Der3/Hrd1p–Hrd3p complex (Haynes et al., 2002). This finding is controversial because other reports show that degradation of overexpressed CPY* is still dependent on Der3/Hrd1p (Friedländer et al., 2000). New data from Taxis et al. (2002) lead to yet another interpretation: they propose that efficient ER degradation simply requires an intact ER with all its components, which is, in turn, dependent on the existence of a normal ER–Golgi traffic. Future experiments are clearly necessary to solve this somewhat controversial issue.
Degradation by the 26S proteasome
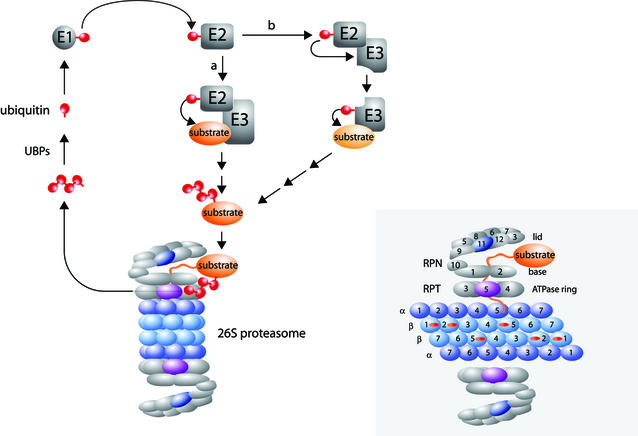
Proteins selected for degradation are tagged with ubiquitin. This 76 amino acid polypeptide is bound in an isopeptide linkage through its C-terminal glycine to the N-terminus or to the ε-amino group of internal lysine residues of target proteins. First, ubiquitin is activated by an ATP-dependent trans-esterification reaction and attached to a cysteine residue of a ubiquitin-activating enzyme (E1, Uba). Next, ubiquitin is transferred to a cysteine of a ubiquitin-conjugating enzyme (E2, Ubc) and finally, to the substrate directly or via the intervention of a third class of enzymes, ubiquitin–protein ligases (E3, Ubl). Proteasomal degradation requires polyubiquitylation of the substrate, accomplished by attaching additional ubiquitin moieties usually to Lys48 of the preceding ubiquitin (Pickart, 2000; Thrower et al., 2000). The 26S proteasome binds the polyubiquitylated substrate, unfolds and degrades it (Figure 2) (Heinemeyer et al. 1991; Baumeister et al., 1998). This is a complex proteolytic assembly that consists of two parts, the 20S core and the 19S regulatory particle. The 20S core is a cylinder composed of four stacked rings, each containing seven different α or β subunits, with an overall α7β7β7α7 geometry. The three different active sites are located inside the cylindrical core within the β-subunit rings (Figure 2) (Groll et al., 1997; Baumeister et al., 1998). N-terminal stretches of the external α-subunits regulate the entry of substrates into the proteolytic core (Groll et al., 2000). The 19S cap is involved in the recognition, binding and unfolding of ubiquitylated proteins, and in the regulation of the opening of the 20S core. It is composed of 17 different subunits, functionally divided into two parts: the base and the lid. The base consists of a ring of six ATPases (Rpt1–Rpt6), which dock onto the α-rings of the 20S core, and three non-ATPase subunits (Rpn1, Rpn2 and Rpn10) (Figure 2). The specific functions of the ATPase subunits in binding and unfolding are slowly emerging (Braun et al., 1999). Rpt5 binds the ubiquitylated substrate (Lam et al., 2002). Rpt2 is believed to control both substrate entry and product release from the 20S channel (Köhler et al., 2001). Rpn1 interacts with Rad23 and Dsk2, two proteins harboring ubiquitin-like domains (Ubl) and capable of binding and delivering ubiquitylated cargo to the proteasome. Recent data suggest that Rpn10 contributes to the binding of ubiquitin chains as well (Elsässer et al., 2002). The lid is composed of eight subunits: Rpn3, Rpn5 to Rpn9, Rpn11 and Rpn12 (Glickman et al., 1998) (Figure 2). Rpn11 contains a highly conserved metalloisopeptidase motif and this activity is necessary for de-ubiquitylation and proteasomal proteolysis of substrates. It is currently believed that Rpn11 de-ubiquitylates the substrate after it has been threaded into the 20S channel, thereby resulting in an irreversible commitment to proteolysis. Failure to de-ubiquitylate probably causes a sterical block to further insertion of the substrate into the proteolytic core (Verma et al., 2002; Yao and Cohen, 2002). Following release from the substrate, the polyubiquitin chain is hydrolyzed into single ubiquitin moieties which can take part in a new round of protein degradation (Figure 2).
