Abstract
In eukaryotes, enzymes of different subcellular compartments participate in the assembly of membrane lipids. As a consequence, interorganelle lipid transfer is extensive in growing cells. A prominent example is the transfer of membrane lipid precursors between the endoplasmic reticulum (ER) and the photosynthetic thylakoid membranes in plants. Mono- and digalactolipids are typical photosynthetic membrane lipids. In Arabidopsis, they are derived from one of two pathways, either synthesized de novo in the plastid, or precursors are imported from the ER, giving rise to distinct molecular species. Employing a high-throughput robotic screening procedure generating arrays of spot chromatograms, mutants of Arabidopsis were isolated, which accumulated unusual trigalactolipids. In one allelic mutant subclass, trigalactosyldiacylglycerol1, the primary defect caused a disruption in the biosynthesis of ER-derived thylakoid lipids. Secondarily, a processive galactosyltransferase was activated, leading to the accumulation of oligogalactolipids. Mutations in a permease-like protein of the outer chloroplastic envelope are responsible for the primary biochemical defect. It is proposed that this protein is part of a lipid transfer complex.
Keywords: chloroplast envelopes/lipid arrays/lipid permease/lipid trafficking/thylakoid lipids
Introduction
In eukaryotic cells, the biosynthesis of membrane lipids often occurs in subcellular compartments different from their final destination. This raises a fundamental cell biological question: how are membrane lipids transported between organelles and across membranes? The intricate compartmentalization of plant lipid metabolism provides a promising opportunity towards answering this question. In plants, the endoplasmic reticulum (ER) and the chloroplastic envelopes participate in the biosynthesis of the lipids of photosynthetic membranes. As a consequence, substantial trafficking of lipid precursors occurs between these two membrane systems. The galactolipids mono- and digalactosyldiacylglycerol (MGDG and DGDG) are the predominant polar lipids of photosynthetic thylakoid membranes (Joyard et al., 1998; Dörmann and Benning, 2002).
In many plants, two pathways are involved in the synthesis of thylakoid lipids (Roughan and Slack, 1982; Frentzen, 1986). Thylakoid glycerolipids are assembled either de novo in the plastid (plastid pathway) or from diacylglycerol precursors originating in the ER. The ER pathway requires the export of fatty acids from the plastid, their incorporation into phosphatidylcholine (PC) at the ER and the return of PC-derived diacylglycerol moieties to the plastid. Many plants, e.g. grasses, lack the plastid pathway for thylakoid lipid biosynthesis (Mongrand et al., 1998), and thylakoid lipid biosynthesis in these plants relies entirely on the transfer of precursors from the ER to the plastid. In the model plant Arabidopsis, ∼50% of thylakoid lipids are derived from the ER pathway (Browse et al., 1986). Such an estimation is possible because plastid- and ER-derived galactolipids can be distinguished based on their diagnostic fatty acid composition. Lipids derived from the plastid pathway contain 18-carbon fatty acids attached to the first carbon of the glycerol backbone (sn-1 position) and 16-carbon fatty acids attached to the second carbon (sn-2 position), while ER-derived lipids preferentially contain 18-carbon fatty acids in both positions.
The ats1(act1) mutant of Arabidopsis is deficient in the plastidic pathway for thylakoid lipid biosynthesis, because the plastidic glycerol-3-phosphate acyltransferase is mutated in this line (Kunst et al., 1988). As a conse quence, the flux through the ER pathway of thylakoid lipid biosynthesis is nearly doubled without impact on growth. On the other hand, the unicellular green alga Chlamydomonas relies exclusively on the plastid pathway for thylakoid lipid assembly (Giroud et al., 1988). In view of this apparent flexibility of lipid metabolism in plants and algae, we reasoned that in Arabidopsis, a block in the ER pathway of thylakoid lipid biosynthesis could be compensated for by the plastid pathway without a major loss of viability.
We have developed a method to screen for mutants of Arabidopsis deficient in the biosynthesis of ER-derived thylakoid lipids. It has been a long-standing observation that isolated chloroplasts or chloroplast envelopes exhibit a processive galactosyltransferase (PGT) activity leading to the formation of trigalactosyldiacylglycerol (TGDG) and other oligomeric galactolipids (van Besow and Wintermans, 1978). However, the presence of these lipids in plant tissues has been documented only rarely (Kojima et al., 1990). We reasoned that the accumulation of oligogalactolipids observed for isolated chloroplast preparations in essence reflects a disruption of the interaction between the ER and the plastid leading to the activation of a PGT. As a consequence, we predict that the accumulation of novel oligogalactolipids in mutants of Arabidopsis could serve as a diagnostic indicator for a disruption of ER to plastid lipid transfer in vivo. This hypothesis became testable after isolating mutants accumulating oligogalactolipids during a search for suppressors of the dgd1 mutation, which causes a deficiency in the biosynthesis of DGDG (Dörmann et al., 1995).
Results
Isolation of oligogalactolipid-accumulating mutants
Applying high-throughput technology, samples from homozygous dgd1 M2 plants were processed in 96-well plates and spotted by a modified pipetting robot in groups of 384 onto 20 × 20 cm thin-layer chromatography (TLC) plates. Circular chromatograms <1 cm in diameter were developed by application of a solvent mixture to the spots using the robot, and glycolipids were visualized by staining with a sugar-sensitive reagent as shown in Figure 1A. After screening 15 000 M2 dgd1 plants, 23 putative dgd1 suppressor lines were isolated. Following reanalysis by linear TLC (Figure 1B), these fell into two classes: those with a strong increase in the amount of DGDG compared with dgd1 (two lines), and those showing an additional glycolipid band, but only slightly increased amounts of DGDG (21 lines). On repeating this mutant screen in the wild-type genetic background, two additional lines containing the extra lipid were isolated by screening 10 000 M2 plants. The TGDG-accumulating mutants were designated tgd for trigalactosyldiacylglcyerol. Allelism testing of the tgd mutant class is still in progress. However, among nine lines analyzed thus far, six independent loci, tgd1–tgd6, were identified, with tgd1 represented by three independent alleles. The focus here will be on the tgd1-1 allele, the analysis of the accumulating lipid, the primary biochemical defect and the characterization of the affected gene product.
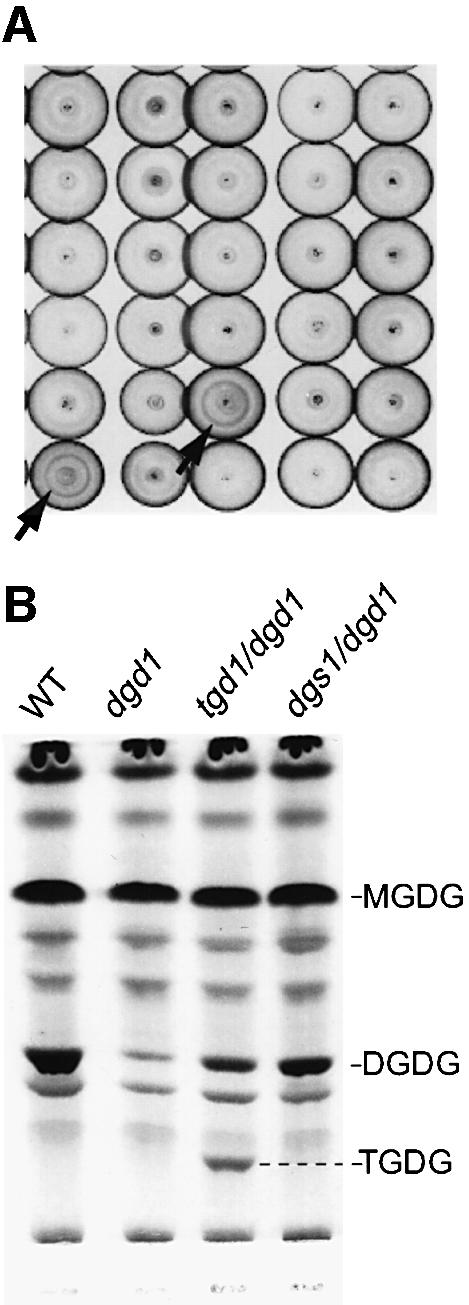
Fig. 1. Array of spot chromatograms (A) and lipid phenotypes (B). (A) Circular chromatograms (∼1 cm in diameter) generated by the robotic procedure are shown. Arrows indicate two suppressor lines with a DGDG ring. (B) Detailed lipid phenotypes for wild type, the homozygous dgd1 mutant, the homozygous tgd1-1/dgd1 double mutant and a more typical dgd1 suppressor (dgs1/dgd1) are shown. Lipids were separated by TLC and visualized using sugar-sensitive stain. DGDG, digalactosyldiacylglycerol; MGDG, monogalactosyldiacylglycerol; TGDG, trigalactosyldiacylglycerol.
