Abstract
A recent observation that female Antarctic fur seals foster pups born to related females raises the fascinating possibility that kin selection may promote altruistic behaviour even on a crowded breeding beach, where individual interactions are frequent and complex. However, the use of genetic markers to identify small numbers of unusually highly related individuals is fraught with difficulty due to the likely presence of genotyping errors and related problems. Consequently, we examined an enlarged dataset where errors had been reduced to an absolute minimum by a combination of close scrutiny and repeat genotyping. We find no support for the idea that females preferentially suckle pups born to female relatives. Instead, the previously reported pattern can be explained by a combination of genotyping errors and de novo mutations. Our study emphasizes the need for caution when interpreting rare events that occur at a rate approaching that expected for normal genotyping errors.
Keywords: relatedness, parental exclusion, microsatellite, genotyping error, pinniped, non-filial nursing
1. Introduction
In general, animals are expected to behave selfishly, promoting their own reproduction at the expense of others. The primary exception to this rule is when individuals are related allowing kin selection to operate (Hamilton 1964). Hamilton's theory provides an appealing explanation for apparently altruistic behaviours but, while there are numerous examples in insects (e.g. Queller & Strassmann 1998), examples in mammals have been mostly confined to communally breeding species where relatedness within groups is high (Griffin & West 2003). An intriguing exception, however, comes from a recent study of fostering behaviour in a pinniped, the Antarctic fur seal Arctocephalus gazella. Here, it was reported that in dense breeding colonies where females sometimes suckle pups that are not their own, there is a tendency to favour pups born to female relatives (Gemmell 2003).
The evolution of mammalian fostering behaviour, where females provide parental care for non-filial young, is particularly challenging to explain because lactation imposes physiological stress on mothers that may reduce their survival and future reproductive success (Konig et al. 1988; Clutton-Brock et al. 1989; Iverson et al. 1993). Furthermore, even if milk production did not entail any fitness penalty, nursing directed towards unrelated offspring should be selected against because it would only serve to increase the fitness of competing individuals in the population. In pinnipeds, such behaviour could be considered especially unlikely because females invest unusually heavily in milk production. Despite this, fostering is widespread, reaching frequencies of up to 90% in some species (Stirling 1975; Riedman & Leboeuf 1982; Lunn 1992; Boness et al. 1998; Perry et al. 1998; Schaeff et al. 1999; Childerhouse & Gales 2001).
There are two principal explanations for why allo-suckling is widespread in pinnipeds. First, in densely populated breeding colonies females may find it difficult to recognize their own offspring and vice versa (Riedman & Leboeuf 1982; Boness 1990; Job et al. 1995; McCulloch et al. 1999). Although frequent mistakes may be costly, occasional errors may be preferable to the risk of under nourishing their own pups. At the same time, unless females are unduly aggressive when approached by an unrelated pup, pups can only benefit by soliciting from any nearby female. Second, there is evidence that some species form kin clusters in which females return to breed close to where they are born (Pomeroy et al. 2000; Pomeroy et al. 2001). Consequently, females that allo-suckle pups born to neighbours may benefit from kin selection, thereby potentially defraying some of the costs.
Using molecular genetic markers it is now quite straightforward to measure relatedness, using relative degrees of allele sharing to estimate the probability that two alleles are identical by descent. Consequently, it is possible to test whether kin selection plays an important role in the suckling fidelity of wild pinnipeds. In previous studies using DNA fingerprinting, no evidence was found that either grey or harbour seal females preferentially suckled pups born to related females (Perry et al. 1998; Schaeff et al. 1999). However, a recent microsatellite study of 183 Antarctic fur seal mother–offspring pairs at Bird Island, South Georgia (Gemmell 2003) reported significantly higher relatedness values among mother–non-filial pup pairs than observed for the total population. On the one hand, this result is plausible because female fur seals show strong site fidelity (Lunn & Boyd 1991) and hence, there is the possibility of strong kin clustering. On the other hand, female fur seals show an impressive ability to recognize their own offspring through vocalizations, suggesting that cases of mistaken identity will be rare (Insley 2000; Charrier et al. 2001; 2003).
An alternative explanation for why non-filial pups might appear closely related to the females who suckle them lies with genotyping errors. Although microsatellites are widely used, they are prone to a wide range of errors that can create mismatches between a parent and its offspring. These include allele non-amplification resulting from primer binding site mutation (‘null alleles’ Callen et al. 1993; Pemberton et al. 1995; Dakin & Avise 2004), stochastic failure of alleles to amplify (‘allelic dropout’ Navidi et al. 1992; Walsh et al. 1992; Gerloff et al. 1995; Taberlet et al. 1996; Gagneux et al. 1997), generation of amplification products that can be misinterpreted as true alleles (Taberlet et al. 1996; Goossens et al. 1998; Bradley & Vigilant 2002) and incorrect scoring of allele banding patterns (Litt et al. 1993; Ginot et al. 1996; Harker 2001; Johansson et al. 2003; Hoffman & Amos 2005). In addition, large datasets inevitably include some level of data-entry and other clerical errors (Hoffman & Amos 2005). Such errors are almost impossible to eliminate yet even very low rates can lead to significant numbers of mismatches between true parent–offspring pairs (Marshall et al. 1998; Dakin & Avise 2004; Hoffman & Amos 2005), thereby generating ‘non-filial’ pups who appear closely related to the female they were suckling.
