Abstract
The Translocase of the Outer Mitochondrial membrane (TOM complex) is centred on a channel, created by Tom40, serving as the only means of entry for proteins into the mitochondrion. Proteins destined for internal mitochondrial compartments interact subsequently with one of the two distinct protein Translocases of the Inner Mitochondrial membrane (TIM23 and TIM54 complexes) or follow specialized paths into the intermembrane space. To investigate the sorting of precursor proteins to these various sub-mitochondrial compartments, we created a library of tom40 mutants and screened for alleles selectively corrupt in protein sorting. One of the tom40 mutants, tom40–97, carries a single point mutation (W243R) resulting in an ineffective transfer of precursors to the TIM23 complex. There is no defect on transfer of precursors to the TIM54 complex or insertion of proteins into the outer membrane. The Tom40 channel is not a passive pore, but plays an active role in protein sorting for all sub-mitochondrial locations.
Keywords: mitochondrion/protein sorting/TIM complex/TOM complex
Introduction
Mitochondria are essential organelles consisting of two discrete membranes: the mitochondrial outer membrane encloses a soluble intermembrane space and the mitochondrial inner membrane surrounding a protein-dense matrix space. Almost all mitochondrial proteins are encoded on nuclear genes, translated in the cytosol and imported by the mitochondria, to be sorted specifically to one of these sub-mitochondrial compartments (Neupert, 1997; Schatz, 1997; Pfanner and Geissler, 2001).
Proteins destined for all sub-mitochondrial compartments are inserted into or translocated across the outer mitochondrial membrane by the TOM complex (Translocase in the Outer Mitochondrial membrane), and can thereafter cross the inner membrane by various TIM complexes (Translocases in the Inner Mitochondrial membrane). The TOM complex consists of distinct membrane protein subunits: these include various receptor subunits that recognize and bind mitochondrial precursor proteins, and a core translocase composed of the central protein Tom40 together with a set of small, tightly associated accessory subunits (Neupert, 1997; Schatz, 1997; Voos et al., 1999; Gabriel et al., 2001). Electron microscopy, electrophysiology and functional assays all suggest protein translocation takes place through an ∼20 Å channel formed by Tom40 (Vestweber et al., 1989; Hill et al., 1998; Ahting et al., 1999; Kanamori et al., 1997; Schwartz and Matouschek, 1999).
The mechanisms for sorting proteins to their intended sub-mitochondrial location remain poorly understood. From the TOM complex, some inner membrane proteins and all proteins destined for the mitochondrial matrix must be translocated to the TIM23 complex (Neupert, 1997; Voos et al., 1999). The receptor domain of the Tim23 subunit of this complex can bind precursor proteins (Bauer et al., 1996; Komiya et al., 1998; Donzeau et al., 2000; Truscott et al., 2001) and might, in principle, actively remove precursors from a passive TOM complex. A distinct set of precursor proteins, including the abundant carrier family of transporters, are instead recruited from the TOM complex by a small TIM shuttle for delivery to the TIM54 complex (Kübrich et al., 1998; Endres et al., 1999; Koehler et al., 1999). An alternate specialized route, such as that used by apo-cytochrome c, can entail transfer from Tom40 to the intermembrane space with assistance from proteins other than the TIM complexes (Diekert et al., 2001).
How are these transfer steps from the TOM complex mediated, and to what extent does the TOM complex actively sort precursors for interaction with subsequent complexes of the import machinery? In vitro assays show that Tom40 can provide binding ‘sites’ for a translocating precursor and assist movement of a precursor from the cis (or cytosolic) to the trans (intermembrane space) face of the TOM complex (Kanamori et al., 1997; Rapaport et al., 1997, 1998). This vectorial movement is also facilitated by the cis and trans receptor domains of Tom22 (Bolliger et al., 1995; Kanamori et al., 1997; Moczko et al., 1997). Subsequent release of the precursor from the TOM complex, and interaction with the appropriate TIM complex, is required to complete translocation across the outer membrane and enable sorting to one of the three inner compartments of the mitochondria.
To test whether active sorting of precursors has already occurred at the level of Tom40, we generated and searched a library of conditional tom40 alleles. The mutant strains have discrete point mutations in Tom40, and tom40–97 mutants (which carry the mutation W243→R243) are defective for the transfer of precursors to the TIM23 complex. However, there is no defect in protein transfer to the TIM54 complex and protein insertion into the outer membrane proceeds efficiently into tom40–97 mitochondria. Thus, a single point mutation in Tom40 selectively affects the ability of the translocase to discriminate precursors destined for one branch of the sorting scheme for import into mitochondria.
Results
A library of conditional tom40 mutants
PCR-mediated mutagenesis was used to generate a library of tom40 mutants. Twenty of these had clear conditional growth defects and were selected for further study (Table I). Some of the alleles result in strict temperature sensitivity in growth on all media tested at 37°C (tom40–132, tom40–139, tom40–170, tom40–182, tom40–185, tom40–270, tom40–288, tom40–302, tom40–350, tom40–357) or at 14°C (tom40–270). Other alleles render respiratory defects that result in failure of cells to grow on plates with glycerol as the carbon source, either at all temperatures tested (Table I, glycerol) or at extreme temperatures only.