A processive galactosyltransferase different from known galactolipid biosynthetic enzymes is responsible for oligogalactolipid formation in tgd1-1
Prior to phenotypic analysis, the tgd1-1 mutant (originally in the dgd1 genetic background) was outcrossed to wild type (Col-2) and subsequently backcrossed three times. Unless mentioned otherwise, all experiments subsequently were carried out with the mutant line carrying the tgd1-1 allele in the wild-type background. Following a test cross between the homozygous tgd1-1 mutant line and the wild type, all F1 plants were indistinguishable. Of 212 F2 plants screened, 58 (27%) plants were recovered which accumulated TGDG, whereas lipid extracts from all other plants were indistinguishable from wild-type extracts. The nearly 3:1 segregation observed for F2 plants was in agreement with a single nuclear recessive mutation that results in the production of TGDG. The homozygous tgd1-1 mutant plants were slightly smaller and had 15% less chlorophyll (w/w fresh weight) as compared with the wild type (data not shown). The newly accumulating lipid in this mutant was isolated from lipid extracts of leaves and purified by repeated TLC. During the process, it became clear that additional glycolipids accumulate in the mutant, which are not present in the wild type and migrate very close to the origin of the TLC plate. It was hypothesized that the tgd1-1 mutant accumulates trigalactolipids, as indicated in Figure 1B, as well as higher oligomeric forms of galactolipids.
What we initially expected to be a routine structural analysis confirming TGDG accumulation in the tgd1-1 mutant surprisingly provided us with important clues to understanding the tgd1-1 phenotype. Compositional analysis of alditol acetates derived from the TGDG head group confirmed the exclusive presence of galactose (data not shown). Fatty acid methylester analysis of the TGDG acyl groups revealed that the major molecular species of TGDG contains equal amounts of 16:3 and 18:3 fatty acids, with 16:3 in the sn-2 position as determined by positional analysis (see below). Positive and negative mode fast atom bombardment mass spectrometry (FAB-MS)/MS of TGDG purified from tgd1-1 yielded a predominant molecular ion and secondary ions consistent with trigalactosyldiacyl-(18:3/16:3)-glycerol. The exact mass determined by positive FAB-MS was 1093.5898 (m/z), in agreement with a composition of C55H90O20Na (2.3 p.p.m. deviation from the theoretical value), the chemical formula of the sodium adduct of trigalactosyldiacyl-(18:3/16:3)-glycerol.
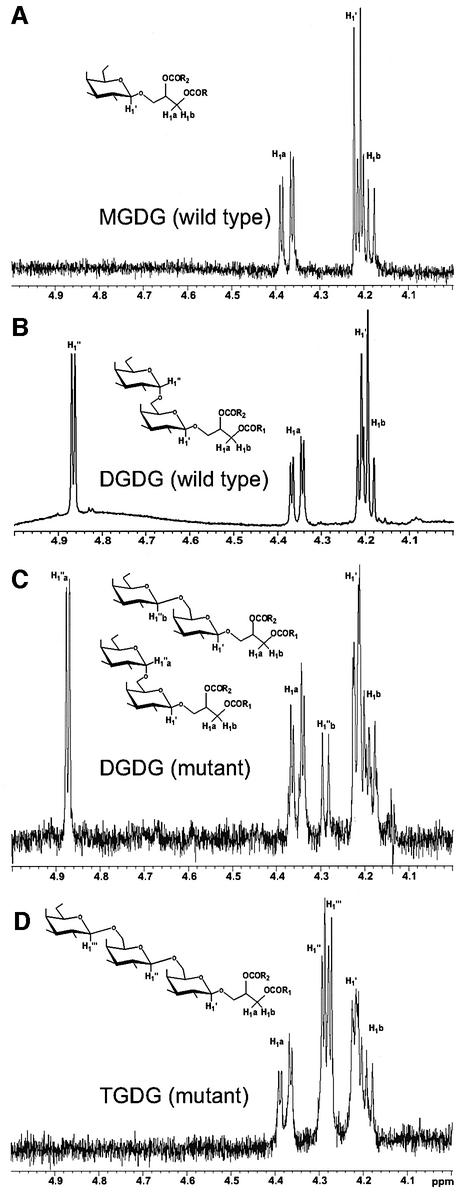
To obtain linkage information for the three galactoses in the TGDG molecule, we acquired a 500 MHz proton NMR for the lipid purified from the tgd1-1 mutant and compared it with spectra recorded for MGDG and DGDG isolated from the wild type and the mutant, respectively (Figure 2). Surprisingly, all putative anomeric protons of TGDG (Figure 2D) were in the β-configuration (4.286 p.p.m., J1,2 7.511 Hz; 4.279 p.p.m., J1,2 7.511 Hz; 4.21 p.p.m., J1,2 7–8 Hz), while the anomeric proton of the first galactose in DGDG from the wild type typically was in the β-configuration and that of the second galactose in the α-configuration (4.220 p.p.m., J1,2 6.185 Hz; 4.871 p.p.m., J1,2 3.756 Hz) as shown in Figure 2B. However, DGDG isolated from the tgd1-1/dgd1 homozygous double mutant (Figure 2C) contained a mixture of the two configurations for the anomeric proton of the second galactose as well as a β-anomeric proton for the first galactose (4.870 p.p.m., J1,2 3.534 Hz; 4.286 p.p.m., J1,2 7.290 Hz; 4.214 p.p.m., J1,2 6.406 Hz). Since it is mechanistically improbable that a retaining glycosyltransferase would suddenly switch in the mutant to become an inverting glycosyltransferase, it seems almost certain that DGD1, which is a retaining glycosyl transferase producing the bulk of DGDG in wild-type leaf tissues (Dörmann et al., 1999), or its paralog DGD2 (Kelly and Dörmann, 2002) were not responsible for the accumulation of TGDG in the tgd1-1 mutant. Furthermore, DGD1 could be ruled out a priori because TGDG biosynthetic activity is present in the dgd1 mutant background. These findings led us to postulate that a distinct processive galactosyltransferase, designated PGT, is activated in the tgd1-1 mutant.
Fig. 2. Proton NMR spectra (500 MHz) of galactolipid species in the wild type and the tgd1-1 mutant. The region shown includes the anomeric protons and protons at the 1-position of glycerol, designated H1a and H1b. Anomeric protons are labeled as H1′, H1″ and H1′″ for the first, second and third galactose moiety, respectively. (A) MGDG isolated from Col-2 wild type, showing a single β-resonance. (B) DGDG isolated from Col-2 wild type, showing a β-signal and an α-resonance from the second galactose. (C) DGDG isolated from the tgd1-1/dgd1 double mutant, representing a mixture of α-β- and β-β-DGDG, as indicated by the H1″a and H1″b labels, respectively. This mutant background was used here, instead of tgd1-1 in the DGD1 wild-type background, to reduce the amount of interfering α-β-DGDG derived from DGD1, leaving only the small amount produced by DGD2. (D) TGDG isolated from plants with a homozygous tgd1-1 mutant background, showing an absence of α-signals and three separate β-resonances.