Fig. 2. Ubiquitylation machinery and the 26S proteasome.
The undisturbed interplay between the folding and retrotranslocation machinery of the ER and the ubiquitin– proteasome system in the cytosol is essential for the maintenance of a healthy cellular life/metabolism both in unicellular and multicellular organisms. We now understand that there are many unique scenarios and substrate-specific details and variations about the way in which cells pursue ER quality control, and a unifying theme, if not for the protein components involved then at least for the sequence of events ending with proteasomal degradation, is emerging. However, there are still a lot of open questions and unresolved issues concerning ER-associated degradation as a whole, which will keep the scientific community busy for quite a long time.
Acknowledgments
Acknowledgements
The authors thank Wolfgang Hilt and Sae Hun Park for help with the figures and Elisabeth Tosta for the preparation of the manuscript. The work of the authors was supported by grants from the Deutsche Forschungsgemeinschaft (Bonn, Germany), the German–Israeli Project Cooperation (DIP) of the German Federal Ministry of Education and Research (BMBF) (Bonn, Germany), and the Fonds der Chemischen Industrie (Frankfurt, Germany).
References
- Aridor M. and Hannan,L.A. (2000) Traffic jam: a compendium of human diseases that affect intracellular transport processes. Traffic, 1, 836–851. [DOI] [PubMed] [Google Scholar]
- Baumeister W., Walz,J., Zuhl,F. and Seemuller,E. (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell, 92, 367–380. [DOI] [PubMed] [Google Scholar]
- Bays N.W., Gardner,R.G., Seelig,L.P., Joazeiro,C.A. and Hampton,R.Y. (2001a) Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell. Biol., 3, 24–29. [DOI] [PubMed] [Google Scholar]
- Bays N.W., Wilhovsky,S.K., Goradia,A., Hodgkiss-Harlow,K. and Hampton,R.Y. (2001b) HRD4/NPL4 is required for the proteasomal processing of ubiquitylated ER proteins. Mol. Biol. Cell, 12, 4114–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T., Volkwein,C. and Sommer,T. (1996) Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin–proteasome pathway. EMBO J., 15, 2069–2076. [PMC free article] [PubMed] [Google Scholar]
- Biederer T., Volkwein,C. and Sommer,T. (1997) Role of Cue1p in ubiquitylation and degradation at the ER surface. Science, 278, 1806–1809. [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S. and Klausner,R.D. (1994) Degradation of proteins retained in the endoplasmic reticulum. Modern Cell Biol., 15, 137–159. [Google Scholar]
- Bordallo J., Plemper,R.K., Finger,A. and Wolf,D.H. (1998) Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol. Biol. Cell, 9, 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun B.C., Glickman,M., Kraft,R., Dahlmann,B., Kloetzel,P.M., Finley,D. and Schmidt,M. (1999) The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat. Cell. Biol., 1, 221–226. [DOI] [PubMed] [Google Scholar]
- Braun S., Matuschewski,K., Rape,M., Thoms,S. and Jentsch,S. (2002) Role of the ubiquitin-selective CDC48UFD1/NPL4 chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J., 21, 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J.L. and McCracken,A.A. (1999) ER protein quality control and proteasome-mediated protein degradation. Semin. Cell. Dev. Biol., 10, 507–513. [DOI] [PubMed] [Google Scholar]
- Brodsky J.L., Werner,E.D., Dubas,M.E., Goeckeler,J.L., Kruse,K.B. and McCracken,A.A. (1999) The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem., 274, 3453–3460. [DOI] [PubMed] [Google Scholar]
- Cabral C.M., Liu,Y. and Sifers,R.N. (2001) Dissecting glycoprotein quality control in the secretory pathway. Trends Biochem. Sci., 26, 619–624. [DOI] [PubMed] [Google Scholar]