Table I. Twenty novel tom40 mutants showing clear growth defects, with their non-permissive growth condition.
| Allele | Non-permissive growth condition | Suppressed by expression of |
||
|---|---|---|---|---|
| TOM6 | TOM7 | TOM22 | ||
| tom40–47 | glycerol at 37°C | + | + | + |
| tom40–92 | glycerol | + | – | + |
| tom40–97 | glycerol | – | – | – |
| tom40–111 | glycerol at 14°C | + | + | – |
| tom40–132 | 37°C | – | – | – |
| tom40–139 | 37°C | + | – | + |
| tom40–149 | glycerol | – | + | + |
| tom40–170 | 37°C | – | – | – |
| tom40–182 | 37°C | – | + | – |
| tom40–185 | 37°C | + | + | + |
| tom40–270 | 14°C | + | + | + |
| tom40–274 | glycerol | + | + | + |
| tom40–288 | 37°C | – | – | – |
| tom40–302 | 37°C | – | – | – |
| tom40–321 | glycerol | + | + | – |
| tom40–333 | glycerol at 37°C | + | – | – |
| tom40–342 | glycerol | + | + | + |
| tom40–347 | glycerol | + | – | + |
| tom40–350 | 37°C | – | – | + |
| tom40–357 | 37°C | – | + | – |
| KKY3.3 | 37°C | + | – | – |
Each of the mutants was transformed with a plasmid inducing over-expression of Tom6, Tom7 or Tom22 and plated for growth at the non-permissive condition. Those cases in which growth was restored are indicated ‘+’. The previously isolated tom40 mutant KKY3.3 [Kassenbrock et al., 1993; also referred to as KKY3.3 (tom40–3) by Krimmer et al., 2001] was included in these experiments.
In a previous study, Kassenbrock et al. (1993) isolated the gene encoding Tom6 because overexpression of the TOM6 gene suppressed a growth defect in the strain KKY3.3, carrying multiple mutations in the TOM40 gene. The growth defects of many of the alleles we generated could be suppressed by overexpression of genes encoding discrete combinations of the Tom6, Tom7 or Tom22 subunits of the TOM core complex (Table I), suggesting these mutations might effect specific protein–protein interactions with Tom40 (Kassenbrock et al., 1993; Krimmer et al., 2001).
To screen the library for selective defects in protein import, mitochondria were prepared from each of the strains and immunoblot analysis was used to determine whether some mitochondrial proteins might have decreased steady-state levels in the tom40 mutants. The mutant tom40–97 had a clear defect in steady-state levels of several mitochondrial proteins and was further analysed in detail. The tom40–97 mutant has a reduced growth rate on media with non-fermentable carbon sources, and the phenotype cannot be restored by over-expression of TOM6, TOM7 or TOM22.
tom40–97 mutants have reduced steady-state levels of some mitochondrial proteins
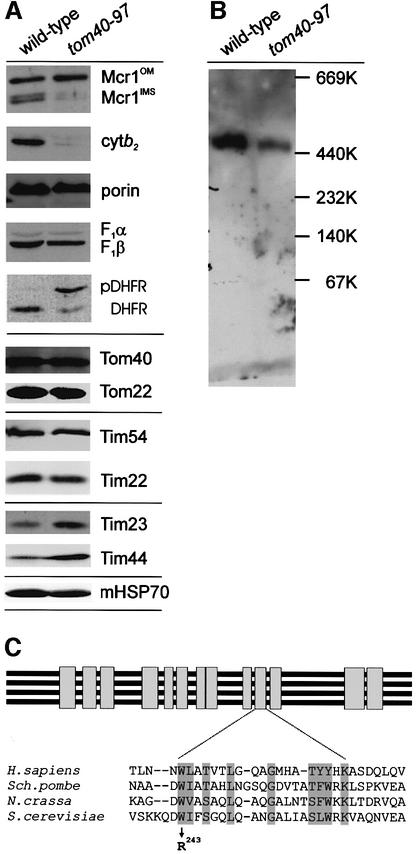
Antibodies to a range of mitochondrial proteins were used in immunoblot analysis. Most proteins, including porin components of the TIM54 complex (Tim54 and Tim22) and the matrix-located HSP70 (mHSP70) were present at wild-type levels in tom40–97 mitochondria (Figure 1A). While components of the TIM23 complex (Tim23 and Tim44) are slightly increased in the mutants, two proteins of the intermembrane space, cytochrome b2 and the processed form of NADH–cytochrome b5 reductase (Mcr1IMS), were reduced in the tom40–97 mutant (Figure 1A). The unprocessed form Mcr1OM in the outer membrane is accumulated in the mutants, where the translocated form Mcr1IMS is barely detectable. There is a very small, but consistently observed, decrease in the steady-state level of the matrix-localized protein F1β, and a clear defect in import of an artificial matrix-targeted version of dihydrofolate reductase (DHFR, Figure 1A). Importantly, the steady-state level of Tom40 itself is not reduced in the tom40–97 mutants, suggesting the mutant form of the protein is stable. Limited proteolysis of isolated mitochondria in the presence of detergent revealed the mutant form of Tom40 to be as stable as wild-type Tom40 (data not shown). Alternatively, when the isolated mitochondria were solubilized in digitonin and committed to blue native PAGE analysis, the detergent-solubilized TOM complex from tom40–97 cells is as soluble in digitonin (data not shown) but observed to be less stable during electrophoresis (Figure 1B).