In view of the large potential impact of even very low error rates, we use a greatly enlarged version of Gemmell's (2003) dataset and employ a variety of approaches to drive error rates as low as possible. When this is done, we find little or no support for the idea that females preferentially suckle pups born to female relatives.
2. Material and methods
(a) Study site and field methods
This study was conducted at a small fur seal breeding beach at Bird Island, South Georgia (54 °00′ S, 38 °02′ W) and is based on a dataset spanning seven consecutive pupping seasons (Hoffman et al. 2003; Hoffman & Amos 2005) that was subsequently expanded to incorporate an additional season (Hoffman et al. 2004). Tissue samples and observational data were collected during the austral summers of 1994/1995–2001/2002 (hereafter, breeding seasons are referred to by the year in which they began). The study beach was separated from adjacent breeding sites by a cliff on the east side, open sea on the west and rocky ridges to the north and south. It covered an area of 440 m2 at high tide (Lunn & Boyd 1993) and on average, 649 pups were born there annually. An elevated scaffold walkway (Doidge et al. 1984) provided access to all parts of the beach while minimizing disturbance to animals.
As part of long-term ongoing demographic study of fur seals, approximately 550 adult females were tagged using plastic cattle ear tags (Dalton Supplies, Henley-on-Thames, UK) placed in the trailing edge of the foreflipper. These females were then captured opportunistically at the end of the breeding season (December to March) and tissue-sampled from the interdigital margin of the foreflipper using piglet ear notching pliers (Majluf & Goebel 1992). Throughout each season, twice-daily surveys were made of all pups born on the beach (newly born pups had wet and shiny fur indicating that they had been born only a few hours previously). Those pups that appeared to have been born to tagged females, either because of close proximity or because they were observed suckling, were captured and tissue sampled in the same way. Additionally, samples were collected from the majority of adult males holding territories on the beach (n=464 individuals, following removal of duplicate genotypes that represented resampled individuals). All sampling equipment was sterilized using ethanol between uses. Samples were stored individually in the preservative buffer 20% dimethyl sulphoxide saturated with salt (Amos & Hoelzel 1991) and stored at −20 °C.
(b) Microsatellite genotyping
Total genomic DNA was extracted using an adapted Chelex 100 protocol (Walsh et al. 1991). All samples were then genotyped using a panel of nine dinucleotide-repeat microsatellite loci, previously isolated from a variety of pinniped species. These loci possessed a mean of 12 alleles (range=5–18 alleles per locus), had an average expected heterozygosity of 0.796 (range=0.450–0.921) and did not deviate significantly from Hardy–Weinberg equilibrium (Hoffman et al. 2003). Polymerase chain reaction (PCR) products incorporating [α32P]-dCTP were resolved by electrophoresis on standard 6% acrylamide sequencing gels and detected by autoradiography. To drive the genotyping error rate as low as practicably possible, any reactions yielding uncertain genotypes (e.g. with faint or unclear bands) were then repeated. Data were also incorporated from a previous study (animals sampled during 1994 and 1995, genotyped at loci Aa4, Hg1.3, Hg8.10, M11a, PvcA and PvcE by Gemmell et al. 2001). To compensate for differences between observers, autoradiographs for all previously genotyped samples were re-scored and whenever an uncertain score was encountered, the sample was genotyped again at that locus. Finally, the overall genotyping error rate was assessed using a variety of approaches, including independently re-genotyping approximately 10% of all samples. Resulting estimates of the error rate were very low, ranging between 0.0013 and 0.0074 per single-locus PCR (Hoffman & Amos 2005). For details of genotyping methods, locus polymorphism characteristics and error-checking procedures, see Hoffman & Amos (2005).