- Caldwell S.R., Hill,K.J. and Cooper,A.A. (2001) Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J. Biol. Chem., 276, 23296–23303. [DOI] [PubMed] [Google Scholar]
- Dai R.M. and Li,C.C. (2001) Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin–proteasome degradation. Nat. Cell. Biol., 3, 740–744. [DOI] [PubMed] [Google Scholar]
- Deak P.M. and Wolf,D.H. (2001) Membrane topology and function of Der3/Hrd1p as a ubiquitin–protein ligase (E3) involved in endoplasmic reticulum degradation. J. Biol. Chem., 276, 10663–10669. [DOI] [PubMed] [Google Scholar]
- Dürr G., Strayle,J., Plemper,R., Elbs,S., Klee,S.K., Catty,P., Wolf,D.H. and Rudolph,H.K. (1998) The medial-Golgi ion pump Pmr1 supplies the yeast secretory pathway with Ca2+ and Mn2+ required for glycosylation, sorting and endoplasmic reticulum-associated protein degradation. Mol. Biol. Cell, 9, 1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L., Molinari,M. and Helenius,A. (1999) Setting the standards: quality control in the secretory pathway. Science, 286, 1882–1888. [DOI] [PubMed] [Google Scholar]
- Elsässer S. et al. (2002) Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell. Biol., 4, 725–730. [DOI] [PubMed] [Google Scholar]
- Fagioli C., Mezghrani,A. and Sitia,R. (2001) Reduction of interchain disulfide bonds precedes the dislocation of Ig-µ chains from the endoplasmic reticulum to the cytosol for proteasomal degradation. J. Biol. Chem., 276, 40962–40967. [DOI] [PubMed] [Google Scholar]
- Fang S., Ferrone,M., Yang,C., Jensen,J.P., Tiwari,S. and Weissman, A.M. (2001) The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl Acad. Sci. USA, 98, 14422–14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebiger E., Story,C., Ploegh,H.L. and Tortorella,D. (2002) Visualization of the ER-to-cytosol dislocation reaction of a type I membrane protein. EMBO J., 21, 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer R., Jarosch,E., Urban,J., Volkwein,C. and Sommer,T. (2000) A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat. Cell. Biol., 2, 379–384. [DOI] [PubMed] [Google Scholar]
- Gardner R.G., Swarbrick,G.M., Bays,N.W., Cronin,S.R., Wilhovsky,S., Seelig,L., Kim,C. and Hampton,R.Y. (2000) Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J. Cell Biol., 151, 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillece P., Luz,J.M., Lennarz,W.J., de la Cruz,F.J. and Romisch,K. (1999) Export of a cysteine-free misfolded secretory protein from the endoplasmic reticulum for degradation requires interaction with protein disulfide isomerase. J. Cell Biol., 147, 1443–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B.S. (2002) The secretory pathway. In Dalbey,R.E. and von Heijne,G. (eds), Protein Targeting, Transport and Translocation. Academic Press, London, UK, pp. 358–376.
- Glickman M.H., Rubin,D.M., Coux,O., Wefes,I., Pfeifer,G., Cjeka,Z., Baumeister,W., Fried,V.A. and Finley,D. (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell, 94, 615–623. [DOI] [PubMed] [Google Scholar]
- Groll M., Ditzel,L., Lowe,J., Stock,D., Bochtler,M., Bartunik,H.D. and Huber,R. (1997) Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature, 386, 463–471. [DOI] [PubMed] [Google Scholar]
- Groll M., Bajorek,M., Kohler,A., Moroder,L., Rubin,D.M., Huber,R., Glickman,M.H. and Finley,D. (2000) A gated channel into the proteasome core particle. Nat. Struct. Biol., 7, 1062–1067. [DOI] [PubMed] [Google Scholar]
- Haigh N.G. and Johnson,A.E. (2002) Protein sorting at the membrane of the endoplasmic reticulum. In Dalbey,R.E. and von Heijne,G. (eds), Protein Targeting, Transport and Translocation. Academic Press, London, UK, pp. 74–106.