Fig. 1. The W243R mutation in tom40–97 cells results in changes in the steady-state levels of some mitochondrial proteins. (A) Mitochondria (100 µg protein) isolated from wild-type and tom40–97 mutant cells were analyzed by SDS–PAGE and immunoblotting to determine steady- state levels of the intermembrane space proteins cytochrome b2 (Cyt b2) and the processed form of NADH–cytochrome b5 reductase (Mcr1), the outer membrane protein porin and the matrix-located proteins F1β, F1α and CoxIV–DHFR (George et al., 1998), and for various subunits of the TOM complex (Tom40, Tom22 and Tom20), the TIM23 complex (Tim23 and Tim44) and the TIM54 complex (Tim54 and Tim22). (B) Mitochondria (200 µg protein) were isolated from wild-type or tom40–97 mutant cells, solubilized in digitonin and analysed by blue native PAGE. The migration of high molecular weight markers (Amersham) is indicated and the presence of the 450 kDa TOM holocomplex is shown after immunoblotting with antibodies recognizing Tom40. (C) ClustalW sequence analysis of Tom40 from various organisms (Macasev et al., 2000). Tom40 homologues were identified in plants, fungi, invertebrate and vertebrate animals by iterative BLAST analysis with short segments of NcTom40 and ScTom40. The sequence cluster revealed 13 blocks of homology as represented diagram matically, and expanded in the four species (Homo sapiens, Schizosaccharomyces pombe, N.crassa and S.cerevisiae) shown. The arrow denotes W243 in the sequence of ScTom40, converted to R243 in the tom40–97 mutant.
DNA sequencing revealed a single mutation, converting W243→R243, in the coding sequence from the tom40–97 mutants (see Materials and methods). Comparative analysis of Tom40 from the three species in which the protein has been functionally analysed, Saccharomyces cerevisiae (Baker et al., 1990), Neurospora crassa (Kiebler et al., 1990) and Rattus norvegicus (Suzuki et al., 2000), and a number of highly homologous sequences from plants and invertebrate animals reveals 13 blocks of highly conserved primary structure (represented graphically in Figure 1C). In a membrane-embedded protein like Tom40, these discrete blocks of homology might represent repetitive transmembrane strands or inter-strand loops and the mutation in tom40–97 maps to a tryptophan residue (W243 in the S.cerevisiae sequence) adjacent to one of the blocks of conserved sequence. Our preliminary sequencing in this region reveals only two of the other 20 alleles have neighbouring mutations: N252S in tom40–270 and Q250R in tom40–347. Western blots of cell extracts and protein import assays into mitochondria isolated from tom40–270 and tom40–347 cells revealed only minor defects; however, a severe and select defect was seen in tom40–97.
A select protein import defect can be measured in mitochondria isolated from tom40–97 cells
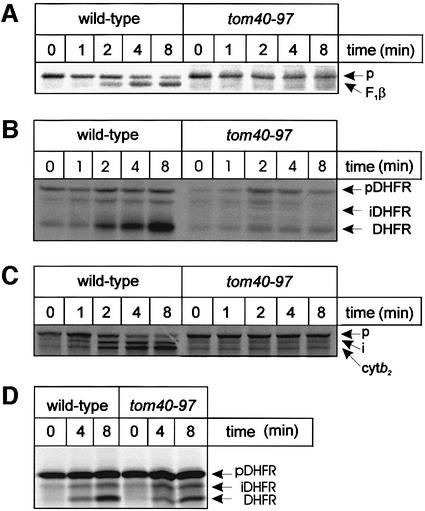
Mitochondria were isolated from wild-type and tom40–97 mutant cells and incubated with proteins destined for the various mitochondrial sub-compartments. The precursor form of F1β is imported into the mutant mitochondria at <10% the wild-type rate (Figure 2A). Similarly, import of Su9–DHFR, measured from the processing of the precursor to its intermediate and mature forms shows a severe defect in the mutants (Figure 2B). Cytochrome b2 is translocated across the outer membrane to the TIM23 complex and subsequently processed for release into the intermembrane space (Glick et al., 1992), and import of cytochrome b2 is similarly diminished in mitochondria isolated from tom40–97 cells (Figure 2C).
Fig. 2. Precursors using the TIM23 complex import slowly into mitochondria from tom40–97 cells. (A) Mitochondria (25 µg of protein) were incubated with 35S-labeled precursor form of F1β for the time indicated (min) and import stopped with CCCP. Samples were treated with 100 µg/ml trypsin and analyzed by SDS–PAGE and fluorography. F1β precursor (p) is cleaved to the mature form. (B) Mitochondria (25 µg of protein) were incubated with 35S-labeled Su9–DHFR for the time indicated (min) and analysed by SDS–PAGE and fluorography. The protein is processed from precursor (p) to and intermediate form (i) before being processed again to reveal a mature protein. (C) Mitochondria (25 µg of protein) were incubated with 35S-labeled precursor form of cytochrome b2 for the time indicated (min) and analyzed by SDS–PAGE and fluorography, to resolve the precursor (p), intermediate form (i) and mature protein. (D) Mitochondria were converted to mitoplasts in hypotonic buffer, and the mitoplasts (25 µg protein) incubated with 35S-labeled Su9–DHFR for the time indicated (min).
To be certain that the TIM23 complex of these tom40–97 mutants is fully functional for later stages of protein translocation, ‘mitoplasts’ were prepared where the outer membrane of mitochondria is ruptured by osmotic shock, allowing precursor proteins direct access to the TIM complexes (Ohba and Schatz, 1987). The kinetics of import of Su9–DHFR into mitoplasts was similar in both wild-type or tom40–97 preparations (Figure 2D). Thus, the transmembrane potential is intact and the TIM23 complex is functional in mitochondria of tom40–97 cells, and the protein translocation defect observed is at the level of the TOM complex.