(c) Data analyses
Putative mother–offspring pairs (n=832) were checked for mismatches (i.e. genotypes that do not share a common allele) using the program Newpat XL (Worthington Wilmer et al. 1999). Genetically matching pups were then classified as being filial, while those mismatching at one or more loci were classified as non-filial. Next, we derived distributions of pairwise relatedness values for: (i) 750 females with filial pups, (ii) 82 females with non-filial pups and (iii) 832 pairs comprising randomly selected individuals from the population. An additional distribution comprising pairs of randomly selected mothers and pups was also generated, but since this was essentially identical to the former randomized distribution, it was not used for subsequent analyses. To enable us to compare our results with those of Gemmell (2003), we estimated pairwise relatedness using Queller and Goodnight's statistic, r (Queller & Goodnight 1989), which has the desirable property of being approximately normally distributed about a mean value of 0.5 for first-order relatives and zero for unrelated individuals. Calculations were implemented using population allele frequencies within the program Kinship version 1.3.1 (Goodnight & Queller 1999). To test whether our analyses were sensitive to the particular relatedness estimator used, we also calculated two other commonly used measures, Lynch and Ritland's (1999) and Wang's (2002) statistics, using SPAGeDi version 1.1 (Hardy & Vekemans 2002). However, since the overall patterns obtained did not differ significantly among estimators (data not shown), only analyses using Queller and Goodnight's r statistic are presented. The resulting distributions of r-values all deviated significantly from normality (Shapiro Wilk tests, filial pairs W=0.990, n=750, p <0.0001; non-filial pairs W=0.969, n=82, p=0.045; random pairs W=0.983, n=832, p <0.0001). Consequently, data were conservatively analysed using non-parametric tests, although equivalent unpaired t-tests yielded virtually identical results (data not shown). To compensate for non-normality, we also calculated 95% confidence intervals about the mean of each distribution using non-parametric bootstrapping with 10 000 replicates (Efron & Tibshirani 1993). These calculations were implemented within Ihaka & Gentleman (1996) using code written by P. Harrison.
Finally, we investigated the extent to which genotyping errors present in the dataset influenced our analyses. First, we examined autoradiographs for all mother–offspring pairs that mismatched at one locus and identified a small number of genotyping errors (n=15; Hoffman et al. 2003; Hoffman & Amos 2005). We then extended this approach to include all pairs mismatching at two loci. Finally, as an additional check, DNA was re-extracted from these samples using a modified phenol : chloroform : isoamyl alcohol protocol (Sambrook et al. 1989) and re-genotyped at all nine loci. Mother–offspring pairs that mismatched due to genotyping errors were then, together with genetically matching pups, classified as being filial. Relatedness values were recalculated and data analyses were repeated. In addition, post hoc calculations of statistical power (1−β) were undertaken by estimating the type II error rate β associated with our analyses using the program G*Power (Erdfelder et al. 1996) for a medium effect size (d=0.5) and α=0.05. Although our data were not normally distributed, because the difference between p-values for parametric and non-parametric tests was negligible, power was calculated based on the convenient t-test equivalent.
3. Results
A total of 832 putative mother–pup pairs were genotyped at nine highly polymorphic microsatellite loci and checked for genotypic mismatches. Of these, 750 pairs matched at all nine loci, suggesting that these pups were filial. The remaining pups (n=82, 9.9%) mismatched their putative mothers at between one and seven loci (mean=3.4 loci) and were consequently classified as putatively non-filial. Following the approach of Gemmell (2003), we tested the hypothesis that female-non-filial pup pairs were more related than random pairs of individuals from the population by calculating relatedness statistics for 750 mother–filial pup pairs, 82 female–non-filial pup pairs and 832 random pairs of individuals. The resulting distributions of Queller and Goodnight's r statistic (Queller & Goodnight 1989) are given in figure 1a. As expected, filial mother–pup pairs have r-values distributed symmetrically around approximately 0.5 and random pairs of individuals have r-values distributed around zero (mean r=0.4898 and −0.0029, respectively; see table 1 for 95% confidence intervals). In contrast, r-values for non-filial pairs appear right-skewed with the majority distributed about zero but with a secondary hump at around 0.4, as found by Gemmell (2003). Consequently, non-filial pairs appear to have relatedness values significantly higher than those of unrelated individuals belonging to the population (Mann–Whitney U test, U=44 711, n1=832, n2=82, p < 0.0001).
Figure 1.
