Abstract
Based on archaeological evidence, the spread of agropastoralism across Europe followed two main paths: the Danubian route, along which Neolithic farmers expanded north across the central European plains; and the Mediterranean route, where migration occurred along the coast of the Mediterranean sea. Here we examine 20 cattle breeds from the continent and assess the genetic diversity levels and relationships among the breeds using 19 microsatellite markers. Additionally, we show evidence that concords with two distinct cattle migrations from the Near East, and also demonstrate that Mediterranean cattle breeds may have had more recent input from both the Near East and Africa.
Keywords: Mediterranean, Bos taurus, microsatellite, domestication, diversity
1. Introduction
At the end of the last ice age (approximately 10 000 years BP), the first domesticated plants and animals appeared. A body of genetic and archaeological evidence strongly argues for a separate domestication of the two main types of cattle, Bos indicus (zebu) and Bos taurus (taurine) from two different strains of Bos primigenius (Grigson 1980; Meadow 1993; Loftus et al. 1994). Taurine cattle were undoubtedly domesticated in the Near East while Baluchistan, in present day Pakistan, is a possible centre for zebu domestication (Meadow 1993). There is also evidence pointing towards a separate domestication for African taurine cattle (Gautier 1984; Wendorf & Schild 1994; Bradley et al. 1996).
The most widely held view contends that European cattle derive from cattle domesticated in the Near East and genetic evidence has supported this hypothesis (Troy et al. 2001). After its origin in the Near East, agropastoralism rapidly spread to neighbouring areas and was already present in Cyprus around 8200 years BP (Guilaine et al. 2000). In mainland Europe, the arrival of farming is thought to have occurred in the floodplains of Thessaly, in the south of the Balkan Peninsula, approximately 7800 years BP (Van Andel & Runnels 1995).
Archaeological evidence indicates that farming spread across Europe following two major paths from the Near East. One group of early farmers expanded north, moving along the rivers of the Balkans into the plains of central and northern Europe—in what is known as the Danubian route. They settled in riverine habitats where conditions would have been most suitable for agriculture and farming (Bogucki 1996) and established settlements in Germany by 7400 BP and Belgium by 5400 BP (Gkliasta et al. 2003).
The other current of Neolithic farmers migrated west, following the coast of the Mediterranean and crossing the sea to the major islands (Bogucki 1996)—the Mediterranean route. Along this route, the onset of farming is linked with the arrival of small Neolithic seafaring groups settling in areas not occupied by local hunter–gatherers (Zilhao 1993). This pattern of diffusion rapidly led to the appearance of farming in a widely spread area, and from its initial settlement in the Balkans and southern Italy, farming reached Corsica around 7900 years BP (Vigne 1999), the French Languedoc by 7800 years BP (Guilaine 2003) and the coast of eastern Spain approximately 7700 years BP (Bernabeu & Marti Olivier 1992).
In this study, 20 cattle populations from Europe and nine from the Near East were analysed. The results presented here show a strong decline in the genetic diversity from the Near East to Europe, this pattern is indicative of the expansion of cattle out of the Near East. The data also demonstrate a difference between cattle breeds from northern and Mediterranean regions, consistent with the movement of cattle occurring along two distinct routes. Finally, traces of both Near Eastern and African influence were detected in different Mediterranean breeds.
2. Material and methods
Data were included from previous studies on the genetic diversity of cattle breeds (Loftus et al. 1999; Schmid et al. 1999; Edwards et al. 2000; Kumar et al. 2003). Three hundred and forty-eight new samples, representing 10 distinct European breeds from five countries were collected. These novel populations are marked with an asterisk in table 1. Every attempt was made to ensure that samples were from unrelated animals, typical of the breed, and from a broad geographic area. For the purpose of some of the following analyses, breeds were divided into regional groups based on their geographic location (table 1).
Table 1.
The origins of the breeds sampled are indicated, together with the number of animals sampled per breed and pooled samples are underlined.
