Abstract
RNA interference (RNAi) is a powerful method for specific gene silencing which may also lead to promising novel therapeutic strategies. It is mediated through small interfering RNAs (siRNAs) which sequence-specifically trigger the cleavage and subsequent degradation of their target mRNA. One critical factor is the ability to deliver intact siRNAs into target cells/organs in vivo. This review highlights the mechanism of RNAi and the guidelines for the design of optimal siRNAs. It gives an overview of studies based on the systemic or local application of naked siRNAs or the use of various nonviral siRNA delivery systems. One promising avenue is the the complexation of siRNAs with the polyethylenimine (PEI), which efficiently stabilizes siRNAs and, upon systemic administration, leads to the delivery of the intact siRNAs into different organs. The antitumorigenic effects of PEI/siRNA-mediated in vivo gene-targeting of tumor-relevant proteins like in mouse tumor xenograft models are described.
INTRODUCTION
Altered expression levels of certain genes play a pivotal role in several pathological conditions. For example, in many cancers the upregulation of certain growth factors or growth factor receptors, or the deregulation of intracellular signal transduction pathways, represents key elements in the process of malignant transformation and progression of normal cells towards tumor cells leading to uncontrolled proliferation and decreased apoptosis. Since these processes may result in the direct, autocrine stimulation of the tumor cell itself as well as the paracrine stimulation of other cells, including the stimulation of tumor-angiogenesis, many novel therapeutic strategies focus on the reversal of this effect, that is, the inhibition of these proteins or the downregulation of their expression. Likewise, several other diseases have been firmly linked to the (over-)expression of endogenous wildtype or mutated genes. Taken together, in addition to strategies based on the inhibition of target proteins, for example, by low molecular weight inhibitors or inhibitory antibodies, this opens an avenue to gene-targeting approaches aiming at decreased expression of the respective gene.
The first method to be introduced for the specific inhibition of gene expression was the use of antisense oligonucleotides in the late 1970s [1, 2]. Upon their introduction into a cell, antisense ODNs are able to hybridize to their target RNA leading to the degradation of the RNA-DNA hybrid double strands through RNAase H, to the inhibition of the translation of the target mRNA due to a steric or conformational obstacle for protein translation and/or to the inhibition of correct splicing. In the early 1980s, the discovery of ribozymes, that is, catalytically active RNAs which are able to sequence-specifically cleave a target mRNA, further expanded gene-targeting strategies [3–5]. Subsequently, both methods were extensively studied and further developed with regard to the optimization of targeting efficacies and antisense-ODN/ribozyme delivery strategies in vitro and in vivo.
Most recently, another naturally occurring biological strategy for gene silencing has been discovered and termed RNA interference (RNAi). Since RNAi represents a particularly powerful method for specific gene silencing and is able to provide the relatively easy ablation of the expression of any given target gene, it is now commonly used as a tool in biological and biomedical research. This includes the RNAi-mediated targeting in vitro and in vivo for functional studies of various genes whose expression is known to be upregulated as well as the development of novel therapeutic approaches based on gene targeting.
RNA INTERFERENCE
RNAi is an evolutionarily conserved, sequence-specific, posttranscriptional gene silencing phenomenon. It is triggered by double-stranded RNA molecules as described first in C elegans by Fire et al [6] who then introduced the name RNA interference. These findings also explained earlier observations in petunias which turned white rather than purple upon the introduction of the “purple gene” in form of dsRNA [7], and on gene silencing by antisense oligonucleotides as well as by sense oligonucleotides in C elegans [8]. Subsequent studies demonstrated that RNAi, while described under different names (posttranscriptional gene silencing (PTGS), cosuppression, quelling), is present in most eukaryotic organisms with the response to dsRNA, however, being more complicated in higher organisms.
