Abstract
Aggregated misfolded proteins are hallmarks of most neurodegenerative diseases. In a chronic disease state, including pathologic situations of oxidative stress, these proteins are sequestered into inclusions. Accumulation of aggregated proteins can be prevented by chaperones, or by targeting their degradation to the UPS. If the accumulation of these proteins exceeds their degradation, they may impair the function of the proteasome. Alternatively, the function of the proteasome may be preserved by directing aggregated proteins to the autophagy-lysosome pathway for degradation. Sequestosome 1/p62 has recently been shown to interact with polyubiquitinated proteins through its UBA domain and may direct proteins to either the UPS or autophagosome. P62 is present in neuronal inclusions of individuals with Alzheimer's disease and other neurodegenerative diseases. Herein, we review p62's role in signaling, aggregation, and inclusion formation, and specifically as a possible contributor to Alzheimer's disease. The use of p62 as a potential target for the development of therapeutics and as a disease biomarker is also discussed.
INTRODUCTION
Many neurodegenerative diseases such as polyglutamine repeat diseases, Lewy bodies in Parkinson disease, Alzheimer's disease, tauopathies, and others share abnormal accumulation of ubiquitinated proteins into aggregates and inclusions as a hallmark feature of the disease pathology. The molecular basis for the accumulation of these inclusions remains poorly defined; however the aberrant accumulation of aggregated proteins and disturbances in protein degradation suggest a common underlying mechanism. These inclusions share amyloid-like structure and several biochemical features such as: ubiquitin, aggregated proteins, proteasome subunits, chaperones, and other proteins that become trapped through their association with aggregated proteins. Here we review the role of aggregates, protein turnover, and the ubiquitin proteasome system (UPS), and focus on the role of a recently discovered proteasomal shuttling protein, sequestosome 1/p62, and its role in neurodegeneration. We also discuss the potential of employing p62 as a biomarker for neurodegenerative disease and as a potential target for therapeutic development.
MISFOLDING AND THE UPS PATHWAY
In normal cells, large amounts of newly synthesized proteins are defective “off-pathway” products. Even with abundant molecular chaperones, nearly 30% of nascent proteins are misfolded due to mutations or inefficient assembly [1]. To correct these mistakes, misfolded proteins can either be degraded via the ubiquitin proteasome pathway (UPS) shortly after their synthesis or they may form aggregates of high molecular weight oligomers [2]. The ultimate fate of misfolded proteins depends on kinetic partitioning between these two competitive pathways [3]. Because aggregates are more stable than the improperly folded protein, to degrade misfolded substrates effectively the proteasome must win the competition for the misfolded substrates before they have an opportunity to aggregate. Under normal conditions, accumulated proteins are removed promptly before any damage can be caused to the cell. However under certain situations in nerve cells, accumulated proteins are prone to form inclusion bodies which are the hallmarks of several neurodegenerative diseases [4, 5]. Increasingly it is becoming apparent that these inclusions/aggresomes may be the sites for sequestration of aggregated proteins. Herein, we will attempt to clarify the function and toxicity of protein aggregates and inclusions.
The UPS is responsible for the degradation of proteins and it serves as a cellular quality control system that tags misfolded proteins with ubiquitin for degradation by the 26S proteasome. Protein degradation via UPS involves two steps: (1) covalent attachment of polyubiquitin chains to target proteins, and (2) degradation of the tagged proteins by 26S proteasome complex with release of free and reusable ubiquitin (Figure 1). Ubiquitin is a protein that forms different chains with itself [6, 7] and serves as a signal through covalent attachment to other proteins. Three enzymes are involved in ubiquitination of substrates, eventually resulting in the formation of a bond between the C-terminus of ubiquitin (Gly76) and the ε-amino group of a substrate lysine residue. Ubiquitin-activating enzyme (E1) forms a thiol ester with the carboxyl group of Gly76, activating the C-terminus of ubiquitin. The activated ubiquitin molecule is carried by ubiquitin-conjugating enzyme (E2) and transferred to the substrate lysine residue by ubiquitin-ligases (E3) (Figure 2) [8]. Additional ubiquitin molecules can be added to form polyubiquitin chains. The terminal carboxyl of each ubiquitin is linked to the ε-amino group of a lysine residue of an adjacent ubiquitin in the chain.Ubiquitin can form chains in vivo at all seven lysine residues (K6, K11, K27, K33, K29, K48, K63) (Figure 3) [9]. Polyubiquitin chains linked through K48 are a primary signal for protein degradation [8]. By comparison, K63-linked chains are involved in DNA repair, ribosome function, mitochondrial DNA inheritance, the stress response and targeting of proteins for endocytosis [8]. However, it should be noted that a model substrate tagged with K63-linked tetra-ubiquitin could effectively signal substrate degradation [10]. A chain of at least four ubiquitin moieties attached to a target protein are required for substrate recognition and subsequent degradation by the 26S proteasome [11].
Figure 1.
The pathway of ubiquitin-linked degradation of proteins and the 26S proteasome (see [84]).
Figure 2.
A schematic representation of substrate ubiquitination (see [85]).
Figure 3.
(a) Ubiquitin can be added as a single molecule at one or more sites. Alternatively, the branched chains of polyubiquitin may be formed. (b) Space-filled model of ubiquitin indicating the seven lysine residues (see [86]).
The ubiquitin conjugation cascade contains a large family of E2s and an even larger set of E3s. For example, in budding yeast there is one E1, eleven E2s, and more than twenty E3s [7]. The large number of E3 enzymes may reflect the extraordinary diversity of the ubiquitinated substrates in eukaryotes. All E3 enzymes belong to three protein families: homologous to E6AP carboxy terminus (HECT), really interesting new gene (RING), and UFD2 homology (U-box) proteins. Those E3s share a common E2-binding domain and a substrate-interacting domain. One remarkable feature of the ubiquitin conjugation pathway is the modulation of target protein selection. The substrate specificity depends mainly on the identity of E3 [7]. On the other hand, biochemical studies have shown that the identity of E2 can influence the recognition of specific structures of a polymeric ubiquitin modification [12], indicating that the specificity of the E2/E3 interaction may determine the final selection of the target substrate.
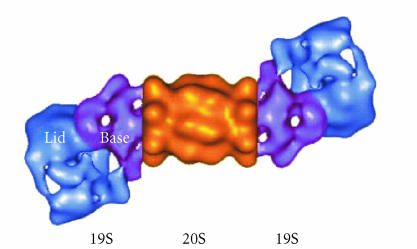
The 26S proteasome is a multimeric protease complex that plays a central role in protein degradation through both ubiquitin-dependent and ubiquitin-independent mechanisms. The 26S proteasome complex consists of a 20S core particle which is proteolytically active, and one or two 19S regulatory caps which are responsible for recognition, unfolding, deubiquitination, and translocation of substrate proteins into the lumen of the core particle (Figure 4) [13].