| population | code | sample size | breed origin | expected | size corrected |
|---|---|---|---|---|---|
| Highland* | HI | 37 | Scotland | 0.52 (0.04) | 3.44 (0.14) |
| Hereford* | HE | 34 | England | 0.6 (0.04) | 3.69 (0.14) |
| Jersey | JE | 44 | Channel Islands | 0.63 (0.04) | 3.89 (0.13) |
| BRITISH ISLES | 115 | POOLED | 0.65 (0.04) | 6.07 (0.06) | |
| Swiss Brown | OB | 50 | Switzerland | 0.65 (0.03) | 4.16 (0.15) |
| Chrolais | CH | 36 | France | 0.66 (0.04) | 4.38 (0.17) |
| Eringer | ER | 50 | Switzerland | 0.57 (0.04) | 3.83 (0.17) |
| Evolenard | EV | 15 | Switzerland | 0.6 (0.03) | 3.64 (0.13) |
| Holstein-Friesian | HO | 50 | Netherlands | 0.65 (0.03) | 4.16 (0.16) |
| Pustertaler | PS | 31 | Germany | 0.69 (0.03) | 4.43 (0.15) |
| Pinzgauer | PG | 30 | Germany | 0.71 (0.03) | 4.9 (0.17) |
| Simmenthal | SI | 50 | Switzerland | 0.58 (0.04) | 3.9 (0.15) |
| Salers* | SL | 38 | France | 0.55 (0.05) | 3.81 (0.15) |
| Vosges | VO | 27 | France | 0.68 (0.03) | 4.29 (0.16) |
| Hungarian Grey | HG | 44 | Hungary | 0.62 (0.04) | 3.97 (0.15) |
| NORTHERN CONTINENTAL | 421 | POOLED | 0.69 (0.03) | 7.97 (0.16) | |
| Alentejana* | AL | 30 | Portugal | 0.66 (0.04) | 4.43 (0.16) |
| Mertolenga* | MT | 37 | Portugal | 0.66 (0.04) | 4.45 (0.17) |
| Arouquesa* | AQ | 34 | Portugal | 0.68 (0.03) | 4.53 (0.17) |
| Romagnola* | RO | 40 | Italy | 0.61 (0.04) | 4.04 (0.15) |
| Meremmana* | MR | 40 | Italy | 0.67 (0.03) | 4.4 (0.16) |
| Modicana* | MD | 37 | Italy | 0.65 (0.04) | 4.59 (0.19) |
| Sykia* | SK | 21 | Greece | 0.69 (0.02) | 4.39 (0.15) |
| MEDITERRANEAN EUROPE | 239 | POOLED | 0.71 (0.03) | 7.62 (0.12) | |
| Turkish Grey | TG | 43 | Turkey | 0.76 (0.02) | 5.81 (0.21) |
| Anatolian Black | AB | 41 | Turkey | 0.78 (0.02) | 5.96 (0.21) |
| East Anatolian Red | EAR | 44 | Turkey | 0.78 (0.02) | 5.96 (0.21) |
| South Anatolian Red | SAR | 43 | Turkey | 0.78 (0.02) | 5.82 (0.21) |
| Damascus | DA | 41 | Syria | 0.74 (0.02) | 5.08 (0.2) |
| Iraqi | IR | 42 | Iraq | 0.78 (0.01) | 6.1 (0.24) |
| Kurdi | KUD | 37 | Iraq | 0.79 (0.02) | 6.06 (0.22) |
| Egyption | EG | 32 | Egypt | 0.77 (0.02) | 5.57 (0.2) |
| Near East | 323 | pooled | 0.79 (0.02) | 10.22 (0.18) | |
| N'Dama | Ngu | 44 | Guinea | 0.55 (0.04) | 4.28 (0.09) |
| AFRICA | 44 | ||||
| Ongole | ON | 32 | India | 0.67 (0.02) | 4.17 (0.18) |
| Nellore | NE | 27 | India | 0.66 (0.03) | 4.11 (0.15) |
| Red Sindhi | RS | 38 | Pakistan | 0.7 (0.03) | 4.79 (0.18) |
| Tharparker | TH | 35 | India | 0.62 (0.03) | 4.02 (0.16) |
| Desi | DE | 26 | India | 0.73 (0.02) | 4.99 (0.18) |
| Hariana | HA | 10 | India | 0.73 (0.03) | 4.6 (0.15) |
| Sahiwal | SA | 10 | India | 0.62 (0.05) | 3.91 (0.14) |
| SOUTH ASIA | 178 | POOLED | 0.75 (0.02) | 8.86 (0.14) | |
Expected heterogozity (He) values are shown. The mean number of alleles (Ā) was calculated by resampling 1000 times from each breed sample.
DNA was extracted from blood using standard procedures (Sambrook et al. 1989). To extract DNA from hair, 100 μl of lysis solution was added to approximately 8–10 follicles, and were incubated at 60 °C for 55 min, followed by 15 min at 95 °C. Lysis solution consists of: 0.2 mM Cresol Red; 1.5 mM Taq buffer (pH 9.0); 1.0 mM MgCl2; 0.55 (v/v) Tween-20; 0.6% IPEGAL DNA extraction buffer (Sigma); 0.48 mg ml−1 Proteinase K.