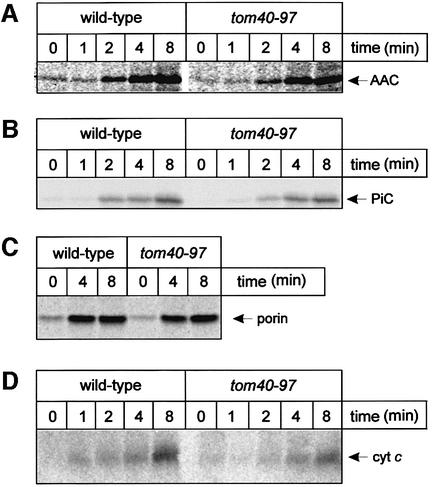
The W243R mutation does not lead to a general block in the translocation pore. Import of the ADP/ATP carrier (AAC) into mitochondria from tom40–97 cells occurs at a rate similar to wild type (Figure 3A). In addition, tom40–97 mitochondria import the phosphate carrier (PiC), another protein of the mitochondrial inner membrane, at a rate close to that of wild-type mitochondria (Figure 3B). Porin also requires Tom40 for insertion into the outer membrane (Krimmer et al., 2001) and porin insertion is equivalent in wild-type or tom40–97 mitochondria (Figure 3C). Apo-cytochrome c requires Tom40 for translocation across the outer membrane (Diekert et al., 2001), and while the W243R mutation does not prevent apo-cytochrome c import (Figure 3D), a decrease of ∼35% is consistently observed in the import rate.
Fig. 3. Import of other precursors into mitochondria from tom40–97 cells is not affected. (A) Mitochondria (25 µg of protein) were incubated with 35S-labeled AAC for the time indicated, then import was stopped with CCCP and samples treated with 100 µg/ml trypsin to remove unimported material. (B) In vitro import of PiC, and (D) import of apo-cytochrome c were similarly assayed. (C) Mitochondria (25 µg of protein) were incubated with 35S-labeled porin for the time indicated, and import stopped with Na2CO3 (pH 11.5). After centrifugation, to collect insoluble material including membrane sheets, and flotation to purify the membranes, porin integrated into the membranes was analysed by SDS–PAGE and fluorography (Krimmer et al., 2001).
tom40–97 and the trans site for protein import into mitochondria
The transmembrane potential across the inner membrane can be dissipated chemically by pretreating mitochondria with CCCP or FCCP, and precursors in transit across the outer membrane are trapped, transiently, in contact with the TOM complex (Bolliger et al., 1995). Figure 4A depicts such a trapping experiment. If the [35S]substrate precursor protein chaperonin 10 (Cpn10) is trapped in this way and the mitochondria then treated with proteinase K, an ∼3 kDa fragment (cpn*) representing the N-terminal presequence can be visualized (the only 35S-labelled methionine residues in cpn10 are Met1 and Met15). The cpn* fragment represents the precursor molecules for which the presequence had entered the TOM complex in wild-type mitochondria to be protected from trypsin. The Cpn10 precursor molecules trapped on mitochondria isolated from tom40–97 cannot be stably trapped in the complex to be protected from trypsin (Figure 4A).
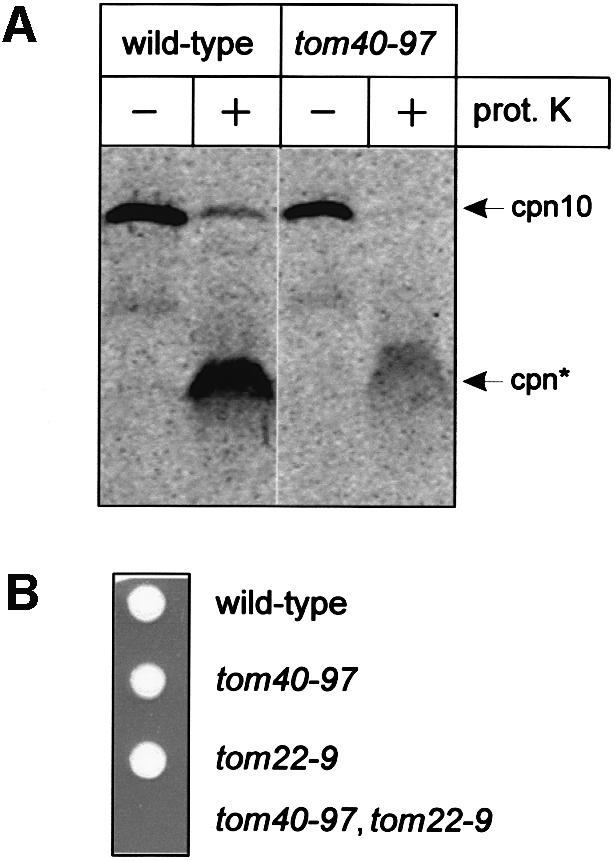
Fig. 4. Mutant tom40–97 mitochondria cannot trap a precursor and require the function of the Tom22 intermembrane space domain. (A) Mitochondria (25 µg protein) isolated from wild-type or tom40–97 mutant cells were pre-incubated with CCCP and then 35S-labeled Cpn10 was added for 15 min. Where indicated (+) samples were treated with 100 µg/ml proteinase K, and then mitochondria were isolated, trapped and the precursor analyzed by SDS–PAGE and fluorography. The position of intact (Cpn10) and the fragment of Cpn10 protected from protease (cpn*) are indicated. (B) A heterozygous diploid strain was induced to sporulated and the meiotic progeny dissected onto plates of rich medium. The identity of the viable colonies was established by marker analysis.
This phenotype is similar to that seen in mitochondria isolated from tom22–9 mutants, where the intermembrane space domain of Tom22 has been deleted (Bolliger et al., 1995). Accordingly, a cross was set up between haploid cells carrying the tom40–97 mutation and haploid tom22–9 mutants. The resulting heterozygous diploid (TOM22/tom22–9, TOM40/tom40–97) yeast strain was sporulated and the meiotic haploid progeny dissected onto rich growth media. While the wild-type daughter cells and each of progeny carrying single mutations germinated and grew with the predicted phenotypes, the cell inheriting the double combination tom40–97, tom22–9 is inviable (Figure 4B).