RNAi relies on a multistep intracellular pathway which can be roughly divided into two phases, that is, the initiation phase and the effector phase. In the initiation phase, double-stranded RNA molecules from endogenous or exogenous origin present in the cell are processed through the cleavage activity of a ribonuclease III-type protein [9–12] into short 21–23 nucleotide fragments termed siRNAs. These effector siRNAs, which contain a symmetric 2 nt overhang at the 3′-end as well as a 5′-phosphate and a 3′-hydroxy group, are then in the effector phase incorporated into a nuclease-containing multiprotein complex called RISC (RNA-induced silencing complex) [13]. Several structural and biochemical studies have shed light on the processing of double-stranded RNA and the formation of the RISC complex (see, eg, [14] for a recent review). Through unwinding of the siRNA duplex by an RNA helicase activity [15], this complex becomes activated with the single-stranded siRNA guiding the RISC complex to its complementary target RNA. Upon the binding of the siRNA through hybridization to its target mRNA, the RISC complex catalyses the endonucleolytical cleavage of the mRNA strand within the target site, which, due to the generation of unprotected RNA ends, results in the rapid degradation of the mRNA molecule. With the RISC complex being recovered for further binding and cleavage cycles, the whole process translates into a net reduction of the specific mRNA levels and hence into the decreased expression of the corresponding gene. For an overview of the RNAi pathway, see Figure 1.
Figure 1.
Mechanism of RNA interference (RNAi) in mammalian systems. Long double-stranded RNA molecules (dsRNA), which are expressed from DNA vectors (left red arrow) or directly enter the cell (center red arrow), are processed by the Dicer complex resulting in the formation of small inhibitory RNAs (siRNAs). Alternatively, to induce RNAi these small 21–23 bp duplexes are directly delivered into the cell (right red arrow). The siRNAs are incorporated into a nuclease-containing multiprotein complex called RISC, which becomes activated upon the ATP-dependent unwinding of the siRNA duplex by an RNA helicase. The now single-stranded siRNA guides the RISC complex to its complementary target mRNA which is then cleaved by the endonucleolytical activity of RISC. While the RISC complex is recovered for further cycles, the cleaved mRNA molecule is rapidly degraded due to its unprotected RNA ends.
While from this mechanism it becomes obvious that siRNA molecules complementary to the target mRNA and thus being able to serve as a guide sequence for the RISC complex play a pivotal role in this process, they need not be derived from long double-stranded precursor molecules. Rather, omitting the initiation phase, they can be delivered directly into the target cell (Figure 1, upper right arrow).
Several studies have led to the development of guidelines for the generation of siRNAs which are optimal in terms of efficacy and specificity [12, 16]. This includes the initial definition of the preferable length (19–25 bp) combined with a low G/C content in the range between 36% and 52% and the requirement of symmetric 2 nt overhangs at the 3′-end [16–18]. Later studies on synthetic siRNA molecules, however, revealed an up to 100-fold higher targeting efficacy in the case of even longer duplexes (25–30 nucleotides) which act as a substrate for Dicer and which therefore allow the direct incorporation of the newly produced siRNAs into the RISC complex [19]. As to be expected, intramolecular foldback structures which can result from internal repeats or palindrome sequences decrease the numbers of functional siRNA molecules with silencing capability [20]. Additional silencing-enhancing criteria include an A in position 3 and a G at position 13 of the sense strand, the absence of a C or G at position 19 and, most importantly, a U in position 10 of the sense strand. Since nucleotides 10–11 represent the site of the RISC-mediated cleavage of the target mRNA, this indicates that RISC is comparable to most other endonucleases in preferentially cleaving 3′ of U rather than any other nucleotide [20, 21]. Furthermore, it was shown more generally that the thermodynamic flexibility of the positions 15–19 of the sense strand correlates with the silencing efficacy and that the presence of at least one A/U base pair in this region improves siRNA-mediated silencing efficacy due to a decreased internal stability of its 3′-end [20].
Still, different siRNA sequences may display differing efficacies, which suggest additional still unknown criteria for optimal siRNA selection and emphasize the influence of target mRNA accessibility. In fact, several studies also correlate the siRNA efficacy with the mRNA secondary structure [18, 22–27].
In conclusion, apart from the selection criteria defined above, the individual screening of different siRNAs for highly efficient and specific duplexes, or the pooling of multiple siRNAs, is the most effective approach to increase siRNA-mediated targeting efficacy.
For the design of effective siRNAs, several algorithms on publicly accessible web sites are available (see [28] for review). To reduce the risk of nonspecific (“off-target”) effects of the siRNAs, a homology search of the targeting sequence against a gene database is necessary and already incorporated in some of these web sites. Nevertheless, it has also been shown that siRNAs may cross-react with targets of limited sequence similarity when regions of partial sequence identity between the target mRNA and the siRNA exist. In fact, in some cases regions comprising of only 11–15 contiguous nucleotides of sequence identity were sufficient to induce gene silencing [29]. The prediction of these off-target activities is difficult so far.