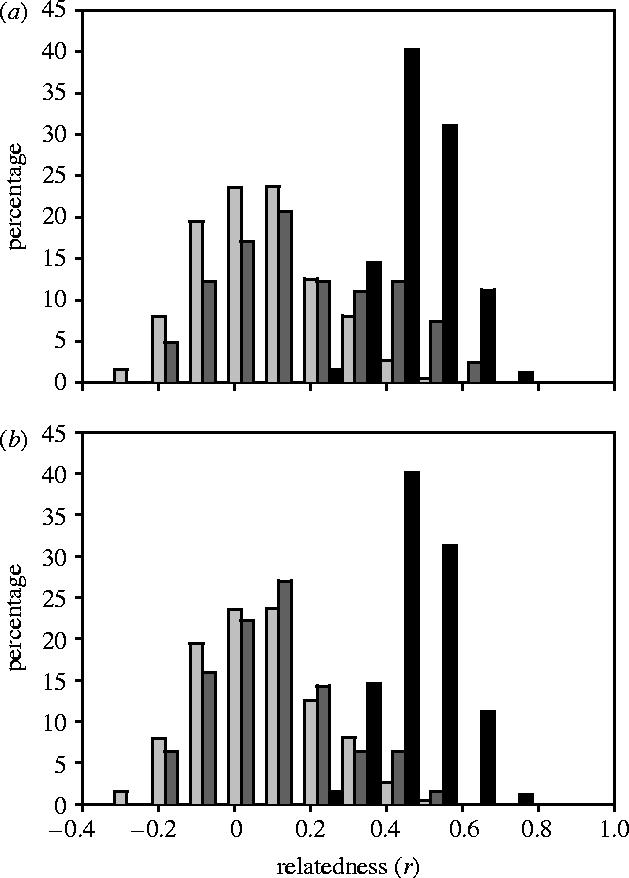
Distributions of pairwise relatedness (r) values for females with filial pups (black bars), females with non-filial pups (dark grey bars) and 832 randomly assigned pairs of individuals from the population (light grey bars), (a) prior to correction of genotyping errors and (b) following error correction.
Table 1.
Mean relatedness (r) and non-parametric 95% confidence intervals calculated pairwise among random individuals from the population, females with filial pups and females with non-filial pups, before and after correction of genotyping errors.
| n | mean r | lower 95% confidence limit | upper 95% confidence limit | |
|---|---|---|---|---|
| randomly paired individuals | 832 | −0.0029 | −0.0137 | 0.0083 |
| filial pairs before error correction | 750 | 0.4898 | 0.4830 | 0.4966 |
| non-filial pairs before error correction | 82 | 0.1101 | 0.0659 | 0.1552 |
| filial pairs after error correction | 769 | 0.4895 | 0.4828 | 0.4962 |
| non-filial pairs after error correction | 63 | 0.0339 | −0.0054 | 0.0750 |
Although the superficial trends we find are broadly in line with Gemmell's conclusions, we have previously shown that error rates as low as 0.01 per allele can lead to a 20% rate of false paternal exclusion (Hoffman & Amos 2005). Consequently, we attempted to drive the error rate as low as possible. First, autoradiographs for all mother–offspring pairs that mismatched at one locus were examined, revealing 15 genotyping errors (Hoffman et al. 2003; Hoffman & Amos 2005). These comprised 10 scoring errors, one data-entry error and four cases of allele dropout. Second, since poor quality samples could potentially give rise to multiple errors, we then re-examined all pairs mismatching at two loci. Third, to remove residual errors we finally re-extracted and re-genotyped all mother–offspring pairs that mismatched at either one or two loci. This yielded an additional seven errors in the genotypes of four pups, comprising four cases of allele dropout, one scoring error and two unknown errors (e.g. pipetting error or contamination).
Figure 2 shows the relationship between pairwise relatedness and the number of mismatches for mother–offspring pairs that were initially classified as non-filial. Many of the highest relatedness values involved pairs of individuals that mismatched at only one or two loci. However, the majority of single-locus mismatches (n=16, 88.8%) and a small number of two-locus mismatches (n=3, 27.3%) were subsequently explained as genotyping errors, suggesting that the high relatedness hump in the distribution of non-filial pups (figure 1a) could be artefactual. Repeating our analyses after reclassifying pups with genotyping errors shows that non-filial and randomly assigned pairs no longer differ in relatedness (figure 1b, table 1; Mann–Whitney U test, U=2944, n1=832, n2=63, p=0.10). This finding is likely to be genuine because our sample size was sufficiently large to discount the possibility of type II error (post hoc power analysis, n1=832, n2=63, α=0.05, β=0.97).
Figure 2.
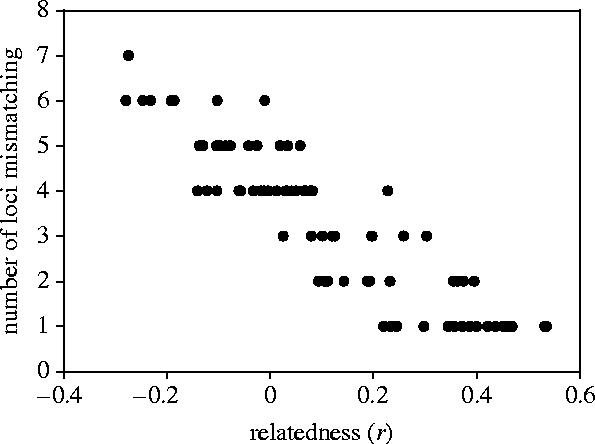
Relationship between pairwise relatedness (r) and the number of mismatching loci for 82 mother–offspring pairs that were initially classified as non-filial.
