Abstract
Barbara McClintock was the first to suggest that transposons are a source of genome instability and that genotoxic stress assisted in their mobilization. The generation of double-stranded DNA breaks (DSBs) is a severe form of genotoxic stress that threatens the integrity of the genome, activates cell cycle checkpoints, and, in some cases, causes cell death. Applying McClintock's stress hypothesis to humans, are L1 retrotransposons, the most active autonomous mobile elements in the modern day human genome, mobilized by DSBs? Here, evidence that transposable elements, particularly retrotransposons, are mobilized by genotoxic stress is reviewed. In the setting of DSB formation, L1 mobility may be affected by changes in the substrate for L1 integration, the DNA repair machinery, or the L1 element itself. The review concludes with a discussion of the potential consequences of L1 mobilization in the setting of genotoxic stress.
THE CELLULAR RESPONSE TO DNA DAMAGE IS COMPLEX
There are many chemical agents and natural processes that have the ability to damage DNA. UV light, X-rays, chemotherapeutic drugs, cigarette smoke, and even cell division have the potential to generate DNA lesions [1]. Depending on the source of DNA damage, the structure of the DNA break and its mechanism of repair may be different. Oxidative damage creates DNA double-strand breaks that are repaired by nonhomologous end joining [2]. Nucleotide base damage and dimer formation induced by UV rays during sun exposure are repaired by base excision repair [3]. Stalled replication forks in dividing cells are repaired by homologous recombination [4].
Shortly after the induction of a DSB, complex signaling pathways are activated [5]. These signaling cascades recruit DNA repair factors to DSBs, alter transcription, and trigger cell fate decisions. Significant damage may trigger cell cycle arrest, or even apoptosis. Various cellular events occurring secondary to DNA damage may affect L1 retrotransposition. Because the cellular response to genotoxic stress can vary depending on the type of lesion and cell type, the effects on L1 retrotransposition could depend on the context of DNA damage.
MOBILIZATION OF TRANSPOSABLE ELEMENTS BY DNA DAMAGE
While direct evidence for the activation of L1 retrotransposition by DNA damage is still sparse, there is a growing body of data that other mobile elements can be activated by DNA damage. Barbara McClintock initially observed Ac/Ds element transposition in response to chromosomal translocations [6, 7]. Indeed, some transposable elements, including P elements in Drosophila and the synthetic Sleeping Beauty element, appear to be activated by DNA damage and repair processes [8–10]. Mobilization is not limited to DNA transposons: various forms of DNA damage activate retrotransposition of long terminal repeat (LTR) and non-LTR retrotransposons including Gypsy and I factor in Drosophila and Ty1 in yeast [11–16]. Even closer to home for L1, transcription and retrotransposition of Alu elements are increased when cells are exposed to etoposide, a topoisomerase II inhibitor that produces DSBs [17, 18]. This is relevant to L1s because Alu elements are thought to co-opt L1 proteins for their mobilization, so increased Alu retrotransposition may reflect increased L1 mobility [19]. In a genome screen of mice exposed to gamma irradiation, new SINE and L1 insertions were detected, but it was unresolved if the frequency of new insertions was significantly different in irradiated compared to unirradiated controls [20].
OVERVIEW OF THE L1 LIFECYCLE
The L1 lifecycle provides ample opportunities for regulation by its host cell (Figure 1). A full-length RNA encoding the ORF1 and ORF2 proteins is transcribed from a retrotransposition-competent L1. L1 mRNA is exported to the cytoplasm where its encoded ORF1 and ORF2 proteins are translated. This protein-RNA complex returns to the nucleus, where the endonuclease domain of ORF2 nicks the target site. The reverse transcriptase domain of ORF2 creates a cDNA copy using the target site's 5′ overhang as a primer. Subsequent displacement of the mRNA by a complementary strand of cDNA and ligation of the breaks are thought to require host machinery.
Figure 1.

DNA damage can affect multiple stages of the L1 life cycle. (1) Transcription of the L1 element is controlled by epigenetic factors and transcription factors. (2) L1 RNA is exported to the cytoplasm, where its copy number influences retrotransposition frequency. (3) Translation of ORF1 and ORF2 proteins. (4) L1 protein and mRNA are imported into the nucleus, where ORF2 endonuclease creates a DNA double-strand break. Induced breaks may be able to serve as alternative substrates for insertion. (5) ORF2 reverse transcribes a cDNA copy of L1 at the insertion site. Host factors are thought to inhibit or assist in resolution of the insertion. The dark square represents the cell nucleus, and the lighter surrounding square represents the cytoplasm.
Any or all steps in the retrotransposition process could be affected by the cellular response to DNA damage. This review will focus on (1) alterations in the activity of the L1 element, primarily by regulation of L1 transcription; (2) alterations in L1 entry into the genome, with emphasis on insertion into pre-existing DSBs, and (3) alterations in cellular factors in response to DNA damage, in particular DNA repair machinery and its effect on L1 retrotransposition.