An additional mechanism that may lead to nonspecific effects in vivo relies on the interferon system [30–33] which is induced when double-stranded RNA molecules enter a cell activating a multi-component signalling complex. This effect is particularly true for long dsRNA molecules and essentially prevents them from being used as inducers of RNA interference in mammalian systems. The development of synthetic siRNAs [10, 12, 33, 34] largely circumvents this problem since they seem to be too small. However, some synthetic siRNAs do induce components of the interferon system which seems to be dependent on their sequence [31, 32, 35] as well as, in the case of in vitro transcribed siRNAs, on the 5′ initiating triphosphate [36]. Thus, strategies to avoid as far as possible the unwanted interferon response upon application of siRNAs in vivo will include a design of siRNAs without known interferon-stimulating sequences, the use of the lowest possible siRNA dose to still achieve the desired effect and optimized siRNA delivery methods.
OLIGONUCLEOTIDE DELIVERY SYSTEMS
Based on the known mechanisms of antisense technology, ribozyme-targeting or RNAi, small oligonucleotides or plasmid-based expression vectors can be used to specifically downregulate the expression of a given gene of interest or of pathological relevance in vitro. In principle, this also applies to the in vivo situation leading to novel, potentially relevant therapeutic approaches.
For the delivery of therapeutic nucleic acids, viral vectors have been used which have the advantage of high transfection efficacy due to the inherent ability of viruses to transport genetic material into cells. On the other hand, however, viral systems show a limited loading capacity regarding that the genetic material are rather difficult to produce in a larger scale and, most importantly, pose severe safety risks due to their oncogenic potential and their inflammatory and immunogenic effects which prevent them from repeated administration [37–40].
In the light of these problems, concerns, and limitations, nonviral systems have emerged as a promising alternative for gene delivery. Main requirements are the protection of their nucleic acid “load” as well as their efficient uptake into the target cells with subsequent release of the DNA or RNA molecules and, if necessary, their transfer into the nucleus. Several strategies can be distinguished, mainly lipofection and polyfection relying on cationic lipids or polymers, respectively (see, eg, [41–43]).
The efficient protection against enzymatic or nonenzymatic degradation is particularly important for RNA molecules including siRNAs. In fact, while the therapeutic potential of siRNAs for the treatment of various diseases is in principle very promising, limitations of transfer vectors may turn out to be rate-limiting in the development of RNAi-based therapeutic strategies. One approach to solve this problem is the use of DNA expression plasmids which encode palindromic hairpin loops with the desired sequence. Upon transcription and folding of the RNA, the double-stranded short hairpin RNAs (shRNAs) are recognized by Dicer and cleaved into the desired siRNAs. Additionally, an in vitro method has been described recently which is based on the expression of shRNAs in E coli and their delivery via bacterial invasion [44]. While all these different DNA-based systems offer the advantage of siRNA expression with a longer duration and a probably higher level of gene silencing, they still rely on (viral or nonviral) delivery of DNA molecules and again raise safety issues in vivo. Hence, the direct delivery of siRNAs molecules, derived from in vitro transcription or chemically synthesized, offers advantages over DNA-based strategies and may be preferable for in vivo therapeutic use.
In the last years, a large body of studies has been published which describe different strategies for the systemic or local application of siRNAs in vivo. Tables 1–3 give an overview. The probably largest number of papers focuses the use of unmodified siRNAs (Table 1) whose administration is often performed IV by hydrodynamic transfection (high pressure tail vein injection). While this method is widely used and in some cases led to efficient target gene inhibition in the liver and, to a lesser extent, in lung, spleen, pancreas, and kidney, it may suffer from certain technical and practical limitations at least in a therapeutical setting since it relies on the rapid IV injection of a comparably large volume (>= 1 ml/mouse/injection, in theory equivalent to a ∼ 3 l IV bolus injection in man). Alternative strategies for the application of naked siRNAs include various delivery routes which, however, often provide an only local administration or rely on an administration at least close to the target tissue or target organ, thus restricting the number of target organs which may not be relevant for certain diseases. It should also be noted that several studies described here and below use rather large amounts of siRNAs and that upon intravenous injection of siRNAs the liver is the primary site of siRNA uptake. As an alternative approach for the application of siRNAs in vivo, their delivery by liposomes/cationic lipids has been described. For liposome-based siRNA formulations, a wide variety of modes of application allowing local or systemic delivery has been used (Table 2). Finally, several other strategies for local or systemic siRNA administration have been explored, including chemical modifications of siRNA molecules, electropulsation, polyamine, or other basic complexes, atelocollagen, virosomes, and certain protein preparations (Table 3).