- Hampton R.Y. (2002) Proteolysis and sterol regulation. Annu. Rev. Cell. Dev. Biol., 18, 345–378. [DOI] [PubMed] [Google Scholar]
- Hampton R.Y. and Bhakta,H. (1997) Ubiquitin-mediated regulation of 3-hydroxy-3-methylglutaryl-CoA reductase. Proc. Natl Acad. Sci. USA, 94, 12944–12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton R.Y., Gardner,R.G. and Rine,J. (1996) Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell, 7, 2029–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes C.M., Caldwell,S. and Cooper,A.A. (2002) An HRD/DER-independent ER quality control mechanism involves Rsp5p-dependent ubiquitylation and ER–Golgi transport. J. Cell Biol., 158, 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W., Kleinschmidt,J.A., Saidowsky,J., Escher,C. and Wolf,D.H. (1991) Proteinase yscE, the yeast proteasome/multicatalytic multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J., 10, 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hill K. and Cooper,A.A. (2000) Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J., 19, 550–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M.M., Finger,A., Schweiger,M. and Wolf,D.H. (1996) ER degradation of a misfolded luminal protein by the cytosolic ubiquitin–proteasome pathway. Science, 273, 1725–1728. [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Wada,I., Hasegawa,K., Yorihuzi,T., Tremblay,L.O., Herscovics,A. and Nagata,K. (2001) A novel ER α-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep., 2, 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob C.A., Burda,P., Roth,J. and Aebi,M. (1998) Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J. Cell Biol., 142, 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob C.A., Bodmer,D., Spirig,U., Battig,P., Marcil,A., Dignard,D., Bergeron,J.J., Thomas,D.Y. and Aebi,M. (2001) Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep., 2, 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosch E., Taxis,C., Volkwein,C., Bordallo,J., Finley,D., Wolf,D.H. and Sommer,T. (2002) Protein dislocation from the ER requires polyubiquitylation and the AAA-ATPase Cdc48. Nat. Cell. Biol., 4, 134–139. [DOI] [PubMed] [Google Scholar]
- Jensen T.J., Loo,M.A., Pind,S., Williams,D.B., Goldberg,A.L. and Riordan,J.R. (1995) Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell, 83, 129–135. [DOI] [PubMed] [Google Scholar]
- Johnson A.E. and van Waes,M.A. (1999) The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell. Dev. Biol., 15, 799–842. [DOI] [PubMed] [Google Scholar]
- Knop M., Finger,A., Braun,T., Hellmuth,K. and Wolf,D.H. (1996a) Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J., 15, 753–763. [PMC free article] [PubMed] [Google Scholar]
- Knop M., Hauser,N. and Wolf,D.H. (1996b) N-Glycosylation affects endoplasmic reticulum degradation of a mutated derivative of carboxypeptidase yscY in yeast. Yeast, 12, 1229–1238. [DOI] [PubMed] [Google Scholar]
- Köhler A., Cascio,P., Leggett,D.S., Woo,K.M., Goldberg,A.L. and Finley,D. (2001) The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol. Cell, 7, 1143–1152. [DOI] [PubMed] [Google Scholar]
- Kopito R.R. (2000) Aggresomes, inclusion bodies and protein aggregation. Trends Cell. Biol., 10, 524–530. [DOI] [PubMed] [Google Scholar]
- Kostova Z. and Wolf,D.H. (2002) Protein quality control in the export pathway: the endoplasmic reticulum and its cytoplasmic proteasome connection. In Dalbey,R.E. and von Heijne,G. (eds), Protein Targeting, Transport and Translocation. Academic Press, London, UK, pp. 180–213.