ALTERED ACTIVITY OF L1s IN THE SETTING OF DNA DAMAGE
The division of potential causes of L1 mobilization in the setting of DNA damage into L1-intrinsic versus extrinsic factors is admittedly an arbitrary one. It is unlikely that an “element intrinsic” property such as the level of L1 RNA or protein is altered without concomitant alterations in cellular factors that influence L1. This section is focused on L1 RNA not because RNA is necessarily the most likely point of regulation (although it is a reasonable target, as discussed below), but because there are more abundant data pointing to a potential role for regulation of L1 RNA in the context of genotoxic stress.
Altering L1 RNA may well alter the abundance of L1 proteins or ribonucleoproteins. Consistent with this idea, cells treated with etoposide exhibited increased Alu RNA, increased reverse transcriptase activity and increased retrotransposition [17, 18]. On the other hand, the expression of some proteins is not directly correlated with element RNA levels. For example, the TyA1 protein of the Ty1 LTR retrotransposon in yeast did not accumulate following exposure to gamma irradiation, whereas mRNA copy number and retrotransposition frequency did increase [15].
The notion that RNA or protein levels correlate with retrotransposition frequency, while direct, may be too simplistic and ignores other, more subtle forms of regulation. For example, the compartmentalization of L1 proteins may be affected by DNA damage. When tagged ORF1 and ORF2 proteins are expressed individually or together from a virus, they localize to the nucleolus [21]. This effect has also been seen in yeast retroelements, where a protein tagged with the Ty3 retrotransposon integrase domain is targeted to the nucleolus [22]. As Goodier and colleagues point out, L1 could traffic through the nucleolus; this idea is supported by the presence of chimeric transcripts of L1s fused to small RNA species such as U6, U3, U5, and 5S (reviewed in [21]). In the setting of DNA damage, nucleolar protein trafficking pathways are altered (reviewed in [23]). For example, PML and Mdm2 are sequestered in the nucleolus following DNA damage [24]. The sequestration of Mdm2 results in enhanced p53 stability [24]. Trafficking of L1 through the nucleolus therefore may be altered in the setting of genotoxic stress and represents a potential pathway for regulating L1 mobility.
L1 RNA LEVELS AND GENOTOXIC STRESS
Several lines of evidence suggest that L1 RNA abundance is critical and rate-limiting for L1 retrotransposition. L1 RNA is required for retrotransposition, not only because it encodes the machinery needed for L1 to retrotranspose, but because the RNA itself serves as a replication intermediate (Figure 1). That L1 transcript abundance is rate-limiting for retrotransposition is suggested by studies in cultured cells with tagged L1 elements showing that decreased L1 mRNA levels result in reduced retrotransposition [25]. Conversely, increased L1 RNA levels have been observed for highly active L1 elements [26]. Furthermore, the correlation between RNA levels and retrotransposition frequency is not unique to L1 retrotransposons: Ty1 elements in yeast appear to retrotranspose in direct proportion to the amount of Ty1 mRNA [15, 27, 28].
Given that RNA is important for L1 mobility, does DNA damage influence L1 transcript abundance? To our knowledge, there are no published data that compare L1 RNA levels in irradiated and unirradiated cells. However, there is evidence that RNA levels of other retrotransposons are influenced by DNA damage. For example, gamma radiation has been shown to increase Ty1 RNA in yeast [15] and IAP RNA in murine myeloid cells [29]. Furthermore, murine and human cell lines expressing the Bcl-2 survival gene exhibit an increase in endogenous Alu mRNA levels following exposure to gamma radiation, UV, etoposide, and cisplatin [17].
Since the induction of DNA damage has an extensive effect on the transcriptional profile of a cell [30], it is plausible that L1 RNA levels are differentially regulated following gamma radiation. One way to regulate L1 expression following DNA damage is to alter transcription factor levels or binding activity. The 5′UTR of the L1 contains an internal promoter element [31–33] with putative binding sites for SRY family members [34], YY1 [35], and RUNX3 [25]. DNA damage could modulate L1 activity by acting through factors that bind these sites.
Binding of the SRY family member, SOX11, to the L1 5′UTR was shown to increase L1 retrotransposition, promoter activity, and RNA copy number [34]. More recently, binding of SOX2 has been shown to inhibit L1 promoter activity in rat hippocampal neuronal stem cells [36]. SOX2 and SOX11 possess high-mobility group domains, which have been shown to bind to cisplatin-DNA adducts [37]. If SRY family members are differentially recruited to the sites of DNA damage, then this could alter the profile of transcription factors at the L1 5′UTR.
Another L1 transcription factor that may be affected by DNA damage is the ubiquitous YinYang1 (YY1) factor. YY1 is thought to facilitate the production of full-length L1 mRNAs [38]. In response to exposure to methyl-N-nitro-N-nitrosoguanidine, YY1 was polyADP-ribosylated in HeLa cells, decreasing its ability to bind its consensus target sequences [39]. YY1 has also been shown to be a negative regulator of p53 activation under conditions of genomic stress in primary and cancer cell lines [40]. This is interesting given that L1 activity is itself thought to be a genomic stressor that induces apoptosis using a p53-dependent mechanism [41]. Under conditions of DNA damage, YY1 could therefore have opposing effects on the retrotransposition frequency: decreased YY1 binding could result in fewer full-length L1 transcripts while YY1's effects on p53 might enhance the survival of cells that harbor new L1 insertions.