Discussion
Tom40 is a sorting station for mitochondrial protein import
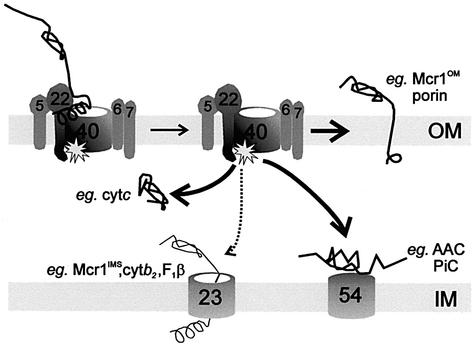
The final events of protein import into mitochondria are mediated by interactions with internal components of the import machinery in processes using ATP hydrolysis and the transmembrane potential across the inner membrane (Pfanner and Geissler, 2001; Neupert and Brunner, 2002), while translocation through the TOM complex does not require ATP hydrolysis or a transmembrane potential (Neupert, 1997; Schatz, 1997; Matouschek et al., 2000). A fundamental question has been whether or not the TOM complex functions only as a passive pore for protein translocation. Our data suggest that Tom40 functions as a sophisticated sorting station, distinguishing substrates for insertion into the outer membrane, translocation into the intermembrane space, translocation and transfer to the TIM54 complex or translocation and transfer to the TIM23 complex (Figure 5). The point mutation W243R discretely blocks one of these pathways, but not the other three.
Fig. 5. Tom40 functions as a protein sorting station. Precursor proteins bound to the cytosolic face of the TOM complex can be inserted into the outer membrane or translocated through Tom40 for transfer to the TIM54 complex, or to the TIM23 complex. A deletion of the intermembrane space domain of Tom22 or conversion of W243R in Tom40 effects transfer of precursors to the TIM23 complex. A combination of these two mutations is lethal. The steady-state levels of Mcr1OM accumulate at the expense of Mcr1IMS as a result of the W243R mutation.
In addition to the in vitro data, the accumulated steady-state levels of the ∼35 kDa Mcr1OM in tom40–97 cells demonstrate the in vivo consequences of failed translocation. In wild-type cells, a proportion of Mcr1 is normally inserted into the outer membrane, while a proportion is translocated through the TOM complex and to the TIM23 complex. After reaching the TIM23 complex, Mcr1 is processed by the Imp protease to release ∼32 kDa Mcr1IMS into the intermembrane space (Hahne et al., 1994; Haucke et al., 1997). Essentially no processed Mcr1IMS is found in tom40–97 cells, with Mcr1OM accumulating instead.
AAC is an example of a precursor sorted from Tom40 to the TIM54 complex. By manipulating in vitro assay conditions, the normal import of AAC can be interrupted, such that AAC is accumulated in the TOM complex almost completely protected from exogenously added protease, yet exposed to the intermembrane space (Ryan et al., 1999). To achieve transfer from this position to the TIM54 complex, AAC does not require the presence of the Tom22 trans domain (Kübrich et al., 1998), nor is this transfer of AAC perturbed by the W243R mutation. Taken together, functional analysis of the tom40–97 mutant suggests the W243R mutation selectively interferes with the contribution Tom40 normally makes to a discrete precursor-binding site for protein transfer to the TIM23 complex.
Consequences of the W243R mutation
The TOM complex is asymmetrical with respect to precursor protein binding sites. On the cytosolic surface, cis receptor domains contributed by Tom20, Tom22 and Tom70 cooperate to bind precursor proteins (Söllner et al., 1992; Kiebler et al., 1993; Lithgow et al., 1994, Bolliger et al., 1995; Mayer et al., 1995; Kanamori et al., 1997; Brix et al., 1998; Kurz et al., 1999; Wiedemann et al., 2001). Once within the channel, targeting sequences gain access to what has been operationally defined as the trans site of the TOM complex. There is strong evidence from cross-linking studies and analysis of deletion mutants that both the C-terminal domain of Tom22 oriented to the intermembrane space, and a surface or domain of Tom40 contribute to this trans site (Bolliger et al., 1995; Kanamori et al., 1997; Moczko et al., 1997; Rapaport et al., 1997, 1998).
As yet, we have no structural details for Tom40 for the final interpretation of how the W243R mutation effects binding and transfer of precursors destined to the TIM23 complex. However, structural and theoretical data from other membrane proteins suggest tryptophan residues are preferentially found as the interfacial residue at the lipid–aqueous interface in α-helix or β-strand transmembrane segments (Reithmeier, 1995; Yuen et al., 2000). Mutational scanning analysis has shown tryptophan can determine the position of a transmembrane segment within the phospholipid plane, in effect pushing the segment into the bilayer in order to register the tryptophan at the lipid–aqueous interface (Braun and von Heijne, 1999; de Planque et al., 2001). Arginine residues can also be readily accommodated towards the ends of transmembrane segments, where the side-chain can be oriented snorkel-like to expose the terminal amine group to the aqueous environment (Monne et al., 1998).