The following 19 microsatellite loci were typed: BM1818, BM1824, BM2113, CSRM60, CSSM66, ETH3, ETH10, ETH225, Hel1, Hel5, Hel9, Hel13, ILSTS005, ILSTS006, INRA005, INRA023, INRA032, INRA035 and INRA063. The loci were selected from a group of 30, which had previously been agreed from a collaborative study of European cattle (http://dad.fao.org/en/refer/library/guidelin/marker.pdf). Microsatellite genotyping was carried out according to Loftus et al. (1999).
Allele frequencies for all loci were determined by direct counting. Unbiased estimates of gene diversity (expected heterozygosity, ) were calculated according to Nei, for all locus/population combinations (Nei 1987). The mean number of alleles per breed was calculated by sampling with replacement 1000 times and averaging over all loci (Park 2001). Pooled was also calculated for the regional groups in table 1. Mann–Whitney tests were implemented using the Spss statistical package (v. 11.0). Interpopulation genetic distances (DA; Takezaki & Nei 1996) were estimated using Populations (http://www.cnrs-gif.fr/pge/bioinfo/populations). Populations was also used to construct a neighbour-joining (NJ) tree (Saitou & Nei 1987) from DA genetic distances. Reliability of the tree obtained was examined by a bootstrap test with 1000 replicate resamplings of loci with replacement. Pairwise geographic distances were calculated using Passage (http://lsweb.la.asu.edu/rosenberg/Passage).
In order to detect possible B. indicus influence in European breeds, we took a population-associated allele (PAA) approach (Kumar et al. 2003). Most microsatellite markers display markedly different and distinctive allelic distributions when analysed in taurine and zebu cattle populations. Alleles that show high frequency differential between the two lineages can be used to assess the ancestry of populations. The presence of a characteristic B. indicus allele in a population may be an indication of introgression from the East.
A scaled frequency differential (SFD) was calculated for each allele by subtracting the average frequency of the allele in a pooled European sample (including British, Northern and Mediterranean breeds) from the average frequency in a pooled sample of the South Asian indicine breeds assayed (Kumar et al. 2003). The absolute value of this frequency differential was divided by the greater of the two values. Alleles that occurred at a frequency of at least 20% in the South Asian B. indicus sample and which have a SFD value of at least 40% were denoted B. indicus PAAs. The average frequency of these B. indicus PAAs was calculated for each breed to detect and assess the relative extent of zebu influence. Average breed values were grouped into geographic regions and plotted as a box plot using Spss (v. 11.0). The box represents the interquartile range and the whiskers extend from the box to the highest and lowest values, excluding outliers. A line across the box indicates the median. Outlier points were values between 1.5 and 3 box lengths from the upper or lower edge of the box. Extreme values were more than 3 box lengths from the upper or lower edge of the box.
3. Results
In total, 249 alleles were detected in the 19 loci surveyed, giving a mean number of 13.1 alleles per locus. The mean number of alleles observed over a range of loci for different populations is considered to be a reasonable indicator of genetic variation, assuming that populations of similar sample size (Loftus et al. 1999). The breed samples in the present study ranged in size from 10 to 50. Therefore, a sample-size corrected mean number of alleles in each of the breeds is given in table 1.
Among the different biogeographical groupings, the Near Eastern breeds displayed the highest allelic diversity (; median=5.89), followed by the breeds from Mediterranean Europe (median=4.43); and the lowest diversity values were present in the breeds from Britain (median=3.69). A Mann–Whitney test showed the difference between the Near East and mainland Europe (Mediterranean and Northern combined) to be highly significant (p<0.001), as was the difference between mainland Europe and Britain (p=0.02). The test also supports greater diversity in Mediterranean breeds than in those from the northern part of the continent (p=0.03). Although less marked, breed values for gene diversity showed a similar pattern with significantly elevated values in the Near East compared to mainland Europe (p<0.001). When samples are pooled into the regional groups delineated in table 1, standard diversity statistics show similar trends, with the exception of the comparison between Mediterranean and Northern Europe where the former shows slightly higher gene diversity (0.69 versus 0.71) but marginally lower allelic diversity (7.62 versus 7.97).
When the allele lists encountered in the regional groupings are compared each shows a number of variants which are only encountered locally; private alleles. Thus the South Asian zebu group exhibits 12 alleles specific to that area. The Near East has the greatest number of private alleles (25) but Northern Europe also displays 10, whereas Mediterranean cattle and British breeds exhibit very few (2 and 0).