As is discussed elsewhere in this issue, L1 RNA levels can also be influenced by epigenetic regulation. Focusing here on CpG methylation as a mode of transcriptional silencing of L1s, negative regulation of L1 retrotransposition by this form of “methylation defense” predicts that L1s are methylated and that demethylation derepresses L1s. Consistent with methylation defense, the L1 5′UTR has been shown to undergo methylation and methylation has a negative effect on L1 promoter activity [42] and retrotransposition using a cultured cell assay [43]. This effect may be mediated by methyl-CpG-binding protein 2 (MeCP2), which inhibits retrotransposition in the cultured cell assay [44]. Oxidative damage has been shown to decrease the affinity of MeCP2 for damaged methylated DNA [45]. DNA damage near an L1 element may therefore release it from negative regulation.
DNA damage may also play a role in regulating global methylation of genomic L1s. Gamma radiation has been shown to induce hypomethylation in cell lines [46] and in mouse livers, and spleens [47]. One potential mechanism for hypomethylation in the setting of irradiation is an alteration in the folate pool. Gamma radiation has been shown to reduce the activity of the enzyme methylenetetrahydrofolate reductase in the livers of mice [48]. A polymorphism associated with reduced activity of this enzyme has been linked to hypomethylation and gastric cancer susceptibility in humans [49]. Another possibility is that irradiation influences the expression of DNA methyltransferases. Hypomethylation in transformed cell lines has been associated with decreased expression of the DNA methyltransferases DNMT1, DNMT3a, and DNMT3b [50] and mobilization of retrotransposons has been linked to methyltransferase deficiency. For example, methylation of the LTR retrotransposon IAP is diminished and transcription is activated in Dnmt1 deficient mouse embryos [51]. Mouse knockouts of Dnmt3L demethylate genomic L1 insertions and exhibit greatly increased levels of L1 mRNA in their germ cells [52]. If by either or both mechanisms widespread demethylation occurs, L1s could be globally activated following DNA damage.
ALTERED L1 ENTRY INTO THE GENOME DURING DNA DAMAGE
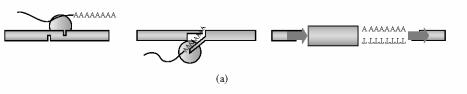
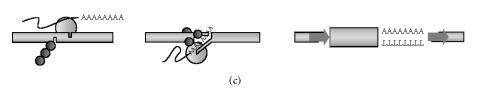
Insertion of an L1 copy into the genome necessitates the creation and repair of broken DNA. Based on the elegant work from Tom Eickbush's group on the non-LTR retrotransposon R2Bm and recent findings using an in vitro L1 system, the L1 endonuclease is believed to nick DNA in a staggered fashion creating overhanging singlestranded DNA [53–55]. After L1 integration, the DNA ends are sealed and filled in, forming the target site duplications that flank a typical L1 insertion (steps 4 and 5, Figure 1). On the other hand, what happens to L1 integration if a cell is subjected to DNA damage (Figure 2)? The presence of broken DNA may allow L1 to integrate into preformed breaks in an endonuclease-independent fashion (Figure 2(b)). Alternatively or in addition, enzymes used by the cell to repair damaged DNA may aid (or inhibit) L1 retrotransposition (Figure 2(c)).
Figure 2.
Potential ways in which DNA damage could influence L1 retrotransposition. (a) Endonuclease-dependent insertion under normal conditions. The L1 endonuclease (○) makes staggered nicks at the target site, creating 3′ overhangs. Filling in generates 7–20 base pair target site duplications (➡) flanking the insertion. (b) Endonuclease-independent insertion at the site of a double-strand break. The preexisting double-strand break shown here lacks staggered nicks or overhangs. L1 entry into this site would therefore also lack target site duplications. Genomic deletions may occur due to processing by cellular DNA repair processes (∗). (c) Endonuclease-dependent insertion potentiated by DNA damage. DNA damage may upregulate cellular cofactors of reverse transcription and integration (●). Insertion via pathway c is endonuclease-dependent, but occurs at an increased efficiency.
Retrotransposons can use artificially induced DNA breaks as substrates for insertion. Yeast with deficiencies in homologous recombination machinery occasionally capture Ty1 cDNA during repair of breaks introduced at the MAT locus [56, 57]. Using a plasmid-based assay in which DNA breaks repaired by captured cDNA are selectively recovered, Yu and Gabriel found that 21 out of 37 captured sequences were derived fromTy1 elements [58]. Furthermore, in mouse cells both LTR retrotransposons and SINE elements were able to repair a break induced by the restriction enzyme I-SceI [59].
Collectively, these studies indicate that retrotransposons can integrate into broken DNA. But does this happen frequently? The previously described experiments used genetic screens to look for what may have been rare events. Under the conditions of the cell culture L1 retrotransposition assay, mutation of the L1 endonuclease active site reduced the retrotransposition frequency to ∼ 1% of wild-type levels [60]. This result suggests that L1 usually uses its own endonuclease to gain entry into the genome. However, in the setting of DNA repair enzyme deficiency (DNA-PKcs or XRCC4 deficiency in particular) L1s lacking endonuclease exhibited greatly increased rates of retrotransposition [61]. L1s lacking endonuclease generated genomic insertions in repair deficient cells with atypical structures (including large deletions at the site of integration), while fully functional L1s generated fewer of these “atypical” insertions [61–64].