Given that there are no effects on AAC import or porin insertion into tom40–97 mitochondria, no effect on the stability of Tom40(W243R) as judged by protease-susceptibility and only a partial decrease in stability of the 450 kDa holocomplex after detergent solubilization of tom40–97 mitochondria, we suspect that the W243R mutation does not grossly effect the structure of Tom40. Instead, we suggest R243 might directly disturb a precursor protein binding surface and we propose the Tom40 subunit of the translocase can discriminate the intended destinations for the wide array of precursors imported into mitochondria. The mutant alleles of tom40 described here provide a means to map various sites functionally important within the TOM complex, both for precursor passage and the docking of the accessory subunits Tom6, Tom7 and Tom22.
Interestingly, the steady-state levels of subunits of the TIM23 complex, Tim23 and Tim44, are always increased in tom40–97 mutants and yet not in other tom40 alleles we have analysed. This regulation of the relative numbers of translocase complexes is presumably an attempt by the mutant cells to compensate for the decrease in protein traffic to the TIM23 complex. The mechanism for such regulation, and the extent to which the TOM and TIM23 complexes physically communicate with each other, remains to be studied.
Materials and methods
PCR mutagenesis on the TOM40 gene
Yeast tom40 mutants were constructed using low-fidelity PCR to mutate a fragment of DNA corresponding to the TOM40 gene followed by recombination of the mutant allele onto a plasmid in vivo (Muhlrad et al., 1992; Staples and Dieckmann, 1993; Koehler et al., 1998). A plasmid encoding the TOM40 gene (including 5′ and 3′ flanking regions) in pBluescript was a kind gift from Kevin Baker. The fragment was amplified by PCR in independent reactions containing 0.6–1.0 mM MgCl2 and 0.1–0.2 mM MnCl2, using primers complementary to regions ∼200 nucleotides away from either side of the multiple cloning region (primer sequences CK3145: 5′-GGCTGCGCAACTGTTG-3′ and CK3143: 5′-CTGAGCGCAACGCAAT-3′). The amplified fragments were combined and co-transformed with linearized centromeric vector pRS315 into the yeast strain YKB14–1a [tom40::HIS4, his4-519, leu2-3 112, Δura3, ade2, YEplac42R (TOM40::URA3)]. Leu+ transformants were selected at 25°C and screened for growth at 14, 25 and 37°C on minimal glucose media containing 5-fluoroorotic acid and appropriate growth supplements. A collection of ∼100 mutants was screened for growth defects. Complete sequencing of three independent clones of the pRS315-tom40–97 plasmid determined the mutation coding for a single amino acid substitution (W243R).
Protein import
Mitochondria were isolated according to published procedures (Daum et al., 1982), but with 18.5% (w/v) Nycondenz used at the bottom of the density gradient. Mitoplasts were prepared as described by Glick et al. (1992). Mitochondria (25 µg protein) isolated from either wild-type or mutant strains were resuspended in 100 µl of import buffer (0.6 M sorbitol, 50 mM HEPES, 2 mM potassium phosphate, 25 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, 1 mM DTT). Precursor proteins were translated in nuclease-treated rabbit reticulocyte lysate (Promega) containing [35S]methionine. 35S-labelled precursor proteins were added to mitochondria in import buffer and incubated at 25°C for the appropriate time (Glick et al., 1992). Where indicated, import was terminated by addition of CCCP to a final concentration 100 µM and trypsin or proteinase K added to remove protein that failed to be imported.
Miscellaneous
Strains of S.cerevisiae were grown at 30°C on YPAD [2% (w/v) glucose, 1% (w/v) yeast extract, 2% (w/v) peptone supplemented with adenine sulfate] grown until late log phase and harvested by centrifugation. Samples of mitochondrial protein (100 µg) were separated by Tris–glycine SDS–PAGE, Tris–tricine SDS–PAGE or blue native PAGE, and western blots were carried out according to published methods (Beilharz et al., 1998; Ryan et al., 1999; Sambrook and Russell, 2001).
Acknowledgments
Acknowledgements
We thank Carla Koehler, Tina Junne-Bieri, Klaus Pfanner, Kevin Baker and Jeff Schatz for plasmids and antisera, Michael Douglas for the KKY-3 mutant, Amy Dalgleish for sequence data on the tom40–270 and tom40–347 mutants and Lena Burri for critical suggestions on the manuscript. This work was supported by a grant from the Australian Research Council (to T.L.) and an Australian Postgraduate Research Award (to K.G.).