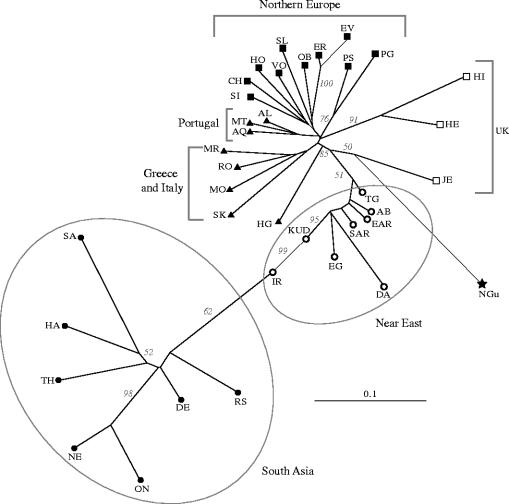
An NJ phylogeny was constructed from a matrix of DA genetic distances (figure 1). The longest branch in the tree separates the B. indicus from the B. taurus populations. The Near Eastern breeds are positioned proximal to the B. taurus radiation; many of these breeds exhibited truncated branch lengths, a feature associated with admixed populations (Mountain et al. 1992). The most striking feature of this tree is geographic integrity; breeds from the same region cluster together. These broad geographical groupings are supported by high bootstrap values. Within the European radiation, the populations of Mediterranean Europe are positioned proximal to the Near East breeds, followed by the breeds from Northern Europe, while the British cattle populations occupy the most distal position.
Figure 1.
An unrooted neighbour-joining tree summarizing DA genetic distances among 21 European, eight Near Eastern, one African and seven South Asian cattle populations. Numbers in grey represent bootstrap values based on 1000 replications. Some of the biogeographical groupings illustrated by the tree are highlighted.
DA genetic distances were plotted against pairwise geographic distances for 19 European breeds (figure 2). While there is a highly significant relationship between the two variables (p=2.46×10−9), as is commonly seen (Cavalli-Sforza & Feldman 1990), the fit of the model, as assessed by r2(adjusted) (0.25) is weak. The fit is greatly improved by dividing the populations into two groups; first, the northern continental and British breeds and second, the Mediterranean breeds. The regression line accounting for the Mediterranean populations (r2(adj)=0.55) has a slope that is considerably different from those from Northern European (r2(adj)=0.56; figure 2). If the British breeds are grouped with the Mediterranean breeds, and the northern continental breeds are plotted separately, the fit of both regression lines is decreased (r2(adj)=0.15 and 0.22, respectively). Thus the former grouping seems most appropriate and indicates that, over equivalent geographic distance, populations on the Mediterranean coast show reduced genetic differentiation compared to those in the north of the continent and Britain.
Figure 2.
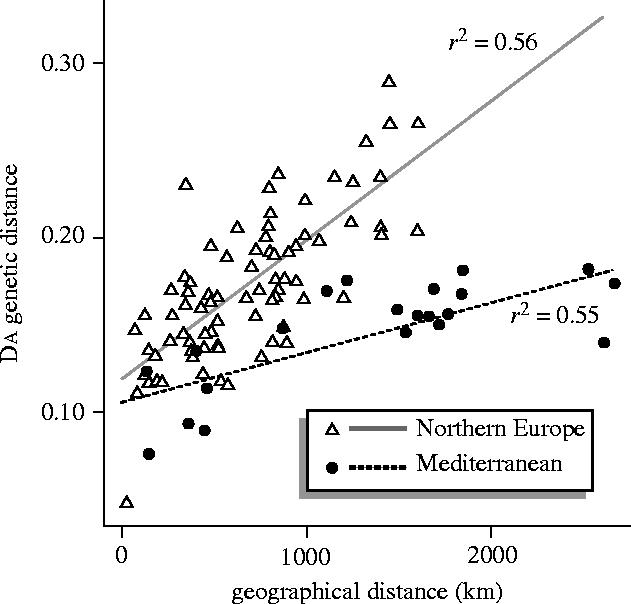
DA genetic distances plotted against pairwise geographic distances for European cattle populations, showing different distributions between the populations of Northern Europe and those found along the Mediterranean coast. Triangles represent pairwise comparisons among the northern European groups (northern continental and British breeds). Filled circles represent pairwise comparisons among the Mediterranean breeds. Best-fit least squares regression lines are shown with associated r2-values.