CELLULAR COFACTORS AND INHIBITORS OF RETROTRANSPOSITION
Cellular proteins involved in the response to DNA damage, particularly those of the nonhomologous end joining cascade (NHEJ), may act as cofactors or inhibitors of retrotransposition. Transcription of NHEJ factors including Ku70 and its partner Ku80 are up-regulated following exposure to gamma radiation [65]. Furthermore, many of these repair factors colocalize at the sites of double-strand breaks [66] and have altered bioavailability following DNA damage [67]. Therefore it seems reasonable to propose that modulation and altered subcellular distribution of DNA repair enzymes in the setting of genotoxic stress could influence L1 retrotransposition.
The contribution of DNA repair factors to the mobilization of DNA transposons has been investigated by several groups. In Drosophila, the P element transposase possesses putative phosphorylation sites for the ataxia telangectasia mutation protein (ATM), a master control kinase of the DNA damage response [68]. Mutation of specific ATM sites increased or in some cases decreased excision of these elements. The DNA repair protein Ku70 and the Bloom helicase, both downstream of ATM [69, 70], have been shown to be important for repair of P element excision sites [71]. Ku70 is also important for repair of Sleeping Beauty excision in mammalian cells [9]. In a survey of multiple repair factors, deficiencies in the NHEJ factors Ku80, DNA-PKcs, and XRCC4 and the homologous recombination factors Rad51C and XRCC3 decreased Sleeping Beauty mobility in mammalian cells [10]. Reconstitution of the knockout reversed the phenotype, and even increased transposition above wild-type levels for DNA-PKcs [10].
DNA repair factors also influence the mobility of retrotransposons. A mutagenesis screen for inhibitors of Ty1 retrotransposition revealed genes that help maintain genomic integrity including telomerase, a yeast homologue of Bloom, and components of the NBS complex [72]. Rad3 and Ssl2, helicases involved in nucleotide excision repair, appear to inhibit Ty1 retrotransposition post-translationally [73]. Potential cofactors for Ty1 retrotransposition are the Ku repair factors. Ku70 protein coprecipitates with Ty1 cDNA, cofractionates with Ty1 retrotransposition intermediates, and deficiency in both Ku70 and Ku80 dramatically decreases retrotransposition [74].
There is also evidence linking NHEJ machinery to the regulation of L1s. Ku70/80 binding sites have been identified in murine L1s: L1s make up 19% of the mouse genome, but account for 26% of the Ku70/80 binding sites [75]. Cell lines deficient in DNA-PKcs permit lower rates of endonuclease-dependent L1 retrotransposition than their wild-type parentals, while XRCC4 mutants permit higher rates of L1 retrotransposition [61]. Repair enzyme deficiency could affect L1 retrotransposition via multiple pathways. Increased persistence of unrepaired double-strand breaks could serve as substrates for insertion and increase endonuclease-independent insertion (Figure 2(b)). On the other hand, a dearth of DNA repair enzymes might hinder the resolution of L1 insertions. The loss or altered availability of inhibitors could, conversely, promote retrotransposition.
POTENTIAL CONSEQUENCES OF L1 ACTIVATION DURING GENOTOXIC STRESS
Increased retrotransposition in the setting of genetic damage could have a beneficial effect on the cell: L1 insertion into the site of a DSB could form a bridge between chromosome fragments, sealing an otherwise irreparable break [76]. Consistent with this idea, activity of the L1-like NL1Tc element resulted in decreased unrepaired DNA breaks and enhanced survival of Trypanosoma cruzi exposed to daunorubicin [77]. Moreover, retrotransposons may have been coopted over the course of evolution to play a role in specialized DNA repair functions. An example of this is the preferential insertion into and maintenance of telomere ends. Mobile elements with an insertion site preference for telomere or subtelomeric regions have been identified in Saccharomyces cerevisiae, Chlorella vulgaris, Bombyx mori, Allium cepa, and Giardia lamblia [78–82]. In Drosophila, the non-LTR retrotransposons HeT-A and TART not only preferentially insert at chromosome ends, but play a direct role in telomere maintenance [83–85]. In other animals including humans, telomere maintenance relies on telomerase and DNA repair molecules such as WRN and Artemis [86, 87]. Evidence from Caenorhabditis elegans (mut7) and mammals (Artemis), indicates that deficiencies of these enzymes can mobilize transposable elements [88], (E.A. Farkash and E.T. Luning Park, unpublished data). Enhanced mobility, coupled with opportunity may cause mobile elements to assist with telomere maintenance under conditions of genotoxic stress.