The activity of PGT was measured in isolated wild-type and tgd1-1 chloroplasts using UDP-Gal as a substrate (Figure 3). The activity was sensitive to thermolysin, a protease which readily digests surface-exposed proteins of the outer chloroplast envelope (Cline et al., 1984). This result suggested that PGT is located at the outer envelope in the same manner as DGD1 (Froehlich et al., 2001). Furthermore, TGDG formation was observed when labeled MGDG was added to isolated tgd1-1 chloroplasts in the absence of UDP-Gal (data not shown). Thus, based on the available in situ evidence, PGT activated in tgd1-1 appears to be identical to the MGDG:MGDG galactosyltransferase originally described by van Besow and Wintermanns (1978). However, PGT location and substrate specificity can only be determined for certain once the respective protein has been identified.
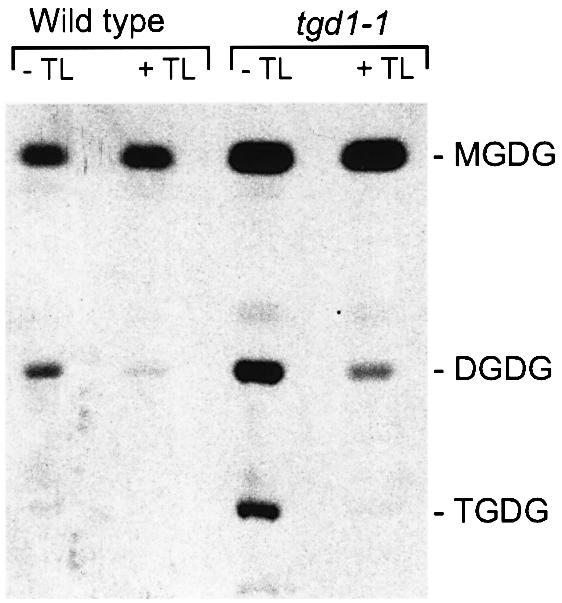
Fig. 3. Biosynthesis of galactolipids from labeled UDP-Gal in isolated chloroplasts of the wild type and the tgd1-1 mutant. Samples treated with the protease thermolysin are indicated (+TL). An autoradiograph of a TLC is shown. Abbreviations are as for Figure 1.
The import of lipid molecular species from the ER is impaired in tgd1-1
The first indication that lipid transfer from the ER to the chloroplast may be the primary defect in the tgd1-1 mutant came from compositional and positional fatty acid analysis of the galactolipids. Overall, the tgd1-1 mutant showed a moderate reduction in the relative amount of the galactolipids MGDG (∼5 mol% reduction) and DGDG (∼20 mol% reduction). The relative amount of TGDG in the mutant was ∼0.5 mol%. A comparison of the fatty acid composition of the galactolipids MGDG, DGDG and TGDG in the wild type and tgd1-1 mutant is shown in Figure 4A. It is apparent that the 16- to-18-carbon fatty acid ratio of MGDG and DGDG has increased. This result was consistent with a strong increase in the relative amount of plastid pathway-derived lipid species characterized by an 18-carbon fatty acid in the sn-1 position and a 16-carbon fatty acid in the sn-2 position of the glycerol backbone. This finding was supported further by specific analysis of the fatty acids at the sn-2 position of the individual galactolipids (Figure 4B). The sn-2 position in the galactolipids from tgd1-1 was highly enriched in 16-carbon fatty acids. Some residual 18-carbon fatty acids were still present in the sn-2 position of the different lipids in the mutant, suggesting that the mutation in the tgd1-1 allele may be leaky. It should also be noted that the 16-carbon fatty acids in the mutant were more saturated, presumably due to increased flow through the plastid pathway, thereby overloading plastidic desaturases. Thus, in tgd1-1, all galactolipids including TGDG were enriched in more saturated molecular species derived from the plastid pathway of galactolipid biosynthesis. Interestingly, the reverse changes in lipid molecular species composition were observed for the ats1(act1) mutant (Kunst et al., 1988), which has a bona fide defect in a reaction of the plastid pathway, leading to a compensatory increase in ER pathway activity.
Fig. 4. Total fatty acid composition (A) and composition exclusively at the sn-2 position of the glycerol backbone (B) of galactolipids in wild type (open bars) and tgd1-1 mutant (closed bars). The carbon and double bond number for each fatty acid is indicated. Total and positional analyses were performed on independently grown plant batches. The means of three replicates are given, with standard errors in all cases <5%.
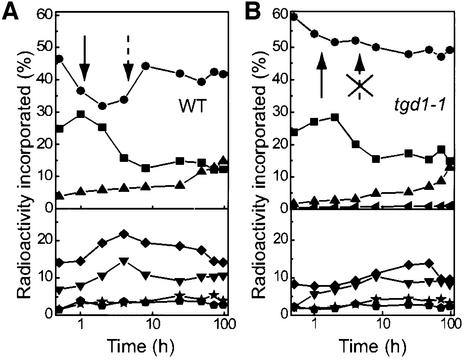
The second indicator that lipid transfer from the ER to the plastid was defective in the tgd1-1 mutant came from in vivo pulse–chase labeling of lipids using acetate. This well-documented approach provides clues about the turnover of different fatty acid pools and gives an indication of movement of lipids through the two pathways (Browse et al., 1986). Thus for these experiments, droplets of buffer containing labeled acetate were applied once to Arabidopsis leaves with a syringe. Leaves were harvested and lipids were analyzed at various intervals. A comparison of the wild type and the tgd1-1 mutant is shown in Figure 5. The labeling of MGDG and PC is most diagnostic in this experiment. In the wild type, MGDG typically is labeled first by the plastid pathway, since fatty acid biosynthesis takes place in the plastid (Figure 5A, solid arrow). However, MGDG labeling declines (as leaves grow and the labeled MGDG pool in the plastid is diluted) until labeled fatty acids have moved through the ER pathway and are returning to the plastid (Figure 5A, broken arrow). In other words, the first (declining) phase of MGDG labeling is indicative of the plastid pathway, while the second (increasing) phase is interpreted as ER pathway activity (Browse et al., 1986). Concomitant with the second phase increase in MGDG labeling, label in PC drops, suggesting that labeled fatty acids move from extraplastidic PC to MGDG. In the ats1(act1) mutant, the plastid pathway is blocked and most of the label appears first in PC, while labeled fatty acids in MGDG slowly increase as species are acquired from the ER pathway (the first phase is missing) (Kunst et al., 1988). The tgd1-1 mutant (Figure 5B) showed yet different labeling kinetics. The MGDG pool was labeled initially much more strongly than in the wild type, consistent with increases in the plastidic pathway activity and increased MGDG turnover due to PGT activity. Label in MGDG decreased steadily over time as de novo synthesized unlabeled fatty acids were introduced into the MGDG pool during leaf growth. There was no second, increasing phase of MGDG labeling as in the wild type. Moreover, PC turnover in the tgd1-1 mutant was altered (less drastic changes in the rate of labeling, different timing), suggesting reduced transfer of fatty acids from PC to MGDG as part of the eukaryotic pathway.
Fig. 5. In vivo pulse–chase acetate labeling of fatty acids associated with individual lipids of the wild type (A) and the tgd1-1 mutant (B). Experiments for the wild type and the tgd1-1 mutant were repeated >3 times with similar results. A representative result is shown. Solid arrows indicate the plastid pathway component of MGDG labeling, the broken arrows the ER pathway component. Lipids are: digalactosyldiacylglycerol (triangles), monogalactosyldiacylglycerol (circles), phosphatidylcholine (squares), phosphatidylethanolamine (inverted triangles), phosphatidylglycerol (diamonds), phosphatidylinositol (pentagons), sulfoquinovosyldiacylglycerol (stars), trigalactosyldiacylglycerol (sideway triangle, upper right panel only).
The third indication that lipid transfer from the ER to the plastid was defective in the tgd1-1 mutant came from a variation of the in vivo acetate labeling experiment. In this experiment, labeled oleic acid was fed to detached leaves for 1 h and the incorporation and redistribution of label in galactolipids and PC was measured as shown in Table I. Oleic acid was chosen because it has been shown to be incorporated almost exclusively into ER pathway-derived lipids when applied exogenously (Roughan et al., 1987). Label in PC was retained in the tgd1-1 mutant at each time point, whereas the incorporation of oleic acid into MGDG and DGDG was decreased, when compared with wild type (Table I). We conclude from this result that transfer of ER-derived PC to the plastid or the incorporation of diacylglycerol backbones derived from PC into the thylakoid lipids MGDG and DGDG is impaired in the mutant.