The presence of B. indicus ancestry in Near Eastern breeds has previously been demonstrated (Loftus et al. 1999). The frequency of B. indicus PAAs (Kumar et al. 2003) was examined to estimate whether this influence has permeated Mediterranean cattle. B. indicus PAAs were selected on the basis of a SFD between pooled European and South Asian populations, and their absolute frequency in the pooled South Asian population. The average frequency of these PAAs was calculated for each breed. The breeds were grouped according to geography and plotted as a boxplot in figure 3.
Figure 3.
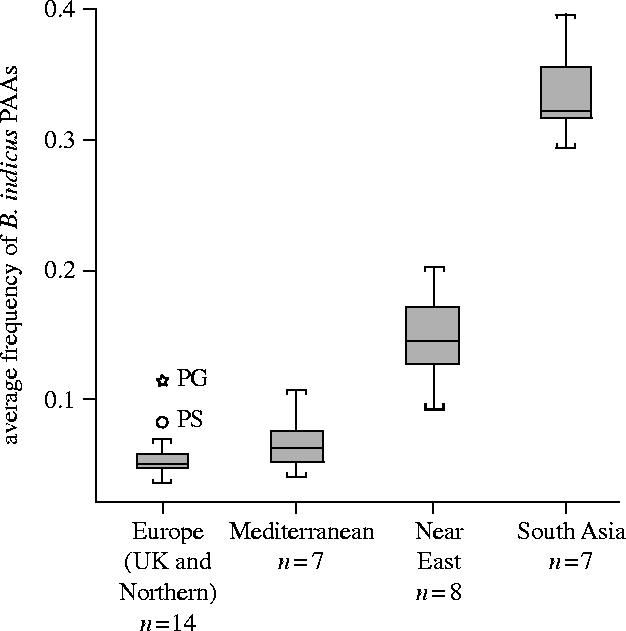
Boxplot showing average frequency of B. indicus population-associated alleles in sample groups. The box represents the interquartile range (50% of values); while the line across the box indicates the median. Whiskers extend to the highest and lowest values, excluding outliers. These are denoted by circles and have values between 1.5 and 3 box lengths from the upper or lower edge of the box. Extreme values (stars) are values that are more than 3 box lengths from the upper or lower edge of the box.
By definition, the South Asian samples have the highest average frequencies of B. indicus PAAs. The average frequency in the Near Eastern group is intermediate between the South Asian sample and the two European groups. The average frequency of B. indicus PAAs in Mediterranean breeds is 6.7%, higher than the average frequency in the rest of Europe (5.7%). The boxplot reveals two distinct outliers in the European population: Pustertaler and Pinzgauer. The Pustertaler is a seriously endangered breed native to South Tyrol, which has suffered dramatic reduction in numbers in recent history (Edwards et al. 2000). Its close relationship to the Pinzgauer breed is supported by the bootstrap value (100) of the NJ tree in figure 1. It is likely that the history of these two strongly associated breeds has contributed to their outlier status. If these two outliers are excluded, the average frequency of PAAs in Europe is reduced to 5.1%. However the difference between the means of the European and Mediterranean groups is not significant (Mann–Whitney, p=0.07).
The locus BM2113 has previously been reported (MacHugh 1996) as diagnostic for West African breeds. This locus showed one allele that was present at a high frequency in the West African N'Dama breed, and was absent from the South Asian and Near Eastern populations and from Northern Europe. This allele here was observed in two southern Portuguese breeds, Alentejana and Mertolenga, with a frequency of 5.4 and 1.4%, respectively.
4. Discussion
Archaeological evidence suggests that the spread of agropastoralism across Europe followed two main paths: the Danubian route, along which Neolithic farmers expanded north across the central Europe plains; and the Mediterranean route, where migration occurred along the coast of the Mediterranean sea (Zilhao 1993). For several reasons involving sea faring, forest densities and relationships between native Mesolithic and immigrant Neolithic human populations (Tresset & Vigne 2001), the expansion of farming and livestock proceeded at a slightly higher rate along the Mediterranean coast than through the continental heartland. The data presented above are in agreement with this hypothesis, and also suggest that Mediterranean cattle breeds may have had more recent input from both the Near East and Africa.
As other authors have previously shown (Loftus et al. 1999; Troy et al. 2001), there is a significant decline of diversity values from the Near East into the rest of Europe with the lowest diversity values found in the breeds from Britain. Regression analysis demonstrates a significant relationship between and geographic latitude (p=0.01). This pattern of genetic variation concurs with the prevailing view that Near Eastern cattle populations display higher genetic diversity because they have retained more ancestral variation from their wild progenitors and that populations in Europe have lost diversity as a consequence of migration and repeated founder events (Jorde et al. 1997; Troy et al. 2001). Furthermore, introgression by B. indicus into the Near Eastern breeds has undoubtedly contributed to the significantly elevated diversity values (Loftus et al. 1999). However, previous work has established that introgression alone does not account for the increased diversity observed (Loftus et al. 1999).