Figure 4.
The structure of the 26S proteasome complex. (http://www.biw.kuleuven.be/dtp/cmpg/pgprb.htm.)
The 20S proteolytic core consists of four stacked rings with two outer α-rings embracing two central β-rings. The outer rings are each composed of seven different alpha subunits and the inner rings of seven different beta subunits (Figure 5). The overall structure of the 20S core resembles a barrel with dimensions of 15 nm in length and 11 nm in diameter. A central proteolytic chamber is formed by two face-to-face β-rings and is separated by 3 nm wide β-annuli. Three subunits β1, β2, and β5 form the catalytic site. β-subunits gain proteolytic activity by autolytic processing of the N-terminal propeptides and the exposure of a critical threonine residue. Access to the chamber requires reorganization of the N-terminal H0 helices of the α-subunits which normally form a seal by interacting with side chains. The N-terminus of the α3-subunit plays a critical role in the seal formation. Addition of the 19S cap can induce channel opening, and the 19S ATPase, Rpt2, plays a key role in this process.
Figure 5.
The structure of the 20S core and 19S regulatory particle. (http://plantsubq.genomics.purdue.edu/plantsubq/html/guide.html.)
Another component of the 26S proteasome, known as the 19S regulator, which is composed of 17 or 18 subunits, is responsible for recognition, unfolding, deubiquitination, and translocation of substrate proteins into the lumen of the core particle, where the substrate is degraded [13]. In high salt concentrations, the 19S regulator breaks down into two subcomplexes; the lid and the base. The base consists of 6 ATPases (Rpt1 to Rpt6) that share a high level of similarity to one another. The ATPases form a six-membered ring that interact directly with the α-ring of the 20S proteasome. Protein substrates need to pass through the center of this ring in order to enter the catalytic chamber of 20S proteasome. In addition, the ATPase ring is involved in the antichaperon activity required to unfold the protein substrates. The base also includes non-ATPase subunits, such as Rpn10 (S5a), which contains an ubiquitin interacting motif (UIM). The lid subcomplex consists of eight non-ATPase subunits, where Rpn11 plays a key role in the recycling of ubiquitin by cleaving the ubiquitin chain from the protein substrate.
In addition to the standard proteasomes, cells are able to produce immunoproteasomes as a transient response to cy-tokines IFN-γ or TNF-α [14]. IFN-γ induces biosynthesis of proteasome maturation protein (POMP) and proteasomal β5i subunit low molecular weight protein 7 (LMP7), accelerating the assembly of the immunoproteasome in which three catalytic subunits are replaced by homologous subunits (β1i, β2i, β5i) [15]. The immunoproteasome may participate in generating antigenic peptides displayed on MHC-class I molecules [16], but are not limited to this function. Recent observations reveal that in familial amyotrophic lateral sclerosis (fALS) patients, impaired degradation of mutant SOD1 is associated with a decrease in the constitutive proteasomes and an increase in the immunoproteasome level, resulting in selective motor neuron degeneration [17]. In Huntington's disease (HD), high levels of immunoproteasome subunits (LMP2 and LMP7) have also been observed, and are associated with neurodegeneration, indicating that immunoproteasomes may play a yet to be defined role in the pathogenesis of neurodegenerative diseases [18].
TARGETING POLYUBIQUITINATED PROTEINS FOR TURNOVER
Polyubiquitin chains are a signal that targets proteins for degradation by the proteasome complex. Recognition of polyubiquitinated proteins by the 26S proteasome plays a critical role in protein degradation. Presentation of a polyubiquitinated substrate to the 26S proteasome takes place through ubiquitin-interacting proteins, such as S5a, Dsk2, Rad23, and p62 [19, 20] (Figure 6). Ubiquitin-interacting proteins involved in ubiquitination/deubiquitination generally have ubiquitin-association (UBA) domains that can directly bind to ubiquitin (Figure 7). Previous studies have shown that most UBA domains bind the polyubiquitin chains rather than the monoubiquitin ones. Some UBA domains even discriminate further binding K63-linked polyubiquitin chains rather than K48-linked chains [21]. However, the interaction between ubiquitin and the UBA domain is a low-affinity interaction. NMR chemical shift mapping shows that ubiquitin specifically, but weakly, binds to a conserved hydrophobic epitope on the UBA domain, while the UBA domains can bind to the hydrophobic patch on the surface of the five-stranded β-sheet of ubiquitin with different orientations [22]. The weak interaction may enable rapid assembly and disassembly between polyubiquitin and the shuttling protein. UBA-containing proteins might contribute to the regulated capture and transient stabilization of proteins that are otherwise constitutively degraded [23]. Recent findings have shown that a functional UBA domain is required for the localization of these shuttling proteins into aggregates [24], suggesting a common mechanism of ubiquitin-mediated sequestration of essential ubiquitin-binding proteins into aggregates.
Figure 6.
Presentation of ubiquitinated substrates to the 26S proteasome (see [87]).
Figure 7.
Proteins containing UBA domains have diverse structure and function (see [86]).
In addition to the UBA domains that bind the polyubiquitin chains, shuttling proteins commonly contain a ubiquitin-like domain (UbL) that binds the proteasome [20, 25, 26]. These proteins are able to shuttle the polyubiquitinated substrates to the 26S proteasome for degradation [27]. Ataxin-3, a proteasome-associated factor, has been shown to interact with the shuttling protein Rad23 to mediate the degradation of ubiquitinated substrates, suggesting an important role for shuttling proteins in the UPS [28]. Since each type of polyubiquitin chain forms a different conformation [29], and each UBA domain may recognize specific types of polyubiquitin chains, shuttling the protein may present chain-specific polyubiquitinated substrates to the proteasome for degradation. A recent study of the polyubiquitin interaction properties of thirty UBA domains reveals that these domains can be classified into four groups [30]: those with linkage specific characteristics, those which bind different chains, those which are nondiscriminatory, and those which do not bind ubiquitin. Moreover, it is possible that non-UBA sequences may modulate interaction properties in the UBA domain [30].