- Lam Y.A., Lawson,T.G., Velayutham,M., Zweier,J.L. and Pickart,C.M. (2002) A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature, 416, 763–767. [DOI] [PubMed] [Google Scholar]
- Lumb R.A. and Bulleid,N.J. (2002) Is protein disulfide isomerase a redox-dependent molecular chaperone? EMBO J., 21, 6763–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer T.U., Braun,T. and Jentsch,S. (1998) Role of the proteasome in membrane extraction of a short-lived ER-transmembrane protein. EMBO J., 17, 3251–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken A.A. and Brodsky,J.L. (1996) Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin and ATP. J. Cell Biol., 132, 291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H.H., Wang,Y. and Warren,G. (2002) Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1–Npl4. EMBO J., 21, 5645–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K., Nishikawa,S., Hosokawa,N., Nagata,K. and Endo,T. (2001) Mnl1p, an α-mannosidase-like protein in yeast Saccharomyces cerevisiae, is required for endoplasmic reticulum-associated degradation of glycoproteins. J. Biol. Chem., 276, 8635–8638. [DOI] [PubMed] [Google Scholar]
- Nishikawa S.I., Fewell,S.W., Kato,Y., Brodsky,J.L. and Endo,T. (2001) Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol., 153, 1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M. (2000) Ubiquitin in chains. Trends Biochem. Sci., 25, 544–548. [DOI] [PubMed] [Google Scholar]
- Pilon M., Schekman,R. and Romisch,K. (1997) Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J., 16, 4540–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper R.K., Bohmler,S., Bordallo,J., Sommer,T. and Wolf,D.H. (1997) Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature, 388, 891–895. [DOI] [PubMed] [Google Scholar]
- Plemper R.K., Bordallo,J., Deak,P.M., Taxis,C., Hitt,R. and Wolf,D.H. (1999a) Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J. Cell Sci., 112, 4123–4134. [DOI] [PubMed] [Google Scholar]
- Plemper R.K., Deak,P.M., Otto,R.T. and Wolf,D.H. (1999b) Re-entering the translocon from the lumenal side of the endoplasmic reticulum. Studies on mutated carboxypeptidase yscY species. FEBS Lett., 443, 241–245. [DOI] [PubMed] [Google Scholar]
- Plemper R.K., Egner,R., Kuchler,K. and Wolf,D.H. (1998) Endoplasmic reticulum degradation of a mutated ATP-binding cassette transporter Pdr5 proceeds in a concerted action of Sec61 and the proteasome. J. Biol. Chem., 273, 32848–32856. [DOI] [PubMed] [Google Scholar]
- Plemper R.K. and Wolf,D.H. (1999) Retrograde protein translocation: ERADication of secretory proteins in health and disease. Trends Biochem. Sci., 24, 266–270. [DOI] [PubMed] [Google Scholar]
- Qu D., Teckman,J.H., Omura,S. and Perlmutter,D.H. (1996) Degradation of a mutant secretory protein, α1-antitrypsin Z, in the endoplasmic reticulum requires proteasome activity. J. Biol. Chem., 271, 22791–22795. [DOI] [PubMed] [Google Scholar]
- Rabinovich E., Kerem,A., Frohlich,K.U., Diamant,N. and Bar-Nun,S. (2002) AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol., 22, 626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M., Hoppe,T., Gorr,I., Kalocay,M., Richly,H. and Jentsch,S. (2001) Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48UFD1/NPL4, a ubiquitin-selective chaperone. Cell, 107, 667–677. [DOI] [PubMed] [Google Scholar]
- Regeimbal J. and Bardwell,J.C.A. (2002) Disulfide bound formation in prokaryotes and eukaryotes. In Dalbey,R.E. and von Heijne,G. (eds), Protein Targeting, Transport and Translocation. Academic Press, London, UK, pp. 131–150.
- Rouiller I., Butel,V.M., Latterich,M., Milligan,R.A. and Wilson-Kubalek,E.M. (2000) A major conformational change in p97 AAA ATPase upon ATP binding. Mol. Cell, 6, 1485–1490. [DOI] [PubMed] [Google Scholar]
- Rutishauser J. and Spiess,M. (2002) Endoplasmic reticulum storage diseases. Swiss Med. Wkly, 132, 211–222. [DOI] [PubMed] [Google Scholar]
- Shamu C.E., Flierman,D., Ploegh,H.L., Rapoport,T.A. and Chau,V. (2001) Polyubiquitylation is required for US11-dependent movement of MHC class I heavy chain from endoplasmic reticulum into cytosol. Mol. Biol. Cell, 12, 2546–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C., Brickner,J.H. and Walter,P. (2002) The unfolded protein response. In Dalbey,R.E. and von Heijne,G. (eds), Protein Targeting, Transport and Translocation. Academic Press, London, UK, pp. 151–179.