Finally, we asked whether the discrepancy between our analysis and that of Gemmell (2003) could have arisen from inter-annual variation in fostering behaviour. Restricting our analyses to the years common to both studies (1994 and 1995) the resulting pattern does not differ from that found using the full dataset, i.e. the uncorrected data show a marginally significant difference between non-filial and random pairs (Mann–Whitney U test, U=2798, n1=236, n2=30, p=0.06) while correction for genotyping errors entirely removes this trend (Mann–Whitney U test, U=2705, n1=236, n2=23, p=0.98). Additionally, the proportion of pups mismatching their mothers following error correction did not vary significantly among years (χ72=6.35, p=0.50), suggesting that each season contributed more or less equally to our analyses.
4. Discussion
We have revisited the surprising observation that female Antarctic fur seals, despite having a remarkable ability to recognize their own offspring's calls, none the less actively choose to suckle the pups of related females. Although the initial trend we find is similar to that found by Gemmell (2003), exhaustive elimination of genotyping errors has a major impact, effectively removing all those pups that had previously been classed as non-filial but closely related to the female they were suckling. This emphasizes the way even small error rates can create misleading patterns.
Use of genetic markers to study kin selection requires considerable care. In essence, the hypothesis being tested is that two interacting individuals are more related to each other than expected by chance. Unfortunately, most studies are only able to employ of the order of 10 microsatellite markers and with this number, as here, it is difficult to distinguish between pups that are non-filial relatives and those that are filial but mismatch their true parents through genotyping errors. Thus, even after substantial experimental effort (Hoffman et al. 2003) that removed many errors present in the original Gemmell dataset, we find essentially the same pattern as Gemmell (2003), the main difference being a slightly lower overall rate of fostering (9.9% compared with 11.5%). It is only after checking all single- and two-locus mismatches and then re-genotyping all cases where errors might be present that the true pattern becomes clear, specifically that fostered pups that are closely related to the female they were suckling are extremely rare.
Even when all efforts have been made to reduce genotyping errors down to a minimum, there remain a few instances where mothers mismatch closely related pups. Given the high rate of microsatellite mutations (between 10−2 and 10−6 mutations per gamete per generation; Dallas 1992; Weber & Wong 1993; Schug et al. 1997; Udupa & Baum 2001) some of these may be explained by de novo mutations. Indeed, the two remaining pups that mismatch their mothers at one locus can together be explained by invoking single repeat unit mutations at an average rate of approximately 3.3×10−4 across loci (two out of 7488 allelic comparisons, but allowing 20% of comparisons to be uninformative due to the paternal allele), eminently within the published range. Of the remaining highly related but genuinely mismatching pups, while there no longer exists any significant difference in relatedness compared with random pups, it should be remembered that kin clustering probably exists in this species. Consequently, we expect to find a few cases of allo-suckling among related females just by chance.
In conclusion, we find that previous reports of allo-suckling by Antarctic fur seal pups are best ascribed to genotyping errors and that there is no evidence that kin selection is operating. Whether this is because most mother–offspring mismatches are due to milk-stealing (Lunn 1992) rather than genuine fostering is unclear. Our study emphasizes the need for caution when interpreting rare events that occur at a rate approaching that expected for normal genotyping errors.
Acknowledgments
We thank D. Briggs, M. Jessop, K. Reid, R. Taylor, T. Walker and N. Warren for help with logistics, field data collection, animal handling and tissue sampling. Also, thanks to P. Harrison for calculating non-parametric confidence intervals. This work contributes to the BAS Dynamics and Management of Ocean Ecosystems (DYNAMOE) science programme. JH was funded by a Natural Environment Research Council (NERC) studentship. Support for the BAS field component was obtained from NERC and the Antarctic Funding Initiative (AFI). Fieldwork was approved by BAS and the University of Cambridge Animal Ethics Board. Samples were collected and retained under permits issued by the Department for Environment, Food and Rural Affairs (DEFRA) and in accordance with the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES).
References
- Amos, W. & Hoelzel, A. R. 1991 Long-term preservation of whale skin for DNA analysis. Report of the International Whaling Commission Special Issue 13, 99–103.