On the other hand, with the exception of HeT-A and TART in Drosophila, preferential insertion into chromosome ends does not necessarily translate into a beneficial function for the element. Insertion into telomeres could be less disruptive than inserting elsewhere, giving elements with this insertion site preference a proliferative advantage. If, as is widely presumed, L1 integration is random, then increasing its mobility will most likely have neutral or negative consequences for the host cell. Even simply upregulating the L1 endonuclease in the absence of successful integration could be toxic to the cell by promoting the formation of additional DSBs, fostering chromosomal rearrangements, and translocations. The consequences of L1 integration into preformed DNA breaks in the setting of genotoxic stress could be severe in that such insertions may be more likely to be accompanied by large deletions [61]. In this regard it is worth noting that pathogenic insertions in chimpanzees and humans have been associated with large deletions [89]. A meta-analysis of human pathogenic insertions found 6 out of 48 (12.5%) were associated with large deletions, compared to 5 out of 145 (3.4%) polymorphic genomic insertions [90] and 6 out of 100 insertions characterized in a cell-culture-based retrotransposition assay [64]. Severe DNA damage can result in cell cycle arrest and apoptosis [91]. Both cell cycle arrest and apoptosis have been seen to accompany retrotransposition in severely stressed cells [28, 41]. Retrotransposition in a cell with damaged DNA could be its final undoing. The potential lethality of genotoxic stress may help to account for the paucity of endonuclease-independent insertions among L1s present in the human genome.
ACKNOWLEDGMENT
This study was supported by NIH grants T32H10791 and CA108812.
References
- 1.Hoeijmakers JHJ. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411(6835):366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23(5):687–696. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- 3.Lankinen MH, Vilpo LM, Vilpo JA. UV- and γ-irradiation-induced DNA single-strand breaks and their repair in human blood granulocytes and lymphocytes. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 1996;352(1-2):31–38. doi: 10.1016/0027-5107(95)00172-7. [DOI] [PubMed] [Google Scholar]
- 4.McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nature Reviews. Molecular Cell Biology. 2002;3(11):859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 5.Ward I, Chen J. Early events in the DNA damage response. Current Topics in Developmental Biology. 2004;63:1–35. doi: 10.1016/S0070-2153(04)63001-8. [DOI] [PubMed] [Google Scholar]
- 6.McClintock B. The origin and behavior of mutable loci in maize. Proceedings of the National Academy of Sciences of the United States of America. 1950;36(6):344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226(4676):792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 8.Handler AM, Gomez SP. P element excision in Drosophila is stimulated by gamma-irradiation in transient embryonic assays. Genetical Research. 1997;70(1):75–78. doi: 10.1017/s0016672397002759. [DOI] [PubMed] [Google Scholar]
- 9.Yant SR, Kay MA. Nonhomologous-end-joining factors regulate DNA repair fidelity during sleeping beauty element transposition in mammalian cells. Molecular and Cellular Biology. 2003;23(23):8505–8518. doi: 10.1128/MCB.23.23.8505-8518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izsvák Z, Stüwe EE, Fiedler D, Katzer A, Jeggo PA, Ivics Z. Healing the wounds inflicted by sleeping beauty transposition by double-strand break repair in mammalian somatic cells. Molecular Cell. 2004;13(2):279–290. doi: 10.1016/s1097-2765(03)00524-0. [DOI] [PubMed] [Google Scholar]
- 11.Georgiev PG, Korochkina SE, Georgieva SG, Gerasimova TI. Mitomycin C induces genomic rearrangements involving transposable elements in Drosophila melanogaster. Molecular and General Genetics: MGG. 1990;220(2):229–233. doi: 10.1007/BF00260486. [DOI] [PubMed] [Google Scholar]
- 12.Bradshaw VA, McEntee K. DNA damage activates transcription and transposition of yeast Ty retrotransposons. Molecular and General Genetics: MGG. 1989;218(3):465–474. doi: 10.1007/BF00332411. [DOI] [PubMed] [Google Scholar]
- 13.Morawetz C, Hagen U. Effect of irradiation and mutagenic chemicals on the generation of ADH2- and ADH4-constitutive mutants in yeast: the inducibility of Ty transposition by UV and ethyl methanesulfonate. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 1990;229(1):69–77. doi: 10.1016/0027-5107(90)90009-s. [DOI] [PubMed] [Google Scholar]
- 14.Morawetz C. Effect of irradiation and mutagenic chemicals on the generation of ADH2-constitutive mutants in yeast. Significance for the inducibility of Ty transposition. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 1987;177(1):53–60. doi: 10.1016/0027-5107(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 15.Sacerdot C, Mercier G, Todeschini A-L, Dutreix M, Springer M, Lesage P. Impact of ionizing radiation on the life cycle of Saccharomyces cerevisiae Ty1 retrotransposon. Yeast. 2005;22(6):441–455. doi: 10.1002/yea.1222. [DOI] [PubMed] [Google Scholar]