- Sommer T. and Jentsch,S. (1993) A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature, 365, 176–179. [DOI] [PubMed] [Google Scholar]
- Sommer T. and Wolf,D.H. (1997) Endoplasmic reticulum degradation: reverse protein flow of no return. FASEB J., 11, 1227–1233. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Park,H., Hollingsworth,N.M., Sternglanz,R. and Lennarz,W.J. (2000) PNG1, a yeast gene encoding a highly conserved peptide: N-glycanase. J. Cell Biol., 149, 1039–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Park,H., Kwofie,M.A. and Lennarz,W.J. (2001) Rad23 provides a link between the Png1 deglycosylating enzyme and the 26S proteasome in yeast. J. Biol. Chem., 276, 21601–21607. [DOI] [PubMed] [Google Scholar]
- Swanson R., Locher,M. and Hochstrasser,M. (2001) A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes Dev., 15, 2660–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C., Vogel,F. and Wolf,D.H. (2002) ER–Golgi traffic is a prerequisite for efficient ER degradation. Mol. Biol. Cell, 13, 1806–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower J.S., Hoffman,L., Rechsteiner,M. and Pickart,C.M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J., 19, 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S. and Weissman,A.M. (2001) Endoplasmic reticulum (ER)-associated degradation of T cell receptor subunits. Involvement of ER-associated ubiquitin-conjugating enzymes (E2s). J. Biol. Chem., 276, 16193–16200. [DOI] [PubMed] [Google Scholar]
- Travers K.J., Patil,C.K., Wodicka,L., Lockhart,D.J., Weissman,J.S. and Walter,P. (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell, 101, 249–258. [DOI] [PubMed] [Google Scholar]
- Tsai B. and Rapoport,T.A. (2002) Unfolded cholera toxin is transferred to the ER membrane and released from protein disulfide isomerase upon oxidation by Ero1. J. Cell Biol., 159, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S., Kim,W., Belden,W.J., Spear,E.D., Barlowe,C. and Ng,D.T. (2001) Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J. Cell Biol., 155, 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Aravind,L., Oania,R., McDonald,W.H., Yates,I.J., Koonin, E.V. and Deshaies,R.J. (2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science, 298, 611–615. [DOI] [PubMed] [Google Scholar]
- Walter J., Urban,J., Volkwein,C. and Sommer,T. (2001) Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p. EMBO J., 20, 3124–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. and Chang,A. (1999) Eps1, a novel PDI-related protein involved in ER quality control in yeast. EMBO J., 18, 5972–5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C.L., Omura,S. and Kopito,R.R. (1995) Degradation of CFTR by the ubiquitin–proteasome pathway. Cell, 83, 121–127. [DOI] [PubMed] [Google Scholar]
- Werner E.D., Brodsky,J.L. and McCracken,A.A. (1996) Proteasome-dependent endoplasmic reticulum-associated protein degradation: an unconventional route to a familiar fate. Proc. Natl Acad. Sci. USA, 93, 13797–13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiertz E.J., Tortorella,D., Bogyo,M., Yu,J., Mothes,W., Jones,T.R., Rapoport,T.A. and Ploegh,H.L. (1996) Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature, 384, 432–438. [DOI] [PubMed] [Google Scholar]
- Wilhovsky S., Gardner,R. and Hampton,R. (2000) HRD gene dependence of endoplasmic reticulum-associated degradation. Mol. Biol. Cell, 11, 1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T. and Cohen,R.E. (2002) A cryptic protease couples deubiquitylation and degradation by the proteasome. Nature, 419, 403–407. [DOI] [PubMed] [Google Scholar]
- Ye Y., Meyer,H.H. and Rapoport,T.A. (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature, 414, 652–656. [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. (2000) Structure of the AAA ATPase p97. Mol. Cell, 6, 1473–1484. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Nijbroek,G., Sullivan,M.L., McCracken,A.A., Watkins,S.C., Michaelis,S. and Brodsky,J.L. (2001) Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell, 12, 1303–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]