PROTEIN AGGREGATES AND NEURODEGENERATION
Protein turnover is dependant on a functional UPS. Failure to remove the polyubiquitinated proteins may lead to the accumulation of aggregated proteins [31]. The capacity of the ubiquitin proteasome pathway can be exceeded either by overexpression of substrates or by a decrease in proteasome activity. In cultured cells, proteasomal inhibitors can cause the aggregation of an overexpressing disease-associated protein [32], indicating that dysfunction of the proteasome might be a factor that initiates the formation of inclusions. A recent study has shown that a wide-range of nondisease-associated proteins is found in inclusions when cells were treated with proteasomal inhibitors. These proteins include ubiquitinated or nitrated α-tubulin, SOD-1, α-synuclein, and 68K neurofilaments [33]. Transient expression of two unrelated aggregation-prone proteins caused nearly complete inhibition of the UPS, indicating that protein aggregation can directly impair the UPS function [34]. A positive-feedback mechanism has been proposed to explain the turnover point of protein aggregation. Impaired proteasome function may result in an increase in protein aggregates, which leads to a further decline of proteasome activity. It is important to note that UPS impairment is not the result of steady-state sequestration of the UPS components, or simple substrate competition [35]. Direct physical interaction between proteasome and aggregates is not required for UPS impairment, indicating that protein aggregates may influence the activity of the proteasome in a currently unknown manner.
Aggregated proteins may sequester to form inclusions also referred to as an aggresome, where molecular chaperones, proteasome subunits, ubiquitin, and intermediate filament (IF) proteins colocalize [3]. The formation of aggresomes occurs at the microtubule organizing center (MTOC) and is considered to be a process distinct from the protein aggregation [36, 37]. The formation of cytoplasmic inclusion bodies requires active transport of misfolded proteins along microtubules, with redistribution of the IF protein to form a cage surrounding the core of aggregated, ubiquitinated protein [31]. Protein misfolding can be prevented or even reversed by chaperones. If chaperone activity fails, then the proteins must be degraded before aggregation takes place. Thus, aggresomes may serve as sites to sequester polyubiquitinated/misfolded proteins. In this regard, the UPS functions as a sensor to control degradation of misfolded proteins that tend to aggregate through exposure of hydrophobic sequences [31]. Should excessive amounts of protein aggregates accumulate, they may negatively impair the function of the UPS [34, 35]. Thus, it is critical to sequester aggregated proteins so as to preserve the UPS function.
Recent evidence has shown that early protein aggregates may be toxic to neuronal cells. Soluble dimers and trimers, and protofibrils or fibrils of amyloid beta (Aβ) peptide are cytotoxic [38]. On the other hand, although the early pre-fibrillar disease-associated protein aggregates are harmful to cells, the mature fibrils are relatively harmless [39]. In addition, a previous study reveals that UPS impairment is independent of inclusion body formation [35]. Altogether, these observations suggest that the inclusions/aggresome pathway are likely to be cytoprotective by recruiting misfolded proteins effectively isolating them within the cell. In further support of this mechanism, it has been reported that inclusion body formation predicts improved survival and leads to decreased levels for the highly aggregating protein huntingtin [40].
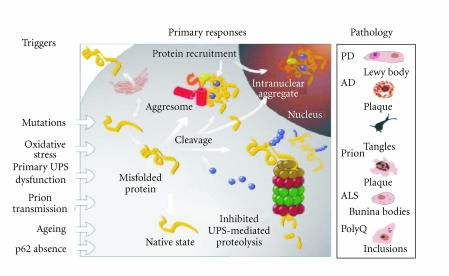
The accumulation of protein aggregates and formation of inclusion bodies are associated with many age-related neurodegenerative diseases, such as Alzheimer's disease (AD), Parkinson's disease (PD), Huntington's disease, and others, suggesting that there are direct links between protein aggregates and the resulting pathology [4] (Figure 8). The accumulation of ubiquitin conjugates may reflect the failed attempt of the UPS to remove damaged proteins [41]. An important component linked to aberrations in UPS and to the pathogenesis of Parkinson's disease is Parkin, an E3 ubiquitin ligase [42]. A previous report reveals that Parkin can associate with Rpn10 (S5a), a subunit of the 26S proteasome, indicating that Parkin may transfer ubiquitin conjugates for proteasomal degradation [43]. Defects in Parkin may result in the accumulation of its substrates, such as α-synuclein, and contribute to the pathogenesis of PD.
Figure 8.
The UPS and pathogenesis of neurodegeneration (see [41]).
There is a strong evidence demonstrating that proteasome inhibition by pharmacological treatment enhances inclusion formation in cellular models. However, there is less support for the notion that aggregates directly inhibit the proteasome in any disease state. More studies are needed in animal models to critically evaluate the role of protein aggregates and inclusions on proteasome function.
SEQUESTOSOME 1/P62, TRAFFICKING, AND INCLUSION FORMATION
Other aggregate-prone proteins participate in neurodegenerative diseases such as Alzheimer's disease (AD) (Figure 8). There are two types of protein deposits in AD: extracellular amyloid plaques rich in Aβ peptides, and intracellular neurofibrillary tangles containing hyperphosphorylated polyubiquitinated tau [44]. Previous study in our lab has shown that p62 can shuttle K63-polyubiquitinated tau for proteasomal degradation. Disturbing tau trafficking may result in the accumulation of insoluble/aggregated tau in the brain, contributing to AD [45]. Aβ peptides are produced by proteolytic cleavage of the amyloid precursor peptide (APP). In solution, Aβ peptides may undergo self-assembly leading to the transient appearance of soluble protofibrils and eventually to insoluble fibrils [46]. Recent proteomic study of amyloid plaques recovered from AD brain revealed that a total of 488 proteins coisolated with plaques. Moreover, 26 proteins were enriched in plaques by comparison with surrounding non-plaque tissues, including proteins involved in cell adhesion, cytoskeleton and membrane trafficking, chaperones, kinase/phosphatase, and regulators [47].
Sequestosome 1/p62 is a highly conserved protein that was initially identified as a phosphotyrosine-independent ligand of the src homology 2 (SH2) domain of p56lck [12]. Sequestosome 1/p62 contains a ubiquitin-associated (UBA) domain at its C-terminus (Figure 9), which can selectively bind K63-polyubiquitinated proteins [20]. The ability of p62 to interact selectively with K63-polyubiquitinated proteins [20] supports the idea that sequences in the holoprotein may modulate the UBA interaction properties [30]. The UBA domain of the human p62 protein forms a compact three-helix bundle. A Pro392 → Leu substitution mutation can modify the UBA domain by extending the N terminus of helix 1. This modification affects interaction of p62's UBA domain with polyubiquitin chain binding, and may contribute to Paget's disease of bone [49]. In addition, recent studies reveal that p62 protein lacking a UBA domain fails to form aggregates in HEK cells with impaired survival responses. This indicates that the UBA domain is critical for sequestering polyubiquitinated proteins [20], which is in keeping with a role for sequestration of polyubiquitinated proteins as an underlying contributor to inclusion formation [24].