References
- Ahting U., Thun,C., Hegerl,R., Typke,D., Nargang,F.E., Neupert,W. and Nussberger,S. (1999) The TOM core complex: the general protein import pore of the outer membrane of mitochondria. J. Cell Biol., 147, 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K.P., Schaniel,A., Vestweber,D. and Schatz,G. (1990) A yeast mitochondrial outer membrane protein essential for protein import and cell viability. Nature, 348, 605–609. [DOI] [PubMed] [Google Scholar]
- Bauer M.F., Sirrenberg,C., Neupert,W. and Brunner,M. (1996) Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell, 87, 33–41. [DOI] [PubMed] [Google Scholar]
- Beilharz T., Suzuki,C.K. and Lithgow,T. (1998) A toxic fusion protein accumulating between the mitochondrial membranes inhibits protein assembly in vivo. J. Biol. Chem., 273, 35268–35272. [DOI] [PubMed] [Google Scholar]
- Bolliger L., Junne,T., Schatz,G. and Lithgow,T. (1995) Acidic receptor domains on both sides of the outer membrane mediate translocation of precursor proteins into yeast mitochondria. EMBO J., 14, 6318–6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun P. and von Heijne,G. (1999) The aromatic residues Trp and Phe have different effects on the positioning of a transmembrane helix in the microsomal membrane. Biochemistry, 38, 9778–9782. [DOI] [PubMed] [Google Scholar]
- Brix J., Rudiger,S., Bukau,B., Schneider-Mergener,J. and Pfanner,N. (1998) Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22 and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J. Biol. Chem., 274, 16522–16530. [DOI] [PubMed] [Google Scholar]
- Daum G., Gasser,S.M. and Schatz,G. (1982) Import of proteins into mitochondria. Energy-dependent, two-step processing of the intermembrane space enzyme cytochrome b2 by isolated yeast mitochondria. J. Biol. Chem., 257, 13075–13080. [PubMed] [Google Scholar]
- de Planque M.R., Goormaghtigh,E., Greathouse,D.V., Koeppe,R.E., Kruijtzer,J.A., Liskamp,R.M., deKruijff,B. and Killian,J.A. (2001) Sensitivity of single membrane-spanning α-helical peptides to hydrophobic mismatch with a lipid bilayer: effects on backbone structure, orientation and extent of membrane incorporation. Biochemistry, 40, 5000–5010. [DOI] [PubMed] [Google Scholar]
- Diekert K., de Kroon,A.I., Ahting,U., Niggemeyer,B., Neupert,W., deKruijff,B. and Lill,R. (2001) Apocytochrome c requires the TOM complex for translocation across the mitochondrial outer membrane. EMBO J., 20, 5626–5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donzeau M., Kaldi,K., Adam,A., Paschen,S., Wanner,G., Guiard,B., Bauer,M.F., Neupert,W. and Brunner,M. (2000) Tim23 links the inner and outer mitochondrial membranes. Cell, 101, 401–412. [DOI] [PubMed] [Google Scholar]
- Endres M., Neupert,W. and Brunner,M. (1999) Transport of the ADP/ATP carrier of mitochondria from the TOM complex to the TIM22.54 complex. EMBO J., 18, 3214–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel K., Buchanan,S.K. and Lithgow,T. (2001) The α and the β: protein translocation across mitochondrial and plastid outer membranes. Trends Biochem. Sci., 26, 36–40. [DOI] [PubMed] [Google Scholar]
- George R., Beddoe,T., Landl,K. and Lithgow,T. (1998) The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proc. Natl Acad. Sci. USA, 95, 2296–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B.S., Wachter,C., Reid,G.A. and Schatz,G. (1992) Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell, 69, 809–822. [DOI] [PubMed] [Google Scholar]
- Hahne K., Haucke,V., Ramage,L. and Schatz,G. (1994) Incomplete arrest in the outer membrane sorts NADH–cytochrome b5 reductase to two different submitochondrial compartments. Cell, 79, 829–839. [DOI] [PubMed] [Google Scholar]
- Haucke V., Ocana,C.S., Hönlinger,A., Tokatlidis,K., Pfanner,N. and Schatz,G. (1997) Analysis of the sorting signals directing NADH–cytochrome b5 reductase to two locations within yeast mitochondria. Mol. Cell. Biol., 17, 4024–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K., Model,K., Ryan,M.T., Dietmeier,K., Martin,F., Wagner,R. and Pfanner,N. (1998) Tom40 forms the hydrophilic channel of the mitochondrial import pore for proteins. Nature, 395, 516–521. [DOI] [PubMed] [Google Scholar]
- Kanamori T., Nishikawa,S., Shin,I., Schultz,P.G. and Endo,T. (1997) Probing the environment along the protein import pathways in yeast mitochondria by site-specific photocrosslinking. Proc. Natl Acad. Sci. USA, 94, 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassenbrock C.K., Cao,W. and Douglas,M.G. (1993) Genetic and biochemical characterization of ISP6, a small mitochondrial outer membrane protein associated with the protein translocation complex. EMBO J., 12, 3023–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler M., Pfaller,R., Söllner,T., Griffiths,G., Horstmann,H., Pfanner,N. and Neupert,W. (1990) Identification of a mitochondrial receptor complex required for recognition and membrane insertion of precursor proteins. Nature, 348, 610–616. [DOI] [PubMed] [Google Scholar]
- Kiebler M., Keil,P., Schneider,H., van der Klei,I.J., Pfanner,N. and Neupert,W. (1993) The mitochondrial receptor complex: a central role of MOM22 in mediating preprotein transfer from receptors to the general insertion pore. Cell, 74, 483–492. [DOI] [PubMed] [Google Scholar]
- Koehler C.M., Jarosch,E., Tokatlidis,K., Schmid,K., Schweyen,R.J. and Schatz,G. (1998) Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science, 279, 369–373. [DOI] [PubMed] [Google Scholar]
- Koehler C.M., Merchant,S. and Schatz,G. (1999) How membrane proteins travel across the mitochondrial intermembrane space. Trends Biochem. Sci., 24, 428–432. [DOI] [PubMed] [Google Scholar]