Table 1.
Studies based on the direct application of siRNAs to induce RNAi in vivo: administration of unmodified siRNAs.
| Administration | Target tissue/organ | Target gene(s) | Target disease/aim of study | Reference |
| Intravenous | ||||
| Hydrodynamic | Liver | caspase-8 | Fas-mediated apoptosis/ | [94] |
| transfection | acute liver failure | |||
| Hydrodynamic | Liver | HBsAg | Inhibition of HBV replication | [95] |
| transfection | ||||
| Hydrodynamic | Liver | HBsAg | Inhibition of HBV replication | [96] |
| transfection | ||||
| Hydrodynamic | Liver | GFP | Downregulation of GFP | [97] |
| transfection | ||||
| Pulse injection | Liver | Fas | Fulminant hepatitis | [98] |
| High or low | Liver | Fas | Fas downregulation in liver | [99] |
| pressure | ||||
| Large-volume, | Liver | mdr1a | Downregulation of mdr1a | [100] |
| high-speed injection | ||||
| High-volume injection | Liver | caspase-8, caspase-3 | Protection against ischemia/ | [101] |
| (with lipiodol) | reperfusion injury | |||
| Hydrodynamic | Liver and limb grafts | DsRed2, GFP | Downregulation of | [102] |
| transfection | target genes | |||
| Metastatic breast cancer cells | CXCR4 | Blockage of breast | [103] | |
| cancer metastasis | ||||
| Hydrodynamic | Coxsackievirus/various organs | CVB 2A | Coxsackieviral cytopathogenicity | [104] |
| transfection | ||||
| Pancreatic adenocarcinoma xenograft | CEACAM6 | Tumor growth inhibition | [105] | |
| Pancreatic adenocarcinoma xenograft | EphA2 | Tumor growth inhibition | [106] | |
| Pancreatic adenocarcinoma xenograft | FAK | Enhanced gemcitabine | [107] | |
| chemosensitivity | ||||
| Hydrodynamic transfection | Kidney | Fas | Renal ischemia- | [108] |
| (renal vein) | reperfusion injury | |||
| Hydrodynamic | Lung | Nucleoprotein, acidic polymerase | Influenza virus infections | [109] |
| transfection | ||||
| Hydrodynamic | Pancreas | Ins2 | Downregulation of the Ins2 gene | [110] |
| transfection | ||||
| Hydrodynamic | Blood-brain barrier | Organic anion transporter 3 | Brain-to-blood transport | [111] |
| transfection | ||||
| Other delivery routes | ||||
| Intraperitoneal | Fibrosarcoma xenografts | VEGF | Tumor growth inhibition | [112] |
| Intraperitoneal | Subcutaneous pancreatic carcinoma xenografts | bcl-2 | Growth inhibition | [113] |
| Local injection | Optic nerve stump | c-Jun, Bax, Apaf-1 | Antiapoptosis in retinal | [114] |
| ganglion cells | ||||
| Intratracheal | Lung | KC, MIP-2 | Acute lung injury | [115] |
| instillation | ||||
| Local into the liver | Liver | Luciferase | Downregulation of cotransfected | [116] |
| luciferase | ||||
| Subretinal | Eye | VEGF | Ocular neovascularization | [117] |
| Local injection | Mouse joint | TNF-α | Collagen-induced arthritis | [118] |
| and electroporation | ||||
| Intradermal | Antigen-presenting cells | Bak, Bax | Cancer vaccine potency | [119] |
| Intranasal | Nose after viral infection | RSV-P, PIV-P | Respiratory viral diseases | [120] |
| Intranasal | Lung | HO-1 | Functional analysis in | [121] |
| lung ischemia-reperfusion injury | ||||
| Intranasal | Lung | SCV | Relief from SARS coronavirus fever | [122] |
| In situ perfusion/ | Pancreatic islet | — | Detection of fluorescing siRNA | [123] |
| Intravenous | ||||
| Intratumoral | Breast carcinoma xenografts | RhoA/RhoC | Inhibition of tumor growth | [124] |