Figure 9.

A schematic diagram showing the domain organization of p62 protein.
Sequestosome 1/p62 also contains a PB1 domain that binds aPKC, a ZZ finger, a binding site for the RING finger protein TRAF6, and two PEST sequences (Figure 9) [48]. With multiple protein-protein interaction motifs, p62 is also considered a scaffold [48], and has been extensively studied in the context of neurotrophin signaling. The N-terminus of p62 protein can directly interact with the proteasome subunit component [20], and the localization of protein substrates to the proteasome is sufficient for degradation [50]. Thus, p62 is also viewed as a shuttling protein, playing an important role in sequestering polyubiquitinated substrates, interacting with ubiquitinated substrates through its UBA domain and the proteasome through its N-terminus [51]. In support of a shuttling role for p62, we have observed that p62 is necessary for both tau and TrkA interaction with the proteasome [45, 51]. In addition, we find that p62 −/− mice possess AD-like neurodegeneration [Babu and Wooten, unpublished]. Likewise depletion of nerve growth factor (NGF) results in AD-like neurodegeneration in anti-NGF transgenic mice [52]. Interestingly, decreased membrane TrkA expression has been correlated with decline in performance on the mini mental state exam and may serve as a marker for late stage AD [53]. The correlation between the phenotype of the p62 −/− mice and the anti-NGF mice is suggestive of an overlap in pathways wherein p62 and NGF function. Further studies will be needed to sort out the exact mechanism whereby p62 regulates the trafficking of TrkA.Likewise depletion of nerve growth factor (NGF) results in AD-like neurodegeneration in anti-NGF transgenic mice [52]. Interestingly, decreased membrane TrkA expression has been correlated with decline in performance on the mini mental state exam and may serve as a marker for late stage AD [53]. The correlation between the phenotype of the p62 −/− mice and the anti-NGF mice is suggestive of an overlap in pathways wherein p62 and NGF function. Further studies will be needed to sort out the exact mechanism whereby p62 regulates the trafficking of TrkA.
The N-terminal PB1 domain of p62 is involved in p62 self-interaction, and in interaction with other proteins that possess a PB1 domain [54]. However, the PB1 domain can assume a ubiquitin fold and this may be the basis of the N-terminus of p62 interacting with the proteasomal subunit S5a [20]. Overexpression of p62 results in large inclusions, while depletion of p62 retards protein degradation and leads to accumulation of nondegraded aggregated polyubiquitinated proteins [20]. We have shown that cells which possess p62 inclusions possess enhanced survival characteristics [55]. This finding supports the growing idea that inclusions are sites for sequestration of misfolded proteins that are being triaged for degradation. In this regard, p62 has been localized to ubiquitin containing inclusions in Alzheimer's disease containing tau [56]. Since p62 has been shown to be necessary for tau interaction with the proteasome [45], it is possible that polyubiquitinated tau may accumulate in the absence of p62. Studies are underway to examine p62's role in tau trafficking. Culture of mouse embryo fibroblasts from wild type or p62 −/− mice challenged with either chloroquine, a lysosomal inhibitor, or MG132, a proteasomal inhibitor, reveals that p62 is necessary for inclusion formation under proteasome impaired conditions (Figure 10). Altogether, these findings suggest that p62 plays a key role in trafficking, regulation of aggregation and inclusion body formation. It is likely that p62 containing inclusions observed in AD and other neurodegenerative diseases contain proteins destined for degradation. In the absence of p62 these proteins would be expected to accumulate in their misfolded polyubiquitinated state and contribute to neurodegeneration.
Figure 10.
Mouse embryo fibroblasts (MEF) from wild type (WT) or p62 −/− mice were treated with chloroquine or MG132 as shown. After 24 hours, the cells were fixed and stained with primary antibody to ubiquitin and secondary antibody to Texas Red.
The genomic structure of p62 reveals the presence of a CpG island and multiple binding sites for SP-1, AP-1 NF-κB, and Ets-1 family transcription factors in the promoter region, suggesting that p62 transcription may be regulated by these factors [57]. In this regard either inhibition of the proteasome or increases in free radicals have been shown to induce p62 expression [58]. Moreover, inhibition of p62 transcription blocked proteasomal-induced sequestration of ubiquitinated proteins, and the enlargement of inclusions [59]. These studies support the idea that p62 is intimately involved in the formation of inclusions and in the protection of cells from the toxicity of misfolded proteins by enhancing inclusion formation [20, 55]. Thus, inclusions may arise as a protective mechanism against stress conditions. Indeed, we find that cells overexpressing p62 possess large inclusions and enhanced survival [20, 55].
Sequestosome 1/p62 may also act as a scaffold of TRAF6 [48, 60]. In this regard, p62 could serve as a site for TRAF6 dependent K63-polyubiquitination of target substrates and in the activation of transcription factor NF-κB. Interestingly, TRAF6 colocalizes into inclusions along with p62 [20]. A recent study in our lab has shown that the p62-UBA domain is required for TRAF6 polyubiquitination, suggesting that p62 may carry ubiquitin chains needed for the activation/autoubiquitination of TRAF6 [60]. When the interaction between p62 and TRAF6 was disrupted by competitive inhibitory peptide, the formation of p62/TRAF6 inclusions in cultured cells was suppressed and survival diminished [20, 45]. Therefore, p62 regulates activation of NF-κB through recognition of TRAF6-catalyzed polyubiquitin chains and/or recruitment of TRAF6 to a microenvironment to enhance protein ubiquitination.
In addition, p62 can form a ternary complex with aPKC and PAR-4 (a stress induced transcript) [61]. Interaction of aPKC with PAR-4 blocks its activity; however, p62 can antagonize PAR-4-induced aPKC inhibition [61]. Moreover, recent studies have shown that p62 can modulate association of Akt with aPKC [62]. Thus, p62 through interaction with aPKC likely regulates cell survival and death signaling through aPKC/PAR-4 [61] and aPKC/Akt [62]. Also, PAR-4 has recently been shown to interact with BACE and regulates the production of Aβ peptide [63].