Table I. Incorporation of radioactivity (% of total lipid ± SE) into lipids of Arabidopsis following labeling of detached leaves with oleic acid.
| Chase time (h) |
||||
|---|---|---|---|---|
| 5 |
24 |
5 |
24 |
|
| Wild type | tgd1-1 | |||
| MGDG | 17.1 ± 0.7 | 28.5 ± 1.2 | 6.4 ± 1.4 | 13.6 ± 1.2 |
| DGDG | 3.9 ± 0.2 | 9.2 ± 0.2 | 1.4 ± 0.2 | 3.5 ± 0.5 |
| PC | 60.3 ± 0.5 | 37.7 ± 0.5 | 68.8 ± 4.5 | 50.6 ± 1.8 |
In summary, the preliminary biochemical analysis of the tgd1-1 mutant revealed two phenotypic phenomena, first the accumulation of new oligogalactolipids, and secondly, a severe reduction in the biosynthesis of thylakoid lipids by the ER pathway. How these two aspects of the mutant phenotype are related, and which might be the primary biochemical phenotype, was addressed further by identifying and characterizing the TGD1 protein.
TGD1 encodes a permease-like protein
The cloning of the TGD1 gene was accomplished by fine mapping of the tgd1-1 mutant locus on chromosome 1 followed by complementation and sequencing of the three tgd1 mutant alleles. The tgd1-1 map-based cloning strategy is summarized in Figure 6. Using a small F2 mapping population of ∼50 plants (100 chromosomes), the mutant locus was placed in the 5 cM interval on chromosome 1 between markers F15H18 and G2395. These two markers subsequently were used to score a mapping population of 1215 F2 homozygous mutant plants (2430 chromosomes) for recombinants in this interval. Of these, 28 were recovered with the marker on the left and 29 with the marker on the right of tgd1-1. Taking advantage of the Cereon genomic sequence of the ecotype Landsberg erecta and the published AGI sequence for ecotype Columbia (The Arabidopsis Genome Initiative, 2000), we were able to develop new dCAPS (derived cleaved amplified polymorphic sequences) and SSLP (simple sequence length polymorphisms) markers for fine mapping and scoring of 57 recombinants in the interval of interest. Ultimately, we placed the gene between markers F15571 and F6F9-3, an interval containing <10 genes. Using probes from this area, cosmids containing genomic DNA between the T-DNA borders were isolated and used for complementation analysis. A set of three overlapping complementing cosmids narrowed the search down to two genuine genes and one pseudogene. The cDNAs were obtained by RT–PCR and sequenced, and three different mutations were found in the three independent tgd1 allelic mutants in gene At1g19800, identifying it as TGD1.
Fig. 6. Map-based cloning of TGD1 (At1g19800). Recombinants per total chromosomes analyzed in the F2 population are indicated above the respective markers. Complementing cosmids are marked by (+) following their designation. The exon (thick bar) intron structure of the TGD1 gene is shown at the bottom. The nucleotide substitutions in the three mutant alleles are indicated.
The cDNA corresponding to At1g19800 was inserted into a T-DNA vector and expressed in the tgd1-1/dgd1 double mutant to test for complementation of the tgd1 mutant locus (Figure 7). Genotyping was used to confirm the presence of tgd1-1 and dgd1 homozygous mutant loci and the presence of the transgene (Figure 7B and C). Expression of the cDNA in the tgd1-1/dgd1 double mutant background suppressed the formation of TGDG, consistent with complementation of tgd1 (Figure 7D). In addition, the intermediate growth of the tgd1-1/dgd1 double mutant was reversed to the slow growth typically observed for the dgd1 homozygous mutant (Figure 7A), suggesting that growth phenotype correlates with the expression of TGD1.
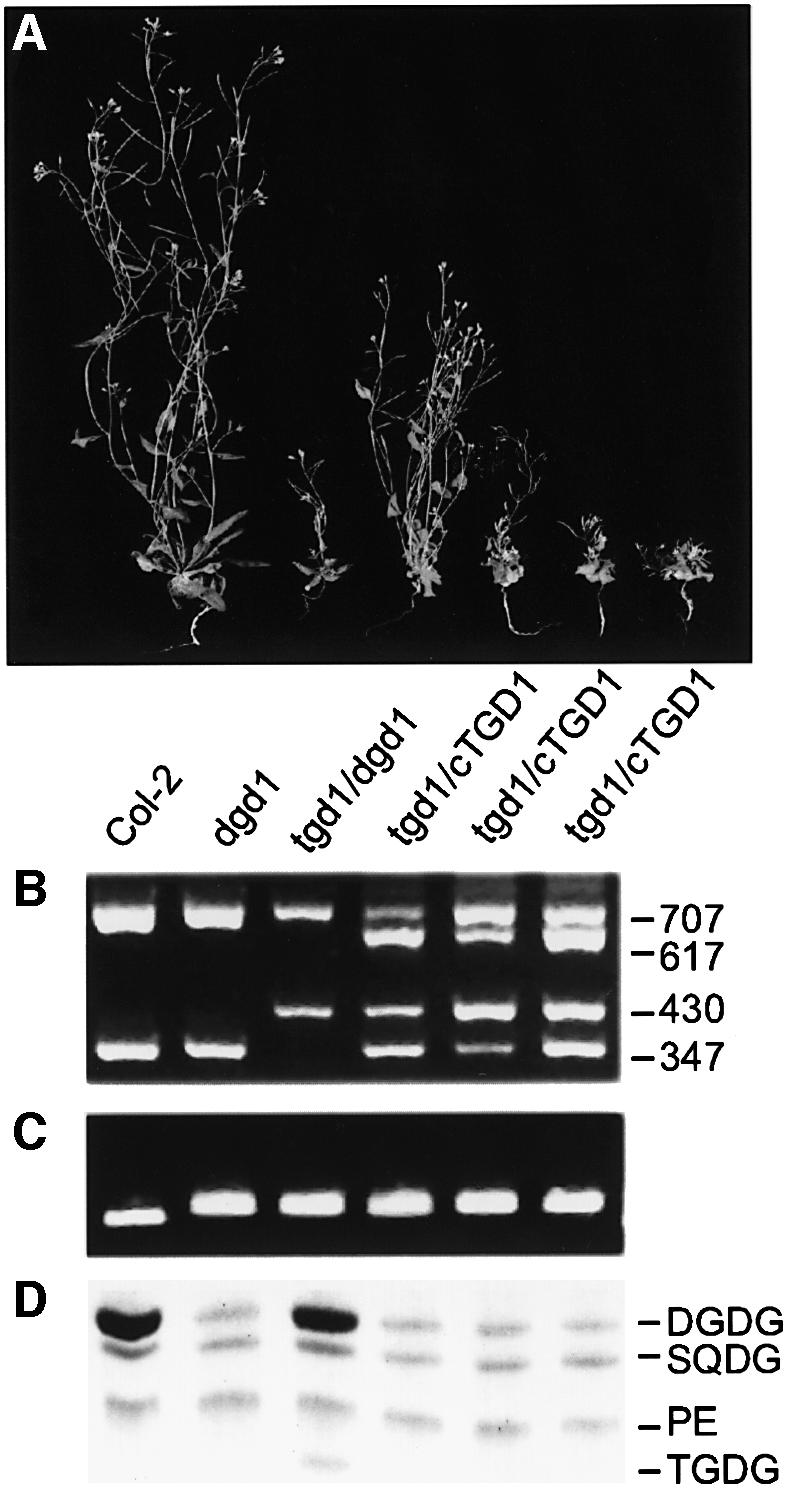
Fig. 7. Complementation of the tgd1-1 mutation by the TGD1 cDNA. (A) Growth habit of wild type, the dgd1 homozygous mutant, the homozygous tgd1-1/dgd1 double mutant and three homozygous tgd1-1/dgd1 double mutants carrying the TGD1 cDNA in trans (from left to right as indicated). (B) Genotyping at the TGD1 locus using a CAPS marker. Fragment sizes are indicated and are diagnostic as follows: 707 bp genomic locus-derived fragment; 617 bp cDNA-specific fragment; 430 bp fragment specific for the mutant tgd1-1 genomic locus; and 347 bp fragment specific for the genomic wild-type locus, but also present in cDNA. (C) Genotyping at the DGD1 locus using a dCAPS marker. The longer fragment is diagnostic for the presence of the dgd1 mutation. (D) Lipid phenotype. A section of a linear TLC is shown. Lipids are: DGDG, digalactosyldiacylglycerol; PE, phosphatidylethanolamine; SQDG, sulfoquinovosyldiacylglycerol; TGDG, trigalactosyldiacylglycerol.