- Komiya T., Rospert,S., Koehler,C., Looser,R., Schatz,G. and Mihara,K. (1998) Interaction of mitochondrial targeting signals with acidic receptor domains along the protein import pathway: evidence for the ‘acid chain’ hypothesis. EMBO J., 17, 3886–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimmer T. et al. (2001) Biogenesis of porin of the outer mitochondrial membrane involves an import pathway via receptors and the general import pore of the TOM complex. J. Cell Biol., 152, 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kübrich M., Rassow,J., Voos,W., Pfanner,N. and Hönlinger,A. (1998) The import route of ADP/ATP carrier into mitochondria separates from the general import pathway of cleavable preproteins at the trans side of the outer membrane. J. Biol. Chem., 273, 16374–16381. [DOI] [PubMed] [Google Scholar]
- Kurz M., Martin,H., Rassow,J., Pfanner,N. and Ryan,M.T. (1999) Biogenesis of Tim proteins of the mitochondrial carrier import pathway: differential targeting mechanisms and crossing over with the main import pathway. Mol. Biol. Cell, 10, 2461–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow T., Junne,T., Suda,K., Gratzer,S. and Schatz,G. (1994) The mitochondrial outer membrane protein Mas22p is essential for protein import and viability of yeast. Proc. Natl Acad. Sci. USA, 91, 11973–11977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macasev D., Newbigin,E., Whelan,J. and Lithgow,T. (2000) How do plant mitochondria avoid importing chloroplast proteins? Components of the import apparatus Tom20 and Tom22 from Arabidopsis differ from their fungal counterparts. Plant Physiol., 123, 811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., Nargang,F.E., Neupert,W. and Lill,R. (1995) MOM22 is a receptor for mitochondrial targeting sequences and cooperates with MOM19. EMBO J., 14, 4204–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouschek A., Pfanner,N. and Voos,W. (2000) Protein unfolding by mitochondria. The Hsp70 import motor. EMBO rep., 1, 404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczko M., Bomer,U., Kübrich,M., Zufall,N., Hönlinger,A. and Pfanner,N. (1997) The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol. Cell. Biol., 17, 6574–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monne M., Nilsson,I., Johansson,M., Elmhed,N. and von Heijne,G. (1998) Positively and negatively charged residues have different effects on the position in the membrane of a model transmembrane helix. J. Mol. Biol., 284, 1177–1183. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Hunter,R. and Parker,R. (1992) A rapid method for localized mutagenesis of yeast genes. Yeast, 8, 79–82. [DOI] [PubMed] [Google Scholar]
- Neupert W. (1997) Protein import into mitochondria. Annu. Rev. Biochem., 66, 863–917. [DOI] [PubMed] [Google Scholar]
- Neupert W. and Brunner,M. (2002) The protein import motor of mitochondria. Nat. Rev. Mol. Cell Biol., 3, 555–565. [DOI] [PubMed] [Google Scholar]
- Ohba M. and Schatz,G. (1987) Disruption of the outer membrane restores protein import to trypsin-treated yeast mitochondria. EMBO J., 6, 2117–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N. and Geissler,A. (2001) Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol., 2, 339–349. [DOI] [PubMed] [Google Scholar]
- Rapaport D., Neupert,W. and Lill,R. (1997) Mitochondrial protein import. Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence. J. Biol. Chem., 272, 18725–18731. [DOI] [PubMed] [Google Scholar]
- Rapaport D., Mayer,A., Neupert,W. and Lill,R. (1998) cis and trans sites of the TOM complex of mitochondria in unfolding and initial translocation of preproteins. J. Biol. Chem., 273, 8806–8813. [DOI] [PubMed] [Google Scholar]
- Reithmeier R.A. (1995) Characterization and modeling of membrane proteins using sequence analysis. Curr. Opin. Struct. Biol., 5, 491–500. [DOI] [PubMed] [Google Scholar]
- Ryan M.T., Muller,H. and Pfanner,N. (1999) Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem., 274, 20619–20627. [DOI] [PubMed] [Google Scholar]
- Sambrook J. and Russell,D.W. (2001) Molecular Cloning: a Laboratory Manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schatz G. (1997) Just follow the acid chain. Nature, 388, 121–122. [DOI] [PubMed] [Google Scholar]
- Schwartz M.P. and Matouschek,A. (1999) The dimensions of the protein import channels in the outer and inner membranes. Proc. Natl Acad. Sci. USA, 96, 13086–13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner T., Rassow,J., Wiedemann,M., Schlossmann,J., Keil,P., Neupert,W. and Pfanner,N. (1992) Mapping of the protein import machinery in the mitochondrial outer membrane by crosslinking of translocation intermediates. Nature, 355, 84–87. [DOI] [PubMed] [Google Scholar]
- Staples R.R. and Dieckmann,C.L. (1993) Generation of temperature-sensitive cbp1 strains of S.cerevisiae by PCR mutagenesis and in vivo recombination: characteristics of the mutant strains imply that CBP1 is involved in stabilization and processing of cytochrome b pre-mRNA. Genetics, 135, 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Okazawa,Y., Komiya,T., Saeki,K., Mekada,E., Kitada,S., Ito,A. and Mihara,K. (2000) Characterization of rat TOM40, a central component of the preprotein translocase of the mitochondrial outer membrane. J. Biol. Chem., 275, 37930–37936. [DOI] [PubMed] [Google Scholar]
- Truscott K.N., Kovermann,P., Geissler,A., Merlin,A., Meijer,M., Driessen,A.J., Rassow,J., Pfanner,N. and Wagner,R. (2001) A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol., 8, 1074–1082. [DOI] [PubMed] [Google Scholar]
- Vestweber D., Brunner,J., Baker,A. and Schatz,G. (1989) A 42K outer membrane protein is a component of the yeast mitochondrial protein import site. Nature, 341, 205–209. [DOI] [PubMed] [Google Scholar]
- Voos W., Martin,H., Krimmer,T. and Pfanner,N. (1999) Mechanisms of protein translocation into mitochondria. Biochim. Biophys. Acta, 1422, 235–254. [DOI] [PubMed] [Google Scholar]
- Wiedemann N., Pfanner,N. and Ryan,M.T. (2001) The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J., 20, 951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen C.T., Davidson,A.R. and Deber,C.M. (2000) Role of aromatic residues at the lipid–water interface in micelle-bound bacteriophage M13 major coat protein. Biochemistry, 39, 16155–16162. [DOI] [PubMed] [Google Scholar]