Therefore, it would not be surprising to observe that in situations where PAR-4 is induced during oxidative stress or injury that low expression of p62 might compromise the neuron and contribute to development of neurogeneration. Excessive accumulation of misfolded proteins is known to provoke oxidative stress and induce PAR-4 expression [61]. Moreover, oxidatively modified proteins are resistant to proteolysis and may further enhance accumulation of aggregated proteins [64]. We hypothesize that decline in expression of p62 could serve as a biomarker for those individuals at risk for developing neurodegenerative disease. In this model, aging and oxidative stress along with diminished expression of p62 would define a threshold where proteins fail to properly fold or triage for degradation, survival signaling is impaired, and the neurodegenerative disease phenotype is manifested. Interestingly mutation in valsolin-containing protein (VCP), a ubiquitin binding protein involved in UPS trafficking, has been linked to frontotemporal dementia [65]. We speculate that p62, as well as other ubiquitin binding proteins, may be candidate-genes for detailed genetic analysis. The goal here would be to examine potential polymorphisms that may serve as risk determinants for neurodegenerative disease. In keeping with this hypothesis, recent genetic analysis of the ubiquilin 1 gene (UBQLN1) revealed that certain genetic variants increase the risk of AD [66]. Similar to VCP and p62, UBQLN1 encodes a protein that serves as a shuttling protein to deliver polyubiquitinated proteins to the proteasome for degradation.
RELATIONSHIP OF MALLORY BODIES TO INCLUSION BODIES
Mallory bodies (MB) are a disease-associated type of aggresomes/inclusion consisting of excessive accumulation of keratin and are characteristic of alcoholic steatohepatitis (ASH) and nonalcoholic steatophepatitis (NASH). These inclusion share abnormal liver morphology observed in Wilson's disease (WD), Indian childhood cirrhosis (ICC), and idiopathic copper toxicosis (ICT) [67]. ICC and ICT individuals develop copper induced cirrhosis of the liver by consumption of contaminated water or milk [67]. WD mutations in ATP7B lead to abnormal copper accumulation in various tissues, particularly the liver [67]. Copper-mediated oxidative stress may also play a pathogenic role in chronic neurodegenerative diseases such as Alzheimer's disease. Sequestosome 1/p62 is an integral component of MB [68] and of the Mallory like-inclusions found in WD, ICC, and ICT, suggesting that p62 may play a role in their formation. The formation of MBs can be induced by prolonged alcohol intoxication and chronic metabolic disturbances [68]. Impaired protein degradation has been implicated as an underlying factor in alcoholic liver disease similar to its role in chronic neurodegenerative disorders [67–69]. Moreover, removal of p62 blocked the formation of MBs, whereas over expression enhanced their formation [69]. MBs contain high molecular weight polyubiquitin conjugates as well [69], which suggest that these are sites for sequestration of polyubiquitinated/misfolded proteins. As previously mentioned, p62 expression is transcriptionally regulated, in particular to agents of oxidative stress [58, 59]. Oxidative stresses causing alterations in mitochondria are well recognized as contributors to Alzheimer's disease [70, 71]. Oxidatively damaged mitochondria are removed by a process of autophagocytosis [72], a process that declines with age. Altogether, these findings strongly suggest that p62 plays a role in the formation of MBs which may have similarity to inclusion bodies observed in neurodegenerative disease.
AUTOPHAGY AS A ROUTE TO PROTEIN DEGRADATION
The cellular trafficking network that takes place involves movement of proteins from one intracellular compartment to another. In some instances receptor proteins in late endosome are deubiquitinated by chain-specific deubiquitinating enzymes at that site [73], while in others it appears that proteins traffic to the proteasome for chain removal and are then transported to the lysosome for degradation [74]. Sequestosome 1/p62 has been shown to be a component of the late endosomes [75, 76], and is able to sort proteins, such as TrkA to the endosome [77]. Under stress conditions late endosomes may fuse with autophagosomes [78], a process that involves bulk phase sequestration of cytoplasmic proteins. Aggregated proteins can be removed by the process of autophagy [79], a process that is impaired in AD [80]. Since the UPS may be impaired by protein aggregates [34, 35], it is reasonable to propose that autophagy could serve as a mechanism that cells hold in reserve for the removal of protein aggregates. In this regard α-synuclein can be degraded by both UPS and autophagy [81]. Therefore as aggregates arise, cells could degrade these proteins by autophagy while preserving the function of the UPS. Numerous studies now indicate that cells may attempt to compensate for impairments in one form of proteolysis (UPS) by dramatically elevating an alternate form of protein degradation (autophagy) [82]. Recently, p62 has been shown to bind light chain 3 (LC3), a protein that is tightly associated with the autophagosomal membrane [83]. Under stress conditions p62 would thereby link polyubiquitinated aggregated proteins to the autophagic machinery, facilitating their clearance. Indeed this has been found to be the case for clearance of mutant huntingtin [83]. Since p62 is localized to late endosomes [75, 76], p62 through interaction with LC3 may be needed in the recruitment of proteins for autophagy. Therefore decline in p62 expression would not only lead to the accumulation of polyubiquitinated proteins, but also to an absence of autophagosomes which in the appropriate genetic environment may further contribute to an absence of inclusions and accumulation of misfolded proteins. Currently we are studying whether neurons isolated from p62 −/− mice fail to form autophagosomes and the effects this may have.
FUTURE DIRECTIONS FOR THERAPEUTIC TARGETS
Recent targets for therapy include reducing protein misfolding and blocking aggregation. Strategies that promote degradation of misfolded proteins, such as: (1) enhanced expression of chaperones; (2) overexpression of E3 ligases; (3) enhanced expression of shuttling proteins, such as p62; and last, (4) up regulation of proteasome activity and/or autophagy Since protein oxidation and aggregation are intimately linked [64, 70], it is likely that more than one approach will be needed to effectively remove aggregated proteins and treat neurodegeneration. Clearly, early diagnostic markers are needed to effectively time intervention and treatment.
CONCLUSIONS
Great strides have been made in the past ten years toward understanding the pathological, cellular, biochemical, biophysical, and molecular bases of targeting proteins for degradation. Two mechanisms have been studied which promote the removal of aggregated/misfolded/polyubiquitinated proteins: the UPS and autophagy. Both mechanisms are regulated by p62. The observation that p62 plays an intimate role in the regulation of protein signaling, polyubiquitination, and trafficking suggests that further study of its role as a regulator of oxidative stress, neuropathology, and neurotoxicity in the brain is warranted. There is clearly a need to learn more about p62 in the context of aging, genetic background, and environmental factors. The convergence of these elements will determine the onset and severity of neurodegeneration. For AD, the greatest risk factor for the disease is age. The known AD genes (PS1, PS2, APOE, APP) account for less than half the genetic variance in the disease, suggesting there are many other risk determinant genes yet to be identified. Altogether, these findings will contribute to the development of more effective means for treating AD and for assessing those who might benefit from therapeutic intervention.