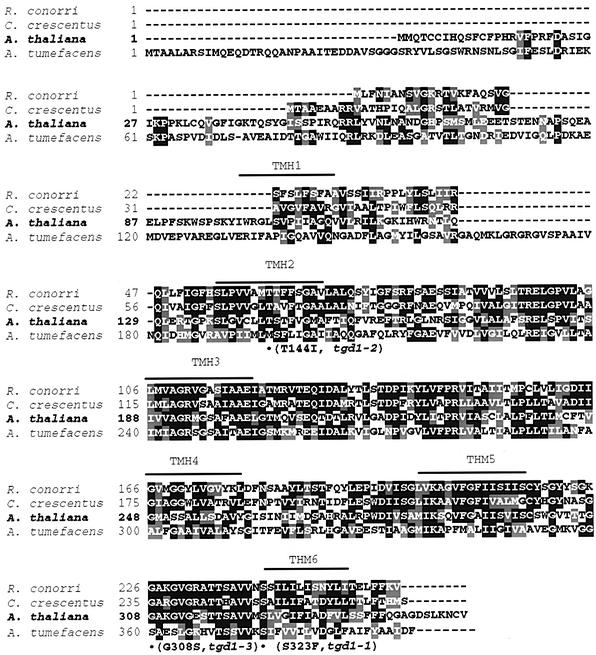
Submitting the predicted amino acid sequence for TGD1 to bioinformatic analysis, it became apparent that this protein is most similar (∼35% identity) to predicted, but still uncharacterized, ABC domain-lacking permease half-molecules of multipartite bacterial ABC transporter complexes (Higgins, 2001). A pileup is shown in Figure 8. As is typical for permease half-molecules, the protein is predicted to contain six putative transmembrane-spanning helices. All three alleles carry substitutions in conserved residues, two of the mutant alleles (tgd1-1, S323F; tgd1-2, T144I) in two of the six membrane-spanning helices, and one (tgd1-3, G308S) outside these helices.
Fig. 8. Comparison of the predicted Arabidopsis TGD1 permease protein and its orthologs in different bacteria. Proteins (GenBank accession Nos) are, from top to bottom: Rickettsia conorii (NP_359766), Caulobacter crescentus (NP_422489), Arabidopsis thaliana (NP_173410) and Agrobacterium tumefaciens (NP_532332). Black boxes indicate identical, gray boxes conserved amino acids. Predicted transmembrane helix domains (THMs) are indicated above the sequence with a bar. Amino acid substitutions in the Arabidopsis tgd1 mutant alleles are shown below the sequence.
TGD1 is localized at the outer chloroplast envelope
Expression of the TGD1 gene was tested by northern analysis and was high in all green tissues, but low in non-green tissues such as roots (data not shown). Young green seedlings showed very high expression of TGD1, consistent with a function of the gene product in providing precursors for photosynthetic membranes.
The predicted TGD1 protein does not contain an obvious transit peptide. To determine a possible chloroplast outer envelope location, which cannot be predicted from the primary sequence, in vitro translation of the wild-type cDNA followed by in vitro import experiments using isolated pea chloroplasts was pursued (Figure 9). In the presence of 0.1 mM ATP, a concentration typically required for binding of proteins to the outer envelope, no association of TGD1 with the membrane fraction was observed (Figure 9A). However, under import conditions in the presence of 4 mM ATP, TGD1 became associated with the chloroplast membrane fraction (Figure 9A). Subsequent experiments demonstrated that TGD1 could not be removed from the membranes by washing with a sodium carbonate solution, but could be readily extracted with detergent (data not shown). TGD1 was not processed upon insertion into the chloroplast membrane. Thermo lysin treatment of the chloroplasts showed that TGD1 is sensitive (Figure 9A). Typically, proteins in the outer chloroplast envelope such as DGD1 are sensitive to this protease (Froehlich et al., 2001), while proteins associated with the inner chloroplast membranes are protected. Indeed, TGD1 is digested by thermolysin, giving rise to a distinct protected proteolytic product, suggesting that portions of the protein are deeply imbedded in the membrane. Based on the presence of six predicted membrane-spanning domains, it seems certain that TGD1 is deeply buried, providing only limited access to the protease. Taken together, these results are in agreement with an outer envelope location of TGD1.
Fig. 9. Binding and insertion of TGD1 to isolated pea chloroplastic membranes. (A) TGD1; (B) the small subunit of rubisco, a typical stroma protein. 35S-labeled TGD1 or the precursor of the rubisco small subunit (pSS) was bound to pea chloroplasts under low ATP conditions or imported under high ATP conditions for 30 min at room temperature. After binding or import, chloroplasts were incubated with (+TL) or without (–TL) thermolysin for 30 min at 4°C. Chloroplasts were recovered by centrifugation through a 40% Percoll cushion and fractionated into a total membrane (P) and soluble (S) fraction. All fractions were isolated and analyzed by SDS–PAGE and fluorography. TP represents 10% of translated product added to a single import assay; mSS, mature form of rubisco small subunit.
Discussion
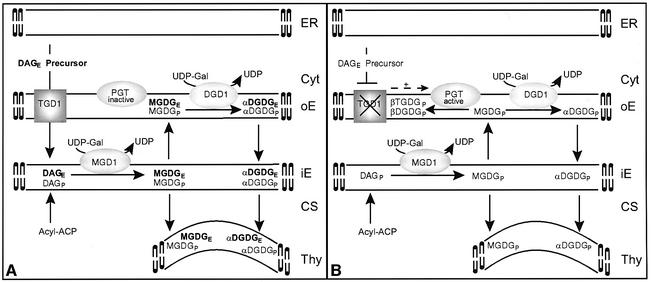
A model explaining the complex tgd1-1 biochemical phenotype and the molecular findings is shown in Figure 10. The facts on which this model is built are as follows: (i) the protein encoded by the locus mutated in tgd1 is similar to the intrinsic half pore protein (permease) of a bacterial ABC transporter complex; (ii) in vitro studies on the import of radiolabeled TGD1 into isolated chloroplast were consistent with TGD1 being an integral membrane protein of the outer chloroplast envelope; (iii) detailed compositional and positional analyses of the fatty acids in chloroplast lipids in the tgd1-1 mutant showed a drastic loss of ER-derived molecular species, such that >80% of lipids are derived from the plastid pathway in the mutant (50% in the wild type); (iv) in vivo pulse labeling of lipids showed that in the tgd1-1 mutant, the flux through the ER pathway of thylakoid lipid biosynthesis is reduced; (v) proton NMR of the new galactolipid accumulating in tgd1-1 revealed a difference in structure as compared with galactolipids typically found in the chloroplast; from that result, we proposed that a PGT different from MGD1 and DGD1 is involved; and finally, (vi) the enzymes responsible for the bulk of galactolipid biosynthesis, MGD1 and DGD1, and their association with the inner and outer envelopes, respectively, are well established (Awai et al., 2001; Froehlich et al., 2001). Thus, we have proposed that TGD1 is part of a lipid transporter in the outer envelope critical for the transfer of ER-derived molecular species to MGD1 (Figure 10A). Impairment of this function in the tgd1-1 mutant (Figure 10B) directly or indirectly activates a PGT, thereby producing TGDG.