- 16.Bregliano JC, Laurencon A, Degroote F. Evidence for an inducible repair-recombination system in the female germ line of Drosophila melanogaster. I. Induction by inhibitors of nucleotide synthesis and by gamma rays. Genetics. 1995;141(2):571–578. doi: 10.1093/genetics/141.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudin CM, Thompson CB. Transcriptional activation of short interspersed elements by DNA-damaging agents. Genes, Chromosomes and Cancer. 2001;30(1):64–71. [PubMed] [Google Scholar]
- 18.Hagan CR, Sheffield RF, Rudin CM. Human Alu element retrotransposition induced by genotoxic stress. Nature Genetics. 2003;35(3):219–220. doi: 10.1038/ng1259. [DOI] [PubMed] [Google Scholar]
- 19.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nature Genetics. 2003;35(1):41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 20.Asakawa J, Kuick R, Kodaira M, et al. A genome scanning approach to assess the genetic effects of radiation in mice and humans. Radiation Research. 2004;161(4):380–390. doi: 10.1667/rr3146. [DOI] [PubMed] [Google Scholar]
- 21.Goodier JL, Ostertag EM, Engleka KA, Seleme MC, Kazazian HH., Jr A potential role for the nucleolus in L1 retrotransposition. Human Molecular Genetics. 2004;13(10):1041–1048. doi: 10.1093/hmg/ddh118. [DOI] [PubMed] [Google Scholar]
- 22.Lin SS, Nymark-McMahon MH, Yieh L, Sandmeyer SB. Integrase mediates nuclear localization of Ty3. Molecular and Cellular Biology. 2001;21(22):7826–7838. doi: 10.1128/MCB.21.22.7826-7838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimber A, Nguyen Q-D, Gespach C. Nuclear bodies and compartments: functional roles and cellular signalling in health and disease. Cellular Signalling. 2004;16(10):1085–1104. doi: 10.1016/j.cellsig.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 24.Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nature Cell Biology. 2004;6(7):665–672. doi: 10.1038/ncb1147. [DOI] [PubMed] [Google Scholar]
- 25.Yang N, Zhang L, Zhang Y, Kazazian HH., Jr An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Research. 2003;31(16):4929–4940. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han JS, Boeke JD. A highly active synthetic mammalian retrotransposon. Nature. 2004;429(6989):314–318. doi: 10.1038/nature02535. [DOI] [PubMed] [Google Scholar]
- 27.Curcio MJ, Hedge AM, Boeke JD, Garfinkel DJ. Ty RNA levels determine the spectrum of retrotransposition events that activate gene expression in Saccharomyces cerevisiae. Molecular and General Genetics: MGG. 1990;220(2):213–221. doi: 10.1007/BF00260484. [DOI] [PubMed] [Google Scholar]
- 28.Staleva LS, Venkov P. Activation of Ty transposition by mutagens. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2001;474(1-2):93–103. doi: 10.1016/s0027-5107(00)00165-2. [DOI] [PubMed] [Google Scholar]
- 29.Ishihara H, Tanaka I, Furuse M, Tsuneoka K. Increased expression of intracisternal A-particle RNA in regenerated myeloid cells after X irradiation in C3H/He inbred mice. Radiation Research. 2000;153(4):392–397. doi: 10.1667/0033-7587(2000)153[0392:ieoiap]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 30.Jen K-Y, Cheung VG. Transcriptional response of lymphoblastoid cells to ionizing radiation. Genome Research. 2003;13(9):2092–2100. doi: 10.1101/gr.1240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Molecular and Cellular Biology. 1990;10(12):6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minakami R, Kurose K, Etoh K, Furuhata Y, Hattori M, Sakaki Y. Identification of an internal cis-element essential for the human L1 transcription and a nuclear factor(s) binding to the element. Nucleic Acids Research. 1992;20(12):3139–3145. doi: 10.1093/nar/20.12.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathias SL, Scott AF. Promoter binding proteins of an active human L1 retrotransposon. Biochemical and Biophysical Research Communications. 1993;191(2):625–632. doi: 10.1006/bbrc.1993.1263. [DOI] [PubMed] [Google Scholar]
- 34.Tchénio T, Casella J-F, Heidmann T. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Research. 2000;28(2):411–415. doi: 10.1093/nar/28.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becker KG, Swergold GD, Ozato K, Thayer RE. Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Human Molecular Genetics. 1993;2(10):1697–1702. doi: 10.1093/hmg/2.10.1697. [DOI] [PubMed] [Google Scholar]
- 36.Muotri AR, Chu VT, Marchetto MCN, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435(7044):903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 37.Trimmer EE, Zamble DB, Lippard SJ, Essigmann JM. Human testis-determining factor SRY binds to the major DNA adduct of cisplatin and a putative target sequence with comparable affinities. Biochemistry. 1998;37(1):352–362. doi: 10.1021/bi971675q. [DOI] [PubMed] [Google Scholar]
- 38.Athanikar JN, Badge RM, Moran JV. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Research. 2004;32(13):3846–3855. doi: 10.1093/nar/gkh698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oei SL, Shi Y. Poly(ADP-ribosyl)ation of transcription factor Yin Yang 1 under conditions of DNA damage. Biochemical and Biophysical Research Communications. 2001;285(1):27–31. doi: 10.1006/bbrc.2001.5115. [DOI] [PubMed] [Google Scholar]
- 40.Grönroos E, Terentiev AA, Punga T, Ericsson J. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(33):12165–12170. doi: 10.1073/pnas.0402283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haoudi A, Semmes OJ, Mason JM, Cannon RE. Retrotrans-position-competent human LINE-1 induces apoptosis in cancer cells with intact p53. Journal of Biomedicine and Biotechnology. 2004;2004(4):185–194. doi: 10.1155/S1110724304403131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hata K, Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene. 1997;189(2):227–234. doi: 10.1016/s0378-1119(96)00856-6. [DOI] [PubMed] [Google Scholar]