The intense nature of animal breeding in Europe over the past 150 years has undoubtedly also contributed to the lower levels of genetic variation present in European breeds. Modern breeding was introduced in northern European countries first, allowing more time for this technology to impact in this region; indeed within-breed statistics give lower diversity in comparison with the south. However, pooled population comparisons give a mixed signal. Interestingly, the north Europe population as a whole displays both more alleles and private alleles than the south.
Comparison of genetic distances with geographic distances reveals two different relationships within Europe (figure 2). Mediterranean populations show less genetic differentiation with geographic distance, suggesting that Mediterranean populations had less time to differentiate from each other as they migrated along the coast. Also, these results may indicate that Mediterranean populations have not been subjected to as much genetic isolation as those in northern Europe. Therefore, these observations can be interpreted either as the result of the recent (i.e. at any time between the Bronze Age and the twentieth century) evolutionary history of European cattle, or may reflect ancient patterns of cattle migration from the Near East to Europe. Note that archaeological evidence supports the hypothesis that the expansion of agriculture through the Mediterranean basin was more rapid than its movement through the continental heartland (Guilaine 2003).
A further influence may have been aurochs introgression. Mitochondrial DNA (mtDNA) evidence shows that the European wild ox was divergent from that domesticated in the Near East and strongly suggests that modern cattle matrilines have no legacy from locally domesticated European aurochs (Troy et al. 2001). However, the potential for male introgression remains and a third source of differentiation between north and south Europe could be different levels of localized input from the wild (Gotherstrom et al. submitted).
It is widely accepted that Anatolia represents a primary centre of domestication for B. taurus cattle and that considerable levels of B. indicus introgression have occurred at this centre (Loftus et al. 1999; Kumar et al. 2003). In the present study, B. indicus influence in Europe was measured systematically using PAAs. These were found at low frequencies in some European breeds (figure 3). The average frequency of B. indicus PAAs is higher in Mediterranean breeds (6.7%) than in the rest of Europe (5.1% without outliers). Within the Mediterranean, the average frequencies of B. indicus PAAs in Italy is the highest (8.1%). The Greek Sykia breed is intermediate (6.3%) and the average frequency in Portugal is 5.4%. The highest absolute values are found in two Italian breeds: Maremanna (8.1%) and Modicana (10.8%). Interestingly, a percentage of individuals of the Modicana breed have bifid processes in the last thoracic vertebrae, traditionally considered a B. indicus anatomical characteristic (Grigson 2000).
The presence of B. indicus associated alleles in Greece and Italy, in otherwise characteristic European taurine cattle, could be interpreted as the result of the geographic and historical association between these two countries and Anatolia. Historically, these areas have been connected since before the Minoan (Bronze Age, third millennium) increase in trade exchanges in the eastern and central Mediterranean Basin (Anon. 1979; Pomey 1997). For hundreds of years after that, active trade in many different commodities was well established, first between the eastern and western Greek colonies, then in the Roman empire (Duby 1995) and later along the famous silk road, stretching from Rome in Italy through Samarkand in Uzbekistan to Luoyang in China (Wood 2002). Within the whole of Europe, there is a tendency to higher B. indicus PAA values in breeds geographically located in the eastern part of the continent: almost certainly the result of proximity to Anatolia. This can be tested by examining the relationship between breed longitude and PAA frequency (regression p=0.04).
A characteristic African B. taurus allele was detected in two southern Portuguese cattle populations. The presence of African mtDNA sequences within these breeds has been reported previously (Cymbron et al. 1999) and was interpreted as a legacy from the long and influential Moorish occupation of the Iberian Peninsula. Our results reinforce the findings of this previous study, showing that the minor African influence in Europe is found in southern Portugal, proximal to the African continent.
In conclusion, our results support the grouping of European B. taurus cattle into two distinct streams, which both migrated out of the Near East and have had divergent relationships with the Near East during subsequent historical times. Archaeological evidence for the presence of distinct agropastoralist cultures in these two regions exists and is reflected in the genetic evidence presented here. Additionally, this study shows that eastern European, and especially Mediterranean, cattle may not have entirely escaped from the wave of B. indicus introgression that is well documented through the Near East. Finally, we show that characteristic African B. taurus microsatellite alleles are present in southern Iberia, supporting the hypothesis of African influence in Portugal.