- Boness D.J. Fostering behavior in Hawaiian monk seals—is there a reproductive cost? Behav. Ecol. Sociobiol. 1990;27:113–122. 10.1007/BF00168454 [Google Scholar]
- Boness D.J, Craig M.P, Honigman L, Austin S. Fostering behavior and the effect of female density in Hawaiian monk seals, Monachus schauinslandi. J. Mammal. 1998;79:1060–1069. [Google Scholar]
- Bradley B.J, Vigilant L. False alleles derived from microbial DNA pose a potential source of error in microsatellite genotyping of DNA from faeces. Mol. Ecol. Notes. 2002;2:602–605. 10.1046/j.1471-8286.2002.00302.x [Google Scholar]
- Callen D.F, Thompson A.D, Shen Y, Phillips H.A, Richards R.I, Mulley J.C, Sutherland G.R. Incidence and origin of “null” alleles in the (AC)n microsatellite markers. Am. J. Hum. Genet. 1993;52:922–927. [PMC free article] [PubMed] [Google Scholar]
- Charrier I, Mathevon N, Jouventin P. Mother's voice recognition by seal pups. Nature. 2001;412:873. doi: 10.1038/35091136. 10.1038/35091136 [DOI] [PubMed] [Google Scholar]
- Charrier I, Mathevon N, Jouventin P. Fur seal mothers memorize subsequent versions of developing pups' calls: adaptation to long-term recognition or evolutionary by-product? Biol. J. Linnean Soc. 2003;80:305–312. 10.1046/j.1095-8312.2003.00239.x [Google Scholar]
- Childerhouse S, Gales N. Fostering behaviour in New Zealand sea lions Phocarctos hookeri. NZ J. Zool. 2001;28:189–195. [Google Scholar]
- Clutton-Brock T.H, Albon S.D, Guinness F.E. Fitness costs of gestation and lactation in wild mammals. Nature. 1989;337:260–262. doi: 10.1038/337260a0. 10.1038/337260a0 [DOI] [PubMed] [Google Scholar]
- Dakin E.E, Avise J.C. Microsatellite null alleles in parentage analysis. Heredity. 2004;93:504–509. doi: 10.1038/sj.hdy.6800545. [DOI] [PubMed] [Google Scholar]
- Dallas J.F. Estimation of microsatellite mutation rates in recombinant inbred strains of mice. Mammal. Genome. 1992;3:452–456. doi: 10.1007/BF00356155. 10.1007/BF00356155 [DOI] [PubMed] [Google Scholar]
- Doidge D.W, Croxall J.P, Baker J.R. Density-dependent pup mortality in the Antarctic fur seal Arctocephalus gazella at South Georgia. J. Zool. 1984;202:449–460. [Google Scholar]
- Efron B, Tibshirani R.J. Chapman & Hall; London: 1993. An introduction to the bootstrap. [Google Scholar]
- Erdfelder E, Faul F, Buchner A. Gpower: a general power analysis program. Behav. Res. Methods Instrum. Comput. 1996;28:1–11. [Google Scholar]
- Gagneux P, Boesch C, Woodruff D.S. Microsatellite scoring errors associated with non-invasive genotyping based on nuclear DNA amplified from shed hair. Mol. Ecol. 1997;6:861–868. doi: 10.1111/j.1365-294x.1997.tb00140.x. [DOI] [PubMed] [Google Scholar]
- Gemmell N.J. Kin selection may influence fostering behaviour in Antarctic fur seals (Arctocephalus gazella) Proc. R. Soc. B. 2003;270:2033–2037. doi: 10.1098/rspb.2003.2467. 10.1098/rspb.2003.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell N.J, Burg T.M, Boyd I.L, Amos W. Low reproductive success in territorial male Antarctic fur seals (Arctocephalus gazella) suggests the existence of alternative mating strategies. Mol. Ecol. 2001;10:451–460. doi: 10.1046/j.1365-294x.2001.01186.x. 10.1046/j.1365-294x.2001.01186.x [DOI] [PubMed] [Google Scholar]
- Gerloff U, Schlötterer C, Rassman K, Rambold I, Hohmann G, Fruth B, Tautz D. Amplification of hypervariable simple sequence repeats (microsatellites) from excremental DNA of wild living bonobos (Pan paniscus) Mol. Ecol. 1995;4:515–518. [Google Scholar]
- Ginot F, Bordelais I, Nguyen S, Gyapay G. Correction of some genotyping errors in automated fluorescent microsatellite analysis by enzymatic removal of one base overhangs. Nucleic Acids Res. 1996;24:540–541. doi: 10.1093/nar/24.3.540. 10.1093/nar/24.3.540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnight K.F, Queller D.C. Computer software for performing likelihood tests of pedigre relationship using genetic markers. Mol. Ecol. 1999;8:1231–1234. doi: 10.1046/j.1365-294x.1999.00664.x. 10.1046/j.1365-294x.1999.00664.x [DOI] [PubMed] [Google Scholar]