Fig. 10. Simplified model for galactolipid biosynthesis in Arabidopsis wild type (A) and the tgd1 mutants (B). The endoplasmic reticulum (ER), the outer (oE) and inner (iE) chloroplast envelope and the thylakoids (Thy) are shown (Cyt, cytosol; CS, chloroplast stroma). Lipid species derived from the plastid and ER pathways are indicated by subscript P or E, respectively. In addition, ER-derived species are emphasized in bold. Molecular species from the two pathways can be distinguished based on their fatty acid composition. Greek letters indicate the anomeric proton configuration of the second galactose in TGDG or DGDG. The configuration is diagnostic for the respective enzyme. For simplicity, the export of fatty acids from the plastid is not shown. Four key proteins and their experimentally determined location are shown: MGD1, the predominant MGDG synthase; DGD1, the predominant DGDG synthase; PGT, a processive galactosyltransferase highly active in tgd1; TGD1, a putative membrane transporter mutated in tgd1. Paralogs of MGD1 (Awai et al., 2001) and DGD1 (Kelly et al., 2002) present in Arabidopsis are not considered here, because they may only be active under certain conditions or in specific tissues. Because the exact nature of the lipid returning from the ER is not known, the generic designation DAGE precursor is chosen. ACP; acyl carrier protein; DAG, diacylglycerol; DGDG, digalactosyldiacylglycerol; MGDG, monogalactosyldiacylglycerol; TGDG, trigalactosyldiacylglycerol; UDP-Gal, UDP-galactose.
It should be noted that the existence of a PGT in chloroplast envelopes that would be separate from the enzyme primarily responsible for DGDG biosynthesis has been discussed previously, however, without experimentally resolving the issue (e.g. Dorne et al., 1982; Heemskerk et al., 1990). It seems likely that the PGT described here is related or identical to the processive MGDG:MGDG galactosyltransferase previously observed in isolated chloroplasts. Its activity can be readily confused in in vitro experiments with the activity of DGD1/DGD2, of which DGD2 has been demonstrated to be non-processive and UDP-Gal dependent (Kelly et al., 2002). As shown here, the activity of PGT can be clearly distinguished from the activity of the major enzyme, DGD1, involved in DGDG biosynthesis, by analyzing the anomeric proton configuration of the products (Figure 2). However, more definitive experiments addressing questions of PGT activity, location and function will have to await the identification of the respective protein.
While all the data are consistent with the presented model shown in Figure 10, many questions remain to be answered. For example, what is the nature, function and activation mechanism of PGT? Interestingly, the growth of the homozygous tgd1-1/dgd1 double mutant is intermediate to that of wild type (Figure 7), and the growth of the homozygous tgd1-1 mutant is only mildly reduced (data not shown). Thus, the presence of TGDG or the increased amount of DGDG in the tgd1-1 mutant, even though DGDG in the mutant has a different structure, seems to compensate for the very low DGDG amounts normally present in the homozygous dgd1 mutant. Another set of important questions concerns the function of the TGD1 protein itself. What is the substrate for TGD1? Is TGD1 part of a larger lipid transport complex involving the ER and the two chloroplast envelopes? Unless all the essential components of the proposed TGD1 transporter complex have been identified, a reconstitution in vitro cannot be attempted and its substrates will not be known for certain. Therefore, at this time, we cannot rule out alternative models for TGD1 function. For example, TGD1 might be part of a receptor or transporter for a signaling molecule indirectly affecting lipid trafficking and PGT. It is anticipated that the answers to these questions can be found by studying the newly isolated TGDG-accumulating mutants and their respective gene products. Among these mutants may be those affected in other components of the transport apparatus, such as an ABC domain protein, directly interacting with TGD1 or a mutated form of PGT, leading to deregulation of the enzyme.
In conclusion, the discovery of a new class of lipid mutants using high-throughput lipid profiling, and the analysis of one of its members, led to a more detailed model for galactolipid biosynthesis and lipid trafficking in plant cells, which now can be rigorously tested. These new mutants also provide a novel means to study interorganelle lipid trafficking in Arabidopsis and possibly other organisms following the identification of orthologs of the respective gene products.
Materials and methods
Plant growth and mutant screening procedure for dgd1 suppressors
Arabidopsis thaliana plants used in this study were of the ecotypes Columbia (Col-2) or Landsberg erecta. Plants were grown as previously described (Xu et al., 2002). A total of 50 000 seeds from plants homozygous for the dgd1 mutant allele were mutagenized with 0.2% ethylmethane sulfonate (EMS, Sigma), as previously described (Dörmann et al., 1995). Following self-fertilization, M2 seeds were harvested as pools, each pool containing M2 seeds from ∼200 EMS-mutagenized M1 seeds. Approximately 15 000 M2 seedlings grown on MS agar medium containing 1% sucrose were screened in a 96-well format using a Tecan Genesis RSP 100 pipetting robot. The robot was equipped with a custom-made 20 × 20 cm TLC plate holder and holders for microtiter plates and solvent delivery containers. It was placed in an enclosed and vented cabinet and operated at room temperature. Single leaf samples were harvested manually into 96-well microplates (polypropylene, 0.5 ml well size with cap strips), which were placed on the robot. To each well, 200 µl of chloroform/methanol/formic acid (2:1:0.1, by vol.) were added, the wells were capped, and the plates were agitated for 5 min using a standard paint shaker. The plates were returned to the robot, uncapped, 100 µl of 1 M KCl, 0.2 M phosphoric acid were added, and the plates were recapped. The plates were centrifuged at ∼500 g for 5 min, uncapped and returned to the robot. A 15 µl aliquot of the lower phase was spotted by the robot at a rate of 2 µl/s onto a 20 × 20 cm TLC plate (Si250, Mallinckrodt Baker, NJ; activated at 120°C for 2 h directly before use) in a 20 × 20 spot grid. The plate was left on the robot for 5 min to allow the solvent to evaporate. For development, 20 µl of chloroform/acetone/methanol/acetic acid/water (50:20:10:10:5, by vol.) were applied to each spot at a rate of 1 µl/s. The plates were left to dry for 5 min and glycolipids were visualized with α-naphthol reagent (Benning et al., 1995).
Galactolipid structural analysis
Samples were purified by repeated TLC, and for NMR spectroscopy, 0.5–2 mg of lipid was dissolved in CDCl3/CD3OD/CD3OOD (10:10:1, by vol.). The addition of CD3OOD shifts the water signal to 5.5–6 p.p.m. Data were collected on a Varian VXR-500 spectrometer at 500 MHz for protons and analyzed with the Varian NMR software package. Chemical shifts were measured in relation to residual methanol as reference, defined as 3.30 p.p.m. For each sample, 128–512 FIDs were collected, depending on concentration. Suppression of the water signal was accomplished using a pre-saturating pulse in all mutant samples, which contained less lipid.
FAB-MS was conducted on a JEOL-HX-110 double focusing mass spectrometer operating in the negative or positive ion mode. Approximately 1 µg of lipid was mixed with 1 µl of matrix (3-nitrobenzyl alcohol). Ions were produced by bombardment with a beam of Xe atoms (6 KeV). The accelerating voltage was 10 kV and the resolution was set at 1000. Exact mass measurements were obtained by peak matching at 10 000 to a reference compound (bradykinin, exact mass of 1060.56900 m/z in positive FAB-MS) added to the sample.
Alditol acetates were prepared from the TGDG sample and analyzed as previously described (Benning et al., 1995).