Acknowledgments
We would like to acknowledge the assistance of Johannes A. Lenstra, Anne Tresset, Alessio Valentini and Isaäc J. Nijman. This paper has been partly discussed in the framework of the European Science Foundation EUROCORES programme ‘The origin of Man, Language and Languages’, especially in the project ‘Early domestic bovids’. A.R.F. is supported by a Wellcome Studentship in Biodiversity number 054275/Z/98/Z.
Footnotes
These authors contributed equally to this work.
References
- Anon. Acts of the International Archaeological symposium. Department of Antiquities; Nicosia: 1979. The relations between Cyprus and Crete, ca. 2000–500 BC. [Google Scholar]
- Bernabeu J, Marti Olivier B. El País Valenciano de la aparición del Neolítico al Horizonte Campaniforme, Aragón/Litoral Mediterrâneo. In: Utrilla P, editor. Intercambios Culturales durante la Prehistoria. Institución Fernando el Católico; Zaragoza: 1992. pp. 213–234. [Google Scholar]
- Bogucki P. The spread of early farming in Europe. Am. Sci. 1996;84:242–253. [Google Scholar]
- Bradley D.G, MacHugh D.E, Cunningham P, Loftus R.T. Mitochondrial diversity and the origins of African and European cattle. Proc. Natl Acad. Sci. USA. 1996;93:5131–5135. doi: 10.1073/pnas.93.10.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L.L, Feldman M.W. Spatial subdivision of populations and estimates of genetic variation. Theor. Popul. Biol. 1990;37:3–25. doi: 10.1016/0040-5809(90)90024-p. [DOI] [PubMed] [Google Scholar]
- Cymbron T, Loftus R.T, Malheiro M.I, Bradley D.G. Mitochondrial sequence variation suggests an African influence in Portuguese cattle. Proc. R. Soc. B. 1999;266:597–603. doi: 10.1098/rspb.1999.0678. 10.1098/rspb.1999.0678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duby G. Larousse; Paris: 1995. Atlas historique. [Google Scholar]
- Edwards C.J, Dolf G, Looft C, Loftus R.T, Bradley D.G. Relationships between the endangered Pustertaler-Sprinzen and three related European cattle breeds as analysed with 20 microsatellite loci. Anim. Genet. 2000;31:329–332. doi: 10.1046/j.1365-2052.2000.00651.x. [DOI] [PubMed] [Google Scholar]
- Gautier A. Archaeozoology of the Bir Kiseiba region, Eastern Sahara. In: Wendorf F, Close A.E, editors. Cattle-keepers of the Eastern Sahara: the Neolithic of Bir Kiseiba. Department of Anthropology, Southern Methodist University; Dallas: 1984. pp. 49–72. [Google Scholar]
- Gkliasta M, Russell T, Shennan S, Steele J. Neolithic transition in Europe: the radiocarbon record revisited. Antiquity. 2003;77:45–62. [Google Scholar]
- Gotherstrom, A., Anderung, C., Hellborg, L., Elburg, R., Smith, C., Bradley, D. G. & Ellegren, H. In press. Cattle domestication in the Near East was followed by hybridization with aurochs bulls in Europe. Proc. R. Soc. B (10.1098/rspb.2005.3243) [DOI] [PMC free article] [PubMed]
- Grigson C. The craniology and relationships of four species of Bos. 5. Bos indicus L. J. Archaeol. Sci. 1980;7:3–32. [Google Scholar]
- Grigson C. Bos africanus (Brehm)? Notes on the archaeozoology of the native cattle of Africa. In: Blench R.M, MacDonald K.C, editors. The origins and development of African livestock: archaeology, genetics, linguistics and ethnography. UCL Press; London: 2000. pp. 38–60. [Google Scholar]
- Guilaine J. Seuil; Paris: 2003. De la vague à la tombe. [Google Scholar]
- Guilaine J, Briois F, Vigne J.-D, Carrère I. Découverte d'un Néolithique précéramique ancien chypriote (fin 9e, début 8e millénaires cal. BC), apparenté au PPNB ancien/moyen du Levant nord (avec “abridged version”) C. R. Acad. Sci., Paris, Sci. de la Terre et des planètes. 2000;330:75–82. [Google Scholar]
- Jorde L.B, Rogers A.R, Bamshad M, Watkins W.S, Krakowiak P, Sung S, Kere J, Harpending H.C. Microsatellite diversity and the demographic history of modern humans. Proc. Natl Acad. Sci. USA. 1997;94:3100–3103. doi: 10.1073/pnas.94.7.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Freeman A.R, Loftus R.T, Gaillard C, Fuller D.Q, Bradley D.G. Admixture analysis of South Asian cattle. Heredity. 2003;91:43–50. doi: 10.1038/sj.hdy.6800277. [DOI] [PubMed] [Google Scholar]
- Loftus R.T, MacHugh D.E, Bradley D.G, Sharp P.M, Cunningham P. Evidence for two independent domestications of cattle. Proc. Natl Acad. Sci. USA. 1994;91:2757–2761. doi: 10.1073/pnas.91.7.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus R.T, Ertugrul O, Harba A.H, El-Barody M.A, MacHugh D.E, Park S.D, Bradley D.G. A microsatellite survey of cattle from a centre of origin: the Near East. Mol. Ecol. 1999;8:2015–2022. doi: 10.1046/j.1365-294x.1999.00805.x. [DOI] [PubMed] [Google Scholar]
- MacHugh, D. E. 1996 Molecular biogeography and genetic structure of domesticated cattle. Ph.D. thesis, Trinity College, University of Dublin, Dublin.