- 43.Woodcock DM, Lawler CB, Linsenmeyer ME, Doherty JP, Warren WD. Asymmetric methylation in the hypermethylated CpG promoter region of the human L1 retrotransposon. The Journal of Biological Chemistry. 1997;272(12):7810–7816. doi: 10.1074/jbc.272.12.7810. [DOI] [PubMed] [Google Scholar]
- 44.Yu F, Zingler N, Schumann G, Strätling WH. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Research. 2001;29(21):4493–4501. doi: 10.1093/nar/29.21.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valinluck V, Tsai H-H, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Research. 2004;32(14):4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalinich JF, Catravas GN, Snyder SL. The effect of gamma radiation on DNA methylation. Radiation Research. 1989;117(2):185–197. [PubMed] [Google Scholar]
- 47.Pogribny I, Raiche J, Slovack M, Kovalchuk O. Dose-dependence, sex- and tissue-specificity, and persistence of radiation-induced genomic DNA methylation changes. Biochemical and Biophysical Research Communications. 2004;320(4):1253–1261. doi: 10.1016/j.bbrc.2004.06.081. [DOI] [PubMed] [Google Scholar]
- 48.Batra V, Kesavan V, Mishra KP. Modulation of enzymes involved in folate dependent one-carbon metabolism by γ-radiation stress in mice. Journal of Radiation Research. 2004;45(4):527–533. doi: 10.1269/jrr.45.527. [DOI] [PubMed] [Google Scholar]
- 49.Graziano F, Kawakami K, Ruzzo A, et al. Methylenetetrahy-drofolate reductase 677C/T gene polymorphism, gastric cancer susceptibility and genomic DNA hypomethylation in an at-risk Italian population. International Journal of Cancer. 2006;118(3):628–632. doi: 10.1002/ijc.21397. [DOI] [PubMed] [Google Scholar]
- 50.Raiche J, Rodriguez-Juarez R, Pogribny I, Kovalchuk O. Sex- and tissue-specific expression of maintenance and de novo DNA methyltransferases upon low dose X-irradiation in mice. Biochemical and Biophysical Research Communications. 2004;325(1):39–47. doi: 10.1016/j.bbrc.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 51.Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nature Genetics. 1998;20(2):116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 52.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431(7004):96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 53.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72(4):595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 54.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. The EMBO Journal. 2002;21(21):5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christensen SM, Eickbush TH. R2 target-primed reverse transcription: ordered cleavage and polymerization steps by protein subunits asymmetrically bound to the target DNA. Molecular and Cellular Biology. 2005;25(15):6617–6628. doi: 10.1128/MCB.25.15.6617-6628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moore JK, Haber JE. Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature. 1996;383(6601):644–646. doi: 10.1038/383644a0. [DOI] [PubMed] [Google Scholar]
- 57.Teng S-C, Kim B, Gabriel A. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature. 1996;383(6601):641–644. doi: 10.1038/383641a0. [DOI] [PubMed] [Google Scholar]
- 58.Yu X, Gabriel A. Patching broken chromosomes with extranuclear cellular DNA. Molecular Cell. 1999;4(5):873–881. doi: 10.1016/s1097-2765(00)80397-4. [DOI] [PubMed] [Google Scholar]
- 59.Lin Y, Waldman AS. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics. 2001;158(4):1665–1674. doi: 10.1093/genetics/158.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87(5):905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 61.Morrish TA, Gilbert N, Myers JS, et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nature Genetics. 2002;31(2):159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 62.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110(3):315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 63.Symer DE, Connelly C, Szak ST, et al. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110(3):327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 64.Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Molecular and Cellular Biology. 2005;25(17):7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brodsky MH, Weinert BT, Tsang G, et al. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Molecular and Cellular Biology. 2004;24(3):1219–1231. doi: 10.1128/MCB.24.3.1219-1231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rapp A, Greulich KO. After double-strand break induction by UV-A, homologous recombination and nonhomologous end joining cooperate at the same DSB if both systems are available. Journal of Cell Science. 2004;117(pt 21):4935–4945. doi: 10.1242/jcs.01355. [DOI] [PubMed] [Google Scholar]
- 67.Drouet J, Delteil C, Lefrançois J, Concannon P, Salles B, Calsou P. DNA-dependent protein kinase and XRCC4-DNA ligase IV mobilization in the cell in response to DNA double strand breaks. The Journal of Biological Chemistry. 2005;280(8):7060–7069. doi: 10.1074/jbc.M410746200. [DOI] [PubMed] [Google Scholar]