ACKNOWLEDGMENT
These studies were funded by NINDS (NS33661) to MWW.
References
- 1.Schubert U, Antón LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404(6779):770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 2.Berke SJS, Paulson HL. rotein aggregation and the ubiquitin proteasome pathway: gaining the UPPer hand on neurodegeneration. Current Opinion in Genetics and Development. 2003;13(3):253–261. doi: 10.1016/s0959-437x(03)00053-4. [DOI] [PubMed] [Google Scholar]
- 3.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends in Cell Biology. 2000;10(12):524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 4.Alves-Rodrigues A, Gregori L, Figueiredo-Pereira ME. Ubiquitin, cellular inclusions and their role in neurodegeneration. Trends in Neurosciences. 1998;21(12):516–520. doi: 10.1016/s0166-2236(98)01276-4. [DOI] [PubMed] [Google Scholar]
- 5.Lang-Rollin I, Rideout H, Stefanis L. Ubiquitinated inclusions and neuronal cell death. Histology and Histopathology. 2003;18(2):509–517. doi: 10.14670/HH-18.509. [DOI] [PubMed] [Google Scholar]
- 6.Jentsch S, Pyrowolakis G. Ubiquitin and its kin: how close are the family ties? Trends in Cell Biology. 2000;10(8):335–342. doi: 10.1016/s0962-8924(00)01785-2. [DOI] [PubMed] [Google Scholar]
- 7.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochimica et Biophysica Acta. 2004;1695(1–3):55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 8.Pickart CM. Ubiquitin in chains. Trends in Biochemical Sciences. 2000;25(11):544–548. doi: 10.1016/s0968-0004(00)01681-9. [DOI] [PubMed] [Google Scholar]
- 9.Peng J, Schwartz D, Elias JE, et al. A proteomics approach to understanding protein ubiquitination. Nature Biotechnology. 2003;21(8):921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 10.Hofmann RM, Pickart CM. In vitro assembly and recognition of Lys-63 polyubiquitin chains. Journal of Biological Chemistry. 2001;276(30):27936–27943. doi: 10.1074/jbc.M103378200. [DOI] [PubMed] [Google Scholar]
- 11.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiological Reviews. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 12.Wu K, Chen A, Tan P, Pan Z-Q. The Nedd8-conjugated ROC1-CUL1 core ubiquitin ligase utilizes Need8 charged surface residues for efficient polyubiquitin chain assembly catalyzed by Cdc34. Journal of Biological Chemistry. 2002;277(1):516–527. doi: 10.1074/jbc.M108008200. [DOI] [PubMed] [Google Scholar]
- 13.Ferrell K, Wilkinson CRM, Dubiel W, Gordon C. Regulatory subunit interactions of the 26S proteasome, a complex problem. Trends in Biochemical Sciences. 2002;25(2):83–88. doi: 10.1016/s0968-0004(99)01529-7. [DOI] [PubMed] [Google Scholar]
- 14.Yewdell JW. Immunoproteasomes: regulating the regulator. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(26):9089–9090. doi: 10.1073/pnas.0504018102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heink S, Ludwig D, Kloetzel P-M, Krüger E. IFN-γ-induced immune adaptation of the proteasome system is an accelerated and transient response. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(26):9241–9246. doi: 10.1073/pnas.0501711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kloetzel P-M, Ossendorp F. Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Current Opinion in Immunology. 2004;16(1):76–81. doi: 10.1016/j.coi.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Cheroni C, Peviani M, Cascio P, DeBiasi S, Monti C, Bendotti C. Accumulation of human SOD1 and ubiquitinated deposits in the spinal cord of SOD1G93A mice during motor neuron disease progression correlates with a decrease of proteasome. Neurobiology of Disease. 2005;18(3):509–522. doi: 10.1016/j.nbd.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 18.Diaz-Hernandez M, Hernandez F, Martin-Aparicio E, et al. Neuronal induction of the immuno-proteasome in Huntington's disease. The Journal of Neuroscience. 2003;23(37):11653–11661. doi: 10.1523/JNEUROSCI.23-37-11653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartmann-Petersen R, Gordon C. Proteins interacting with the 26S proteasome. Cellular and Molecular Life Sciences. 2004;61(13):1589–1595. doi: 10.1007/s00018-004-4132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Molecular and Cellular Biology. 2004;24(18):8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanayama A, Seth RB, Sun L, et al. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Molecular Cell. 2004;15(4):535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Mueller TD, Kamionka M, Feigon J. Specificity of the interaction between ubiquitin-associated domains and ubiquitin. Journal of Biological Chemistry. 2004;279(12):11926–11936. doi: 10.1074/jbc.M312865200. [DOI] [PubMed] [Google Scholar]
- 23.Madura K. The ubiquitin-associated (UBA) domain: on the path from prudence to prurience. Cell Cycle. 2002;1(4):235–244. [PubMed] [Google Scholar]
- 24.Donaldson KM, Li W, Ching KA, Batalov S, Tsai C-C, Joazeiro CAP. Ubiquitin-mediated sequestration of normal cellular proteins into polyglutamine aggregates. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(15):8892–8897. doi: 10.1073/pnas.1530212100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elsasser S, Gali RR, Schwikart M, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nature Cell Biology. 2002;4(9):725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 26.Rao H, Sastry A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. Journal of Biological Chemistry. 2002;277(14):11691–11695. doi: 10.1074/jbc.M200245200. [DOI] [PubMed] [Google Scholar]
- 27.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118(1):99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 28.Doss-Pepe EW, Stenroos ES, Johnson WG, Madura K. Ataxin-3 interactions with Rad23 and valosin-containing protein and its associations with ubiquitin chains and the proteasome are consistent with a role in ubiquitin-mediated proteolysis. Molecular and Cellular Biology. 2003;23(18):6469–6483. doi: 10.1128/MCB.23.18.6469-6483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varadan R, Walker O, Pickart CM, Fushman D. Structural properties of polyubiquitin chains in solution. Journal of Molecular Biology. 2002;324(4):637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 30.Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nature Structural and Molecular Biology. 2005;12(8):708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 31.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. Journal of Cell Biology. 1998;143(7):1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ardley HC, Scott GB, Rose SA, Tan NGS, Markham AF, Robinson PA. Inhibition of proteasomal activity causes inclusion formation in neuronal and non-neuronal cells overexpressing parkin. Molecular Biology of the Cell. 2003;14(11):4541–4556. doi: 10.1091/mbc.E03-02-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyun D-H, Lee M, Halliwell B, Jenner P. Proteasomal inhibition causes the formation of protein aggregates containing a wide range of proteins, including nitrated proteins. Journal of Neurochemistry. 2003;86(2):363–373. doi: 10.1046/j.1471-4159.2003.01841.x. [DOI] [PubMed] [Google Scholar]