- Goossens B, Waits L.P, Taberlet P. Plucked hair samples as a source of DNA: reliability of dinucleotide microsatellite genotyping. Mol. Ecol. 1998;7:1237–1241. doi: 10.1046/j.1365-294x.1998.00407.x. 10.1046/j.1365-294x.1998.00407.x [DOI] [PubMed] [Google Scholar]
- Griffin A.S, West S.A. Kin discrimination and the benefit of helping in cooperatively breeding vertebrates. Science. 2003;302:634–636. doi: 10.1126/science.1089402. 10.1126/science.1089402 [DOI] [PubMed] [Google Scholar]
- Hamilton W.D. The genetical theory of social behaviour I, II. J. Theor. Biol. 1964;7:1–52. doi: 10.1016/0022-5193(64)90038-4. 10.1016/0022-5193(64)90038-4 [DOI] [PubMed] [Google Scholar]
- Hardy O.J, Vekemans X. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes. 2002;2:618–620. 10.1046/j.1471-8286.2002.00305.x [Google Scholar]
- Harker N. Collection, reporting and storage of microsatellite genotype data. In: Henry R.J, editor. Plant genotyping: the DNA fingerprinting of plants. CAB International; Wallingford, UK: 2001. pp. 251–264. [Google Scholar]
- Hoffman J.I, Amos W. Microsatellite genotyping errors: detection approaches, common sources and consequences for paternal exclusion. Mol. Ecol. 2005;14:599–612. doi: 10.1111/j.1365-294X.2004.02419.x. 10.1111/j.1365-294x.2004.02419.x [DOI] [PubMed] [Google Scholar]
- Hoffman J.I, Boyd I.L, Amos W. Male reproductive strategy and the importance of maternal status in the Antarctic fur seal Arctocephalus gazella. Evolution. 2003;57:1917–1930. doi: 10.1111/j.0014-3820.2003.tb00598.x. [DOI] [PubMed] [Google Scholar]
- Hoffman J.I, Boyd I.L, Amos W. Exploring the relationship between parental relatedness and male reproductive success in the Antarctic fur seal Arctocephalus gazella. Evolution. 2004;58:2087–2099. doi: 10.1111/j.0014-3820.2004.tb00492.x. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. J. Comput. Graphical Stat. 1996;5:299–314. [Google Scholar]
- Insley S.J. Long-term vocal recognition in the northern fur seal. Nature. 2000;406:404–405. doi: 10.1038/35019064. 10.1038/35019064 [DOI] [PubMed] [Google Scholar]
- Iverson S.J, Bowen W.D, Boness D.J, Oftedal O.T. The effect of maternal size and milk energy output on pup growth in grey seals (Halichoerus grypus) Physiol. Zool. 1993;66:61–88. [Google Scholar]
- Job D.A, Boness D.J, Francis J.M. Individual variation in nursing vocalizations of Hawaian monk seal pups, Monachus schauinslandi (Phocidae, Pinnipedia) and lack of maternal recognition. Can. J. Zool.-Revue Canadienne De Zoologie. 1995;73:975–983. [Google Scholar]
- Johansson A, Karlsson P, Gyllensten U. A novel method for automatic genotyping of microsatellite markers based on parametric pattern recognition. Hum. Genet. 2003;113:316–324. doi: 10.1007/s00439-003-0973-x. 10.1007/s00439-003-0973-x [DOI] [PubMed] [Google Scholar]
- Konig B, Riester J, Markl H. Maternal care in house mice (Mus musculus): II. The energy cost of lactation as a function of litter size. J. Zool. Lond. 1988;216:195–210. [Google Scholar]
- Litt M, Hauge X, Sharma V. Shadow bands seen when typing polymorphic dinucleotide repeats—some causes and cures. Biotechniques. 1993;15:280. et seq. [PubMed] [Google Scholar]
- Lunn N.J. Fostering behavior and milk stealing in Antarctic fur seals. Can. J. Zool.-Revue Canadienne De Zoologie. 1992;70:837–839. [Google Scholar]
- Lunn N.J, Boyd I.L. Pupping-site fidelity of Antarctic fur seals at Bird Island, South Georgia. J. Mammal. 1991;72:202–206. [Google Scholar]
- Lunn N.J, Boyd I.L. Influence of maternal characteristics and environmental variation on reproduction in Antarctic fur seals. Symp. Zool. Soc. London. 1993;66:115–129. [Google Scholar]
- Lynch M, Ritland K. Estimation of pairwise relatedness with molecular markers. Genetics. 1999;152:1753–1766. doi: 10.1093/genetics/152.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majluf P, Goebel M.E. The capture and handling of female South American fur seals and their pups. Mar. Mammal Sci. 1992;8:187–190. [Google Scholar]
- Marshall T.C, Slate J, Kruuk L.E.B, Pemberton J.M. Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. 10.1046/j.1365-294x.1998.00374.x [DOI] [PubMed] [Google Scholar]