Lipid analysis
Rosette leaves were frozen immediately in liquid nitrogen upon harvesting, and lipids were extracted as previously described (Dörmann et al., 1995). Lipid extracts were analyzed on activated ammonium sulfate-impregnated silica gel TLC plates (Si250 with pre-adsorbent layer, Mallinckrodt Baker, NJ) using a solvent system of acetone/toluene/water (91:30:7, by vol.). Lipids were visualized with iodine vapor and identified by co-chromatography with lipid extracts of known composition. For quantitative analysis, individual lipids were isolated from TLC plates and used to prepare fatty acid methyl esters. The methyl esters were quantified by gas–liquid chromatography (GLC) using myristic acid as internal standard (Rossak et al., 1997). The fatty acid composition at the sn-1 and sn-2 positions of the glycerol backbone was determined by Rhizopus arrhizus lipase digestion as described in Härtel et al. (2000).
Labeling of lipids
For in vivo labeling experiments, sodium [1-14C]acetate was diluted to 1.9 MBq/ml with water and was spread with a syringe in 3 µl droplets over the leaf surface of young plants (4–5 weeks old; 500 µl per 10 plants). [1-14C]Oleic acid (1.96 MBq/mmol) in 100 µl of ethanol was dispersed in 20 ml of MS medium (Murashige and Skoog, 1962) in standard culture dishes, and detached leaves from 4- to-5-week-old plants were incubated for 1 h at room temperature. The leaves were then washed twice in non-radioactive MS medium in order to remove exogenous radioactivity, and incubation in MS medium was continued. At various times after application of the label, samples of ∼0.15 g (fresh weight) were harvested and metabolism was halted by immersion in liquid nitrogen. Lipids were extracted and separated by TLC as described above. The silica zones corresponding to individual lipids were scraped into scintillation vials, and the radioactivity associated with the lipids was determined by liquid scintillation counting.
In vitro galactolipid biosynthesis in isolated chloroplasts
Chloroplasts were isolated by centrifugation through a discontinuous Percoll gradient as previously described (Xu et al., 2002). Pigments were determined as described by Lichtenthaler (1987). The purified chloroplasts were incubated with 0.5 mg/ml thermolysin in 0.33 M sorbitol, 50 mM HEPES pH 7.3 and 1.0 mM CaCl2 in the dark for 1 h at 22°C with a chlorophyll concentration of 1.0 mg/ml. After treatment, intact chloroplasts were repurified by centrifigation through a Percoll gradient as used for the isolation. Intact chloroplasts were resuspended in 0.3 M sorbitol, 50 mM HEPES pH 7.6, 5 mM EDTA, 1 mM MnCl2, 1 mM MgCl2. Total galactolipid synthesis in intact chloroplasts (50 mg Chl) was assayed by incubation with 18.5 KBq of UDP [14C]galactose (1.2 MBq/mmol; Amersham, Piscataway, NJ) in 150 µl of medium containing 0.33 M sorbitol, 50 mM HEPES pH 7.6, 5 mM EDTA, 1 mM MnCl2, 1 mM MgCl2 for 30 min. Lipids were extracted as above and separated by TLC. The lipids were visualized by autoradiography.
Genetic analysis and map-based cloning of the TGD1 locus
Homozygous tgd1-1 mutant plants were used for mapping. To separate the tgd1-1 mutation from the dgd1 mutation, we crossed the tgd1-1/dgd1 double mutant to wild-type Col-2, allowed these F1 plants to self-fertilize, and identified DGD1/DGD1 tgd1-1/tgd1-1 progeny in the F2 or F3 generation. Genotyping at the DGD1 locus throughout the study was accomplished by using a dCAPS marker (Neff et al., 1998) employing the PCR primers 5′-AATTGCTGAAGAGAGATCCCGTGGTGCA-3′ and 5′-CTTTGTCCAGGCATACCACGATCCGGC-3′, and the restriction enzyme ApaLI. Different independent mutant lines were tested for complementation in reciprocal test crosses followed by scoring of the F1 plants for lipid phenotypes.
For mapping purposes, tgd1-1 plants (Col-2 wild-type background) were crossed to wild-type plants of the Landsberg erecta ecotype. The F1 progeny were allowed to self-pollinate, and the F2 plants were scored for the presence of TGDG. DNA was prepared from the parental, F1 and 1215 F2 mutant progeny. The genetic linkage between tgd1-1 and molecular markers was determined by employing SSLP markers (Bell and Ecker, 1994) and cleaved amplified polymorphism (CAPS) markers (Konieczny and Ausubel, 1993). TGD1 was mapped to chromosome 1 between CAPS marker F15H18 and G2395. For fine mapping, nine new markers including six SSLPs and three dCAPS markers were generated: F6F9-1, 5′-TTGTTCTAGAGTGAGATTGGTTTCC-3′ and 5′-GAGTATGAGCCGCGTACGAT-3′; F6F9-2, 5′-TGGTCTCTCTCTTTGTGTTTATTT-3′ and 5′-GATCGGCGGAGGTATCATC-3′; F6A14, 5′-TCACCTAGACACCTACCAAGTGAA-3′ and 5′-GGGAGAGGATGGACCACTTT-3′; F15H18, 5′-AATACACAAAAATTACAGGGGC-3′ and 5′-TTCGTCTCTGTAGCGCGAAT-3′; T20H2-1, 5′-TTTGAGCTCCGGATTTGAGT-3′ and 5′-TGATTAAGGTTAAGCGGGAAA-3′; T20H2-2, 5′-TCGAATGATAAACTGAAATCCA-3′ and 5′-CTTTTAAAACCCTGTGAAATGA-3′; F6F9-3, 5′-AATCGAGCCGCACGCGCTCTTACGCG-3′ and 5′-GGCCGTGACTATTAGAACGACGTCG-3′, cut with MluI; F18O14-1, 5′-TCCACTAGCTTAATAGATGCGC-3′ AND 5′-GCCGGAATGTGAAATCTACAA-3′, cut with FspI; and F18O14-2, 5′-GTAGTTAGTTATCCTTCGA-3′ and 5′-TGAAACTCCAGCTGAATC-3′, cut with BstBI.
Cosmids carrying TGD1 were isolated from an Arabidopsis cosmid library constructed in the binary T-DNA cosmid vector pBIC20 (Meyer et al., 1996). Mutant plants were transformed with various cosmid clones using the floral dip method (Clough and Bent, 1998). The full-length coding sequence of wild-type TGD1 cDNA was amplified by RT–PCR using 5′-AAGGTACCATGATGCAGACTTGTTGTAT-3′ and 5′-CGTCTAGATCAAACACAGTTCTTCAAAG-3′. This fragment was ligated into the pPCR-Script Amp SK+ vector (Stratagene, La Jolla, CA) and the nucleotide sequence was determined by automated sequencing at the MSU Genomics Technology Faciltiy. The insert was excised with KpnI, taking advantage of the site in the vector and in one of the primers, and subsequently was cloned into the respective site of the binary vector pBINAR-Hyg (Dörmann and Benning, 1998). This construct was transformed into a homozygous tgd1-1/dgd1 double mutant as described above. Transformants were selected on agar-solidified MS medium containing 1% (w/v) sucrose and 25 µg/ml hygromycin B. At least 20 transformants were obtained. The lipid phenotype was determined by TLC analysis of lipid extracts. The presence of the transgene construct was confirmed by PCR using the primers 5′-ATGATGCAGACTTGTTGTATCCA-3′ and 5′-CACAGT TCTTCAAAGAATCTCC-3′, which amplified both genomic DNA of At1g19800 and the transformed cDNA. The presence of a point mutation was tested by digesting the PCR fragments with BsmAI, since the mutation in tgd1-1 disrupts one of the two BsmAI digesting sites in At1g19800.
Chloroplast import analysis
Pea plants (Pisum sativum var. Little Marvel; Olds Seed Co., Madison, WI) were grown under natural light in the greenhouse at 18–20°C. Chloroplasts were isolated from 8- to 12-day-old plants as previously described (Bruce et al., 1994). TGD1 and rubisco small subunit genes were transcribed, translated, and labeled with [35S]methionine using the TNT-coupled reticulocyte lysate system (Promega Corp., Madison, WI). Binding or import reactions (adapted from Bruce et al., 1994) received 500 000 d.p.m. of radiolabeled translation product and 0.1 mM ATP for binding or 4 mM ATP for translocation following the addition of intact chloroplasts corresponding to 25 µg of chlorophyll in a final volume of 150 µl. Treatment of binding and import reactions with thermolysin was performed as described by Cline et al. (1984). Briefly, binding and translocation reactions were incubated with thermolysin for 30 min on ice, in the dark and quenched with 5 mM EDTA for 5 min. Intact chloroplasts were then recovered by sedimentation through a 40% (v/v) Percoll cushion. The pellets were resuspended in lysis buffer (25 mM HEPES-KOH pH 8.0, 4 mM MgCl2), and incubated on ice for 15 min. After ultracentrifugation at 100 000 g, total membrane and soluble fractions were obtained. All fractions were analyzed by SDS–PAGE (Laemmli, 1970) and fluorography.