- 34.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292(5521):1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 35.Bennett EJ, Bence NF, Jayakumar R, Kopito RR. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Molecular Cell. 2005;17(3):351–365. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Mata R, Gao Y-S, Sztul E. Hassles with taking out the garbage: aggravating aggresomes. Traffic. 2002;3(6):388–396. doi: 10.1034/j.1600-0854.2002.30602.x. [DOI] [PubMed] [Google Scholar]
- 37.Taylor JP, Tanaka F, Robitschek J, et al. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Human Molecular Genetics. 2003;12(7):749–757. doi: 10.1093/hmg/ddg074. [DOI] [PubMed] [Google Scholar]
- 38.Ellis RJ, Pinheiro TJT. Danger-misfolding proteins. Nature. 2002;416(6880):483–484. doi: 10.1038/416483a. [DOI] [PubMed] [Google Scholar]
- 39.Dobson CM. Principles of protein folding, misfolding and aggregation. Seminars in Cell and Developmental Biology. 2004;15(1):3–16. doi: 10.1016/j.semcdb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431(7010):805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 41.Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40(2):427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 42.Imai Y, Soda M, Takahashi R. Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. Journal of Biological Chemistry. 2000;275(46):35661–35664. doi: 10.1074/jbc.C000447200. [DOI] [PubMed] [Google Scholar]
- 43.Sakata E, Yamaguchi Y, Kurimoto E, et al. Parkin binds the Rpn 10 subunit of 26S proteasomes through its ubiquitin-like domain. EMBO Reports. 2003;4(3):301–306. doi: 10.1038/sj.embor.embor764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 45.Babu JR, Geetha T, Wooten MW. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. Journal of Neurochemistry. 2005;94(1):192–203. doi: 10.1111/j.1471-4159.2005.03181.x. [DOI] [PubMed] [Google Scholar]
- 46.Nichols MR, Moss MA, Reed DK, Hoh JH, Rosenberry TL. Rapid assembly of amyloid-beta peptide at a liquid/liquid interface produces unstable beta-sheet fibers. Biochemistry. 2005;44(1):165–173. doi: 10.1021/bi048846t. [DOI] [PubMed] [Google Scholar]
- 47.Liao LJ, Cheng DM, Wang J, et al. Proteomic characterization of postmortem amyloid plaques isolated by laser capture microdissection. Journal of Biological Chemistry. 2004;279(35):37061–37068. doi: 10.1074/jbc.M403672200. [DOI] [PubMed] [Google Scholar]
- 48.Geetha T, Wooten MW. Structure and functional properties of the ubiquitin binding protein p62. FEBS Letters. 2002;512(1–3):19–24. doi: 10.1016/s0014-5793(02)02286-x. [DOI] [PubMed] [Google Scholar]
- 49.Ciani B, Layfield R, Cavey JR, Sheppard PW, Searle MS. Structure of the ubiquitin-associated domain of p62 (SQSTM1) and implications for mutations that cause Paget's disease of bone. Journal of Biological Chemistry. 2003;278(39):37409–37412. doi: 10.1074/jbc.M307416200. [DOI] [PubMed] [Google Scholar]
- 50.Janse DM, Crosas B, Finley D, Church GM. Localization to the proteasome is sufficient for degradation. Journal of Biological Chemistry. 2004;279(20):21415–21420. doi: 10.1074/jbc.M402954200. [DOI] [PubMed] [Google Scholar]
- 51.Geetha T, Babu JR, Peng J, Wooten MW. Sequestosome 1/p62 involved in ubiquitin proteasome degradation of the nerve growth factor receptor TrkA. In press. [Google Scholar]
- 52.Capsoni S, Ugolini G, Comparini A, Ruberti F, Berardi N, Cattaneo A. Alzheimer-like neurodegeneration in aged antinerve growth factor transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(12):6826–6831. doi: 10.1073/pnas.97.12.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Counts SE, Mufson EJ. The role of nerve growth factor receptors in cholinergic basal forebrain degeneration in prodromal Alzheimer disease. Journal of Neuropathology and Experimental Neurology. 2005;64(4):263–272. doi: 10.1093/jnen/64.4.263. [DOI] [PubMed] [Google Scholar]
- 54.Lamark T, Perander M, Outzen H, et al. Interaction codes within the family of mammalian Phox and Bem1p domaincontaining proteins. Journal of Biological Chemistry. 2003;278(36):34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- 55.Paine MG, Babu JR, Seibenhener ML, Wooten MW. Evidence for p62 aggregate formation: role in cell survival. FEBS Letters. 2005;579(22):5029–5034. doi: 10.1016/j.febslet.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 56.Kuusisto E, Salminen A, Alafuzoff I. Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. NeuroReport. 2001;12(10):2085–2090. doi: 10.1097/00001756-200107200-00009. [DOI] [PubMed] [Google Scholar]
- 57.Vadlamudi RK, Shin J. Genomic structure and promoter analysis of the p62 gene encoding a non-proteasomal multiubiquitin chain binding protein. FEBS Letters. 1998;435(2-3):138–142. doi: 10.1016/s0014-5793(98)01021-7. [DOI] [PubMed] [Google Scholar]
- 58.Kuusisto E, Suuronen T, Salminen A. Ubiquitin-binding protein p62 expression is induced during apoptosis and proteasomal inhibition in neuronal cells. Biochemical and Biophysical Research Communications. 2001;280(1):223–228. doi: 10.1006/bbrc.2000.4107. [DOI] [PubMed] [Google Scholar]
- 59.Nakaso K, Yoshimoto Y, Nakano T, et al. Transcriptional activation of p62/A170/ZIP during the formation of the aggregates: possible mechanisms and the role in Lewy body formation in Parkinson's disease. Brain Research. 2004;1012(1-2):42–51. doi: 10.1016/j.brainres.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 60.Wooten MW, Geetha T, Seibenhener ML, Babu JR, Diaz-Meco MT, Moscat J. The p62 scaffold regulates nerve growth factorinduced NF-κB activation by influencing TRAF6 polyubiquitination. Journal of Biological Chemistry. 2005;280(42):35625–35629. doi: 10.1074/jbc.C500237200. [DOI] [PubMed] [Google Scholar]