- McCulloch S, Pomeroy P.P, Slater P.J.B. Individually distinctive pup vocalizations fail to prevent allo-suckling in grey seals. Can. J. Zool.-Revue Canadienne De Zoologie. 1999;77:716–723. 10.1139/cjz-77-5-716 [Google Scholar]
- Navidi W, Arnheim N, Waterman M.S. A multiple-tubes approach for accurate genotyping of very small DNA samples by using PCR: statistical considerations. Am. J. Hum. Genet. 1992;50:347–359. [PMC free article] [PubMed] [Google Scholar]
- Pemberton J.M, Slate J, Bancroft D.R, Barrett J.A. Nonamplifying alleles at microsatellite loci: a caution for parentage and population studies. Mol. Ecol. 1995;4:249–252. doi: 10.1111/j.1365-294x.1995.tb00214.x. [DOI] [PubMed] [Google Scholar]
- Perry E.A, Boness D.J, Fleischer R.C. DNA fingerprinting evidence of nonfilial nursing in grey seals. Mol. Ecol. 1998;7:81–85. doi: 10.1046/j.1365-294x.1998.00313.x. 10.1046/j.1365-294x.1998.00313.x [DOI] [PubMed] [Google Scholar]
- Pomeroy P.P, Twiss S.D, Redman P. Philopatry, site fidelity and local kin associations within grey seal breeding colonies. Ethology. 2000;106:899–919. 10.1046/j.1439-0310.2000.00610.x [Google Scholar]
- Pomeroy P.P, Worthington Wilmer J, Amos W, Twiss S.D. Reproductive performance links to fine-scale spatial patterns of female grey seal relatedness. Proc. R. Soc. B. 2001;268:711–717. doi: 10.1098/rspb.2000.1422. 10.1098/rspb.2000.1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queller D.C, Goodnight K.F. Estimating relatedness using genetic markers. Evolution. 1989;43:258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. [DOI] [PubMed] [Google Scholar]
- Queller D.C, Strassmann J.E. Kin selection and social insects. Bioscience. 1998;48:165–175. [Google Scholar]
- Riedman M.L, Leboeuf B.J. Mother–pup separation and adoption in northern elephant seals. Behav. Ecol. Sociobiol. 1982;11:203–215. 10.1007/BF00300063 [Google Scholar]
- Sambrook J, Fritsch E.F, Maniatis T. 2nd edn. Cold Spring Harbor Laboratory Press; New York: 1989. Molecular cloning: a laboratory manual. [Google Scholar]
- Schaeff C.M, Boness D.J, Bowen W.D. Female distribution, genetic relatedness, and fostering behaviour in harbour seals, Phoca vitulina. Anim. Behav. 1999;57:427–434. doi: 10.1006/anbe.1998.1001. 10.1006/anbe.1998.1001 [DOI] [PubMed] [Google Scholar]
- Schug M.D, Mackay T.F.C, Aquadro C.F. Low mutation rates of microsatellite loci in Drosophila melanogaster. Nat. Genet. 1997;15:99–102. doi: 10.1038/ng0197-99. 10.1038/ng0197-99 [DOI] [PubMed] [Google Scholar]
- Stirling I. Adoptive suckling in pinnipeds. J. Aust. Mammal Soc. 1975;1:389–391. [Google Scholar]
- Taberlet P, Griffin S, Goosens B, Questiau S, Manceau V, Escaravage N, Waits L.P, Bouvet J. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 1996;24:3189–3194. doi: 10.1093/nar/24.16.3189. 10.1093/nar/24.16.3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udupa S.M, Baum M. High mutation rate and mutational bias at (TAA)n microsatellite loci in the chickpea (Cicer arietinum L.) Mol. Genet. Genomics. 2001;265:1097–1103. doi: 10.1007/s004380100508. 10.1007/s004380100508 [DOI] [PubMed] [Google Scholar]
- Walsh P.S, Metzger D.A, Higuchi R. Chelex100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- Walsh P.S, Ehrlich H.A, Higuchi R. Preferential amplification of alleles: mechanisms ans solutions. PCR Methods Appl. 1992;1:241–250. doi: 10.1101/gr.1.4.241. [DOI] [PubMed] [Google Scholar]
- Wang J. An estimator for pairwise relatedness using molecular markers. Genetics. 2002;160:1203–1215. doi: 10.1093/genetics/160.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J.L, Wong C. Mutation of human short tandem repeats. Hum. Mol. Genet. 1993;2:1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- Worthington Wilmer J, Allen P.J, Pomeroy P.P, Twiss S.D, Amos W. Where have all the fathers gone? An extensive microsatellite analysis of paternity in the grey seal (Halichoerus grypus) Mol. Ecol. 1999;8:1417–1429. doi: 10.1046/j.1365-294x.1999.00705.x. 10.1046/j.1365-294x.1999.00705.x [DOI] [PubMed] [Google Scholar]