- 68.Beall EL, Mahoney MB, Rio DC. Identification and analysis of a hyperactive mutant form of Drosophila P-element transposase. Genetics. 2002;162(1):217–227. doi: 10.1093/genetics/162.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brown KD, Lataxes TA, Shangary S, et al. Ionizing radiation exposure results in up-regulation of Ku70 via a p53/ataxia-telangiectasia-mutated protein-dependent mechanism. The Journal of Biological Chemistry. 2000;275(9):6651–6656. doi: 10.1074/jbc.275.9.6651. [DOI] [PubMed] [Google Scholar]
- 70.Ababou M, Dutertre S, Lécluse Y, Onclercq R, Chatton B, Amor-Guéret M. ATM-dependent phosphorylation and accumulation of endogenous BLM protein in response to ionizing radiation. Oncogene. 2000;19(52):5955–5963. doi: 10.1038/sj.onc.1204003. [DOI] [PubMed] [Google Scholar]
- 71.Min B, Weinert BT, Rio DC. Interplay between Drosophila Bloom's syndrome helicase and Ku autoantigen during non-homologous end joining repair of P element-induced DNA breaks. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(24):8906–8911. doi: 10.1073/pnas.0403000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scholes DT, Banerjee M, Bowen B, Curcio MJ. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159(4):1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee BS, Lichtenstein CP, Faiola B, et al. Posttranslational inhibition of Ty1 retrotransposition by nucleotide excision repair/transcription factor TFIIH subunits Ssl2p and Rad3p. Genetics. 1998;148(4):1743–1761. doi: 10.1093/genetics/148.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Downs JA, Jackson SP. Involvement of DNA end-binding protein Ku in Ty element retrotransposition. Molecular and Cellular Biology. 1999;19(9):6260–6268. doi: 10.1128/mcb.19.9.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Katz DJ, Beer MA, Levorse JM, Tilghman SM. Functional characterization of a novel Ku70/80 pause site at the H19/Igf2 imprinting control region. Molecular and Cellular Biology. 2005;25(10):3855–3863. doi: 10.1128/MCB.25.10.3855-3863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eickbush TH. Repair by retrotransposition. Nature Genetics. 2002;31(2):126–127. doi: 10.1038/ng897. [DOI] [PubMed] [Google Scholar]
- 77.Olivares M, López MC, García-Pérez JL, Briones P, Pulgar M, Thomas MC. The endonuclease NL1Tc encoded by the LINE L1Tc from Trypanosoma cruzi protects parasites from daunorubicin DNA damage. Biochimica et Biophysica Acta (BBA)/Gene Structure and Expression. 2003;1626(1–3):25–32. doi: 10.1016/s0167-4781(03)00022-8. [DOI] [PubMed] [Google Scholar]
- 78.Zhu Y, Zou S, Wright DA, Voytas DF. Tagging chromatin with retrotransposons: target specificity of the Saccharomyces Ty5 retrotransposon changes with the chromosomal localization of Sir3p and Sir4p. Genes & Development. 1999;13(20):2738–2749. doi: 10.1101/gad.13.20.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Noutoshi Y, Arai R, Fujie M, Yamada T. Structure of the Chlorella Zepp retrotransposon: nested Zepp clusters in the genome. Molecular and General Genetics: MGG. 1998;259(3):256–263. doi: 10.1007/s004380050811. [DOI] [PubMed] [Google Scholar]
- 80.Anzai T, Takahashi H, Fujiwara H. Sequence-specific recognition and cleavage of telomeric repeat (TTAG)n by endonuclease of non-long terminal repeat retrotransposon TRAS1. Molecular and Cellular Biology. 2001;21(1):100–108. doi: 10.1128/MCB.21.1.100-108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pich U, Schubert I. Terminal heterochromatin and alternative telomeric sequences in Allium cepa. Chromosome Research. 1998;6(4):315–321. doi: 10.1023/a:1009227009121. [DOI] [PubMed] [Google Scholar]
- 82.Arkhipova IR, Morrison HG. Three retrotransposon families in the genome of Giardia lamblia: two telomeric, one dead. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(25):14497–14502. doi: 10.1073/pnas.231494798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Biessmann H, Mason JM, Ferry K, et al. Addition of telomere-associated HeT DNA sequences “heals” broken chromosome ends in Drosophila. Cell. 1990;61(4):663–673. doi: 10.1016/0092-8674(90)90478-w. [DOI] [PubMed] [Google Scholar]
- 84.Levis RW, Ganesan R, Houtchens K, Tolar LA, Sheen F-M. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75(6):1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- 85.Pardue M-L, DeBaryshe PG. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annual Review of Genetics. 2003;37:485–511. doi: 10.1146/annurev.genet.38.072902.093115. [DOI] [PubMed] [Google Scholar]
- 86.Lee JW, Harrigan J, Opresko PL, Bohr VA. Pathways and functions of the Werner syndrome protein. Mechanisms of Ageing and Development. 2005;126(1):79–86. doi: 10.1016/j.mad.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 87.Rooney S, Alt FW, Lombard D, et al. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. The Journal of Experimental Medicine. 2003;197(5):553–565. doi: 10.1084/jem.20021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99(2):133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 89.Han K, Sen SK, Wang J, et al. Genomic rearrangements by LINE-1 insertion-mediated deletion in the human and chimpanzee lineages. Nucleic Acids Research. 2005;33(13):4040–4052. doi: 10.1093/nar/gki718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen J-M, Stenson PD, Cooper DN, Férec C. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Human Genetics. 2005;117(5):411–427. doi: 10.1007/s00439-005-1321-0. [DOI] [PubMed] [Google Scholar]
- 91.Barzilai A, Yamamoto K-I. DNA damage responses to oxidative stress. DNA Repair. 2004;3(8-9):1109–1115. doi: 10.1016/j.dnarep.2004.03.002. [DOI] [PubMed] [Google Scholar]