| Intratumoral | Mammary tumor xenografts | CSF-1 | Inhibition of tumor growth | [125] |
| Intrathecal | Brain | cation channel P2X3 | Chronic neuropathic pain | [126] |
| Renal artery and | Kidney | TGF-β1 | Glomerulonephritis | [127] |
| electroporation | ||||
| Intratracheal | Lung | Fas | Hemorrhagic shock and sepsis | [128] |
| Stereotactic injection | Brain | Agouti-related peptide | Increased metabolic rate | [129] |
| to hypothalamus | ||||
| Intrathecal infusion | Brain | Pain-related cation channel P2X3 | Decreased mechanical hyperanalgesia | [126] |
| using mini-osmotic pump | ||||
| Infusion into the | Brain | Dopamine transporter | Temporal hyperlocomoter response | [130] |
| ventricular system | ||||
| Infusion into the | Brain | Serotonin transporter | Antidepressant-related | [131] |
| ventricular system | behavioural response | |||
| Intraocular | Retinal cells/terminals in supcolliculus | APP/APLP2 | Alterations of synaptic function | [132] |
| Intraocular | Eye | VEGFA, VEGFR1, VEGFR2 | Inhibition of ocular angiogenesis | [133] |
| Intraocular | Eye | TGF-beta RII | Prevention of ocular | [134] |
| inflammation and scarring | ||||
Table 3.
Studies based on the direct application of siRNAs to induce RNAi in vivo: other strategies of siRNA administration.
| siRNA formulation | Target tissue/organ | Administration | Target gene(s) | Target disease/aim of study | Reference |
| Chemically modified | Liver and jejunum | Intravenous | apoB | Reduction of apoB | [152] |
| and total cholesterol | |||||
| Chemically modified | Liver | Intravenous | HBV | Reduced serum HBV DNA | [153] |
| + lipid incapsulation | |||||
| Electropulsation | Muscle | Intramuscular | GFP | Downregulation of GFP | [154] |
| Histidine-lysine | Breast tumor | Intratumoral | Raf-1 | Breast cancer | [155] |
| complex | xenograft | ||||
| Atelocollagen | Subcutaneous prostate | Intratumoral | VEGF | Tumor growth inhibition | [156] |
| carcinoma xenograft | |||||
| Atelocollagen | Orthotopic germ cell | Intratumoral | HST-1/FGF-4 | Tumor growth inhibition | [157] |
| tumor xenograft in testes | |||||
| Atelocollagen | Bone-metastatic | Intravenous | EZH2 | Inhibition of metastatic | [158] |
| prostate cancer | tumor growth | ||||
| Inactivated HVJ | Subcutaneous HeLa | Intratumoral | Rad51 | Enhancement of cisplatin | [159] |
| suspension | xenografts | anticancer effect | |||
| Protamin-antibody | Subcutaneous melanoma | Intravenous or | c-myc, | Tumor growth inhibition | [160] |
| fusion protein | xenografts | Intratumoral | MDM2, VEGF | ||
| PEI complexation | Subcutaneous ovarian | Intraperitoneal | HER-2 | Tumor growth inhibition | [70] |
| carcinoma xenografts | |||||
| PEI complexation | Lung | Intravenous | Influenza | Influenza virus infections | [74] |
| virus genes | |||||
| Nanoplexes | Subcutaneous N2A neuroblastoma | Intravenous | VEGF R2 | Tumor growth inhibition | [73] |
| (RGD-PEG-PEI) | xenografts | ||||
| TransIT-TKO | Nose after viral infection | Intranasal | RSV-P, PIV-P | Respiratory viral diseases | [120] |
| (polyamine) | |||||
| Polyamine | Myocard | Intraperitoneal | Heat shock factor 1 | Abrogation of HSF- | [161] |
| induced cardioprotection | |||||
| Virosomes + | Peritoneal cavity | Intraperitoneal | GFP | GFP downregulation | [162] |
| cationic lipids | |||||
Table 2.