- Meadow R.H. Animal domestication in the middle east: a revised view from the eastern margin. In: Poeeshl G, editor. Harappan civilisation. Oxford and IBH; New Dehli: 1993. pp. 295–320. [Google Scholar]
- Mountain J.L, Lin A.A, Bowcock A.M, Cavalli-Sforza L.L. Evolution of modern humans: evidence from nuclear DNA polymorphisms. Phil. Trans. R. Soc. B. 1992;337:159–165. doi: 10.1098/rstb.1992.0093. [DOI] [PubMed] [Google Scholar]
- Nei M. Columbia University Press; New York, USA: 1987. Molecular evolutionary genetics. [Google Scholar]
- Park, S. D. 2001 Trypanotolerance in West African cattle and the population genetic effects of selection. Ph.D. thesis, Trinity College, University of Dublin, Dublin.
- Pomey P. Edisud edition; Aix-en-Provence: 1997. La navigation dans l'antiquité. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E.F, Maniatis T. Cold Spring Harbor Laboratory Press; New York: 1989. Molecular cloning: a laboratory manual. [Google Scholar]
- Schmid M, Saitbekova N, Gaillard C, Dolf G. Genetic diversity in Swiss cattle breeds. J. Anim. Breed. Genet. 1999;116:1–8. doi: 10.1046/j.1365-2052.1999.00429.x. [DOI] [PubMed] [Google Scholar]
- Takezaki N, Nei M. Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics. 1996;144:189–399. doi: 10.1093/genetics/144.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresset A, Vigne J.-D. La chasse, principal élément structurant la diversité des faunes archéologiques du Néolithique ancien, en Europe tempérée comme en Méditerranéenne: tentative d'interprétation fonctionnelle. In: Arbogast R.-M, Jeunesse C, Schibler J, editors. Rôle et statut de la chasse dans le Néolithique ancien danubien (5500–4900 av. J.-C.) Rahden/West; Marie Leidorf: 2001. pp. 129–151. [Google Scholar]
- Troy C.S, MacHugh D.E, Bailey J.F, Magee D.A, Loftus R.T, Cunningham P, Chamberlain A.T, Sykes B.C, Bradley D.G. Genetic evidence for Near-Eastern origins of European cattle. Nature. 2001;410:1088–1091. doi: 10.1038/35074088. [DOI] [PubMed] [Google Scholar]
- Van Andel T.H, Runnels C.N. The earliest farmers in Europe. Antiquity. 1995;69:481–500. [Google Scholar]
- Vigne J.-D. The large “true” Mediterranean islands as a model for the Holocene human impact on the European vertebrate fauna? Recent data and new reflections. In: Benecke N, editor. The Holocene history of the European vertebrate fauna. Modern aspects of research. Deutsches Archäologisches Institut, Eurasien-Abteilung; Berlin: 1999. pp. 295–322. Archäologie in Eurasien, 6. [Google Scholar]
- Wendorf F, Schild R. Are the early Holocene cattle in the Eastern Sahara domestic or wild? Evol. Anthropol. 1994;3 [Google Scholar]
- Wood F. The Folio Society; London: 2002. The silk road. [Google Scholar]
- Zilhao J. The spread of agro-pastoral economies across Mediterranean Europe: a view from the Far West. J. Mediterr. Archaeol. 1993;6:5–63. [Google Scholar]