Acknowledgments
Acknowledgements
We are grateful to Beverly Chamberlain for her expert mass spectral analysis at the MSU Mass Spectrometry Facility, and Alex Cernac for his advise during mapping. This work was supported by grants from the US Department of Energy and from the MSU Center for Novel Plant Products.
References
- Awai K., Marechal,E., Block,M.A., Brun,D., Masuda,T., Shimada,H., Takamiya,K., Ohta,H. and Joyard,J. (2001) Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA, 98, 10960–10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C.J. and Ecker,J.R. (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics, 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Benning C., Huang,Z.H. and Gage,D.A. (1995) Accumulation of a novel glycolipid and a betaine lipid in cells of Rhodobacter sphaeroides grown under phosphate limitation. Arch. Biochem. Biophys., 317, 103–111. [DOI] [PubMed] [Google Scholar]
- Browse J., Warwick,N., Somerville,C.R. and Slack,C.R. (1986) Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the ‘16:3’ plant Arabidopsis thaliana. Biochem. J., 235, 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce B.D., Perry,S., Froehlich,J. and Keegstra,K. (1994) In vitro import of protein into chloroplasts. In Gelvin,S.B. and Schilperoort,R.A. (eds), Plant Molecular Biology Manual. Kluwer, Boston, MA, pp. 1–15.
- Cline K., Werner-Washburne,M., Andrews,J. and Keegstra,K. (1984) Thermolysin is a suitable protease for probing the surface of intact pea chloroplasts. Plant Physiol., 75, 675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J. and Bent,A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J., 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dörmann P. and Benning,C. (1998) The role of UDP-glucose epimerase in carbohydrate metabolism of Arabidopsis. Plant J., 13, 641–652. [DOI] [PubMed] [Google Scholar]
- Dörmann P. and Benning,C. (2002) Galactolipids rule in seed plants. Trends Plant Sci., 7, 112–118. [DOI] [PubMed] [Google Scholar]
- Dörmann P., Hoffmann-Benning,S., Balbo,I. and Benning,C. (1995) Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell, 7, 1801–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann P., Balbo,I. and Benning,C. (1999) Arabidopsis galactolipid biosynthesis and lipid trafficking mediated by DGD1. Science, 284, 2181–2184. [DOI] [PubMed] [Google Scholar]
- Dorne A.-J., Block,M.A., Joyard,J. and Douce,R. (1982) The galactolipid:galactolipid galactosyltransferase is located on the outer surface of the outer membrane of the chloroplast envelope. FEBS Lett., 145, 30–34. [Google Scholar]
- Frentzen M. (1986) Biosynthesis and desaturation of the different diacylglycerol moieties in higher plants. J. Plant Physiol., 124, 193–209. [Google Scholar]
- Froehlich J.E., Benning,C. and Dörmann,P. (2001) The digalactosyl diacylglycerol (DGDG) synthase DGD1 is inserted into the outer envelope membrane of chloroplasts in a manner independent of the general import pathway and does not depend on direct interaction with monogalactosyldiacylglycerol synthase for DGDG biosynthesis. J. Biol. Chem., 276, 31806–31812. [DOI] [PubMed] [Google Scholar]
- Giroud C., Gerber,A. and Eichenberger,W. (1988) Lipids of Chlamydomonas reinhardtii. Analysis of molecular species and intracellular site(s) of biosynthesis. Plant Cell Physiol., 29, 587–595. [Google Scholar]
- Härtel H., Dörmann,P. and Benning,C. (2000) DGD1-independent biosynthesis of extraplastidic galactolipids following phosphate deprivation in Arabidopsis. Proc. Natl Acad. Sci. USA, 97, 10649–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemskerk J.W.M., Storz,T., Schmidt,R.R. and Heinz,H. (1990) Biosynthesis of digalactosyldiacylglycerol in plastids from 16:3 and 18:3 plants. Plant Physiol., 93, 1286–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C.F. (2001) ABC transporters: physiology, structure and mechanism—an overview. Res. Microbiol., 152, 205–210. [DOI] [PubMed] [Google Scholar]
- Joyard J., Marechal,E., Miege,C., Block,M.A., Dorne,A.J. and Douce,R. (1998) Structure, distribution and biosynthesis of glycerolipids from higher plant chloroplasts. In Siegenthaler,P.-A. and Murata,N. (eds), Lipids in Photosynthesis: Structure, Function and Genetics. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 21–52.
- Kelly A.A. and Dörmann,P. (2002) DGD2, an Arabidopsis gene encoding a UDP-galactose-dependent digalactosyldiacylglycerol synthase is expressed during growth under phosphate-limiting conditions. J. Biol. Chem., 277, 1166–1173. [DOI] [PubMed] [Google Scholar]
- Kojima M., Seki,K., Ohnishi,M., Ito,S. and Fujino,Y. (1990) Structure of novel glyceroglycolipids in Adzuki bean (Vigna angularis) seeds. Biochem. Cell Biol., 68, 59–64. [PubMed] [Google Scholar]
- Konieczny A. and Ausubel,F.M. (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J., 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Kunst L., Browse,J. and Somerville,C. (1988) Altered regulation of lipid biosynthesis in a mutant Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc. Natl Acad. Sci. USA, 85, 4143–4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (1987) Chlorophylls and carotenoids: pigments of photosynthetic membranes. Methods Enzymol., 148, 350–382. [Google Scholar]
- Meyer K., Benning,G. and Grill,E. (1996) Cloning of plant genes based on the genetic map position. In Patterns,A.H. (ed.), Genome Mapping in Plants. Academic Press, New York, NY, pp. 137–154.
- Mongrand S., Besoule,J.-J., Cabantous,F. and Cassagne,C. (1998) The C16:3/C18:3 fatty acid balance in photosyntheitc tissues from 468 plant species. Phytochemistry, 49, 1049–1064. [Google Scholar]
- Murashige T. and Skoog,F. (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant., 15, 473–497. [Google Scholar]
- Neff M.M., Neff,J.D., Chory,J. and Pepper,A.E. (1998) dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J., 14, 387–392. [DOI] [PubMed] [Google Scholar]
- Rossak M., Schäfer,A., Xu,N., Gage,D.A. and Benning,C. (1997) Accumulation of sulfoquinovosyl-1-O-dihydroxyacetone in a sulfolipid-deficient mutant of Rhodobacter sphaeroides inactivated in sqdC. Arch. Biochem. Biophys., 340, 219–230. [DOI] [PubMed] [Google Scholar]
- Roughan P.G. and Slack,C.R. (1982) Cellular organization of glycerolipid metabolism. Annu. Rev. Plant Physiol., 33, 97–132. [Google Scholar]
- Roughan P.G., Thompson,G.A. and Cho,S.H. (1987) Metabolism of exogenous long-chain fatty acids by spinach leaves. Arch. Biochem. Biophys., 259, 481–496. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- van Besow A. and Wintermans,J.F.G.M. (1978) Galactolipid formation in chloroplast envelopes I. Evidence for two mechanisms in galactosylation. Biochim. Biophys Acta, 529, 44–53. [DOI] [PubMed] [Google Scholar]
- Xu C., Härtel,H., Wada,H., Hagio,M., Yu,B., Eakin,C. and Benning,C. (2002) The pgp1 locus of Arabidopsis encodes a phosphatidylglycerol synthase with impaired activity. Plant Physiol., 129, 594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]