- 61.Chang S, Kim JH, Shin J. p62 forms a ternary complex with PKCζ and PAR-4 and antagonizes PAR-4-induced PKCζ inhibition. FEBS Letters. 2002;510(1-2):57–61. doi: 10.1016/s0014-5793(01)03224-0. [DOI] [PubMed] [Google Scholar]
- 62.Joung I, Kim HJ, Kwon YK. p62 modulates Akt activity via association with PKCζ in neuronal survival and differentiation. Biochemical and Biophysical Research Communications. 2005;334(2):654–660. doi: 10.1016/j.bbrc.2005.06.138. [DOI] [PubMed] [Google Scholar]
- 63.Xie J, Guo Q. PAR-4 is involved in regulation of β-secretase cleavage of the Alzheimer amyloid precursor protein. Journal of Biological Chemistry. 2005;280(14):13824–13832. doi: 10.1074/jbc.M411933200. [DOI] [PubMed] [Google Scholar]
- 64.Grune T, Jung T, Merker K, Davies KJA. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and ‘aggresomes’ during oxidative stress, aging, and disease. International Journal of Biochemistry and Cell Biology. 2004;36(12):2519–2530. doi: 10.1016/j.biocel.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 65.Watts GDJ, Wymer J, Kovach MJ, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nature Genetics. 2004;36(4):377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 66.Bertram L, Hiltunen M, Parkinson M, et al. Family-based association between Alzheimer's disease and variants in UBQLN1. New England Journal of Medicine. 2005;352(9):884–894. doi: 10.1056/NEJMoa042765. [DOI] [PubMed] [Google Scholar]
- 67.Müller T, Langner C, Fuchsbichler A, et al. Immunohistochemical analysis of Mallory bodies in Wilsonian and non-Wilsonian hepatic copper toxicosis. Hepatology. 2004;39(4):963–969. doi: 10.1002/hep.20108. [DOI] [PubMed] [Google Scholar]
- 68.Bardag-Gorce F, van Leeuwen FW, Nguyen V, et al. The role of the ubiquitin-proteasome pathway in the formation of Mallory bodies. Experimental and Molecular Pathology. 2002;73(2):75–83. doi: 10.1006/exmp.2002.2451. [DOI] [PubMed] [Google Scholar]
- 69.Nan L, Wu Y, Bardag-Gorce F, et al. p62 is involved in the mechanism of Mallory body formation. Experimental and Molecular Pathology. 2004;77(3):168–175. doi: 10.1016/j.yexmp.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 70.Reddy PH. Amyloid precursor protein-mediated free radicals and oxidative damage: implications for the development and progression of Alzheimer's disease. Journal of Neurochemistry. 2006;96(1):1–13. doi: 10.1111/j.1471-4159.2005.03530.x. [DOI] [PubMed] [Google Scholar]
- 71.Reddy PH, Beal MF. Are mitochondria critical in the pathogenesis of Alzheimer's disease? Brain Research Reviews. 2005;49(3):618–632. doi: 10.1016/j.brainresrev.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 72.Brunk UT, Terman A. The mitochondrial-lysosomal axis theory of aging: accumulation of damaged mitochondria as a result of imperfect autophagocytosis. European Journal of Biochemistry. 2002;269(8):1996–2002. doi: 10.1046/j.1432-1033.2002.02869.x. [DOI] [PubMed] [Google Scholar]
- 73.Urbé S, Sachse M, Row PE, et al. The UIM domain of Hrs couples receptor sorting to vesicle formation. Journal of Cell Science. 2003;116(20):4169–4179. doi: 10.1242/jcs.00723. [DOI] [PubMed] [Google Scholar]
- 74.Alwan HAJ, van Zoelen EJJ, van Leeuwen JEM. Ligand-induced lysosomal epidermal growth factor receptor (EGFR) degradation is preceded by proteasome-dependent EGFR deubiquitination. Journal of Biological Chemistry. 2003;278(37):35781–35790. doi: 10.1074/jbc.M301326200. [DOI] [PubMed] [Google Scholar]
- 75.Sanchez P, De Carcer G, Sandoval IV, Moscat J, Diaz-Meco MT. Localization of atypical protein kinase C isoforms into lysosome- targeted endosomes through interaction with p62. Molecular and Cellular Biology. 1998;18(5):3069–3080. doi: 10.1128/mcb.18.5.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Samuels IS, Seibenhener ML, Neidigh K, Wooten MW. Nerve growth factor stimulates the interaction of ZIP/p62 with atypical protein kinase C and targets endosomal localization: evidence for regulation of nerve growth factor-induced differentiation. Journal of Cellular Biochemistry. 2001;82(3):452–466. doi: 10.1002/jcb.1177. [DOI] [PubMed] [Google Scholar]
- 77.Geetha T, Wooten MW. Association of the atypical protein kinase C-interacting protein p62/ZIP with nerve growth factor receptor TrkA regulates receptor trafficking and Erk5 signaling. Journal of Biological Chemistry. 2003;278(7):4730–4739. doi: 10.1074/jbc.M208468200. [DOI] [PubMed] [Google Scholar]
- 78.Lucocq J, Walker D. Evidence for fusion between multilamellar endosomes and autophagosomes in HeLa cells. European Journal of Cell Biology. 1997;72(4):307–313. [PubMed] [Google Scholar]
- 79.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306(5698):990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nixon RA, Wegiel J, Kumar A, et al. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. Journal of Neuropathology and Experimental Neurology. 2005;64(2):113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 81.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. α-synuclein is degraded by both autophagy and the proteasome. Journal of Biological Chemistry. 2003;278(27):25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 82.Keller JN, Dimayuga E, Chen Q, Thorpe J, Gee J, Ding Q. Autophagy, proteasomes, lipofuscin, and oxidative stress in the aging brain. International Journal of Biochemistry and Cell Biology. 2004;36(12):2376–2391. doi: 10.1016/j.biocel.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 83.Bjørkøy G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. Journal of Cell Biology. 2005;171(4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wolf DH. From lysosome to proteasome: the power of yeast in the dissection of proteinase function in cellular regulation and waste disposal. Cellular and Molecular Life Sciences. 2004;61(13):1601–1614. doi: 10.1007/s00018-004-4134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weissman AM. Themes and variations on ubiquitylation. Nature Reviews Molecular and Cell Biology. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 87.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nature Reviews Molecular Cell Biology. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 87.Hartmann-Petersen R, Gordon C. Ubiquitin-proteasome system: proteins interacting with the 26S proteasome. Cellular and Molecular Life Sciences. 2004;61(13):1589–1595. doi: 10.1007/s00018-004-4132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]