Studies based on the direct application of siRNAs to induce RNAi in vivo: administration of siRNAs based on liposomes/cationic lipids.
| Administration | Target tissue/organ | siRNA formulation | Target gene(s) | Target disease/aim of study | Reference |
| Intravenous | Liver metastasis | Liposomes | bcl−2 | Metastasis | [135] |
| growth inhibition | |||||
| Intravenous | Kidney | Liposomes | V2R | Role of V2R in water/ | [136] |
| sodium homeostasis | |||||
| Intravenous | Subcutaneous tumor | DOPC liposomes | EphA2 | Tumor growth inhibition | [137] |
| xenograft | |||||
| Intravenous | Lung | Liposomes | caveolin-1 | Increase in lung | [138] |
| vascular permeability | |||||
| Intravenous/intraperitoneal | Various | Liposomes | – | Detection of FITC- | [139] |
| labeled siRNA | |||||
| Intraperitoneal | Peritoneal cavity | Liposomes | IL-12p40 | Inflammation | [140] |
| Intraperitoneal | Peritoneal cavity | Liposomes | β-catenin | Tumor growth Inhibition | [141] |
| Intraperitoneal | Various | Liposomes | TNF-α | Sepsis after | [142] |
| lipopolysaccharide injection | |||||
| Transurethral | Bladder cancer | Liposomes | PLK-1 | Tumor growth inhibition | [143] |
| Local | Ear | Liposomes | GJBR75W | Hearing loss | [144] |
| Subcutaneous | Subcutaneous prostate | Liposomes | bcl-2 | Tumor growth inhibition | [135] |
| carcinoma xenograft | |||||
| Local (tracheal grafts) | Subcutaneous tracheal grafts | Liposomes | MIF | Decreased formation of | [145] |
| obstructive bronchiolitis | |||||
| Intracardiac | Developing vascular | Lipoplexes | GFP | Downregulation of GFP | [146] |
| network of chicken embryo | |||||
| Systemic | Prostate cancer | Cationic cardiolipin liposomes | Raf-1 | Inhibition of | [147] |
| xenografts | tumor growth | ||||
| Intravenous | Subcutaneous breast | Cationic cardiolipin analogue | c-raf | Tumor growth inhibition | [148] |
| cancer xenografts | |||||
| Intrathecal | Spinal cord/ | i-Fect (cationic lipid) | Delta opioid receptor | DELT antinociception | [149] |
| dorsal root ganglia | |||||
| Intratumoral | Subcutaneous HeLa xenograft | Cytofectin GSV | GFP | Downregulation of GFP | [150] |
| Intra-cerebroventricular | Brain | JetSI (+ DOPE) | Luciferase | Downregulation of luciferase | [71] |
| Intravaginal | Vagina | Oligofectamine | HSV-2 proteins | Protection from HSV-2 infection | [151] |
An alternative approach relies on the complexation of unmodified siRNA molecules with a cationic polymer, polyethylenimine (PEI).
POLYETHYLENIMINES: FROM DNA TRANSFECTION TO siRNA DELIVERY IN VITRO AND IN VIVO
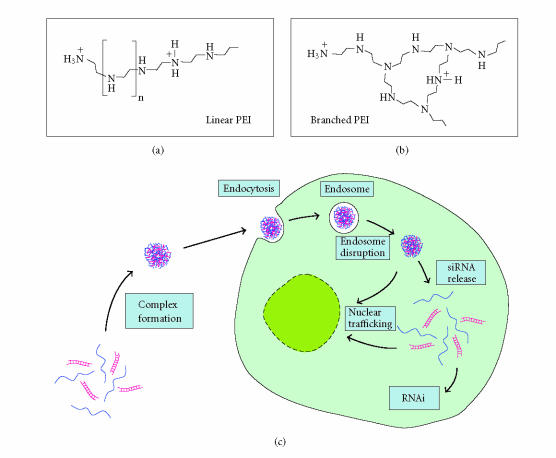
Polyethylenimines (PEIs) are synthetic polymers available in branched or linear forms (Figure 2, upper panels) and in a broad range of molecular weights from < 1000 Da to > 1000 kd. Commercial PEI preparations, although labelled with a defined molecular weight, consist of PEI molecules with a broad molecular weight distribution [45–47]. PEIs possess a high cationic charge density due to a protonable amino group in every third position [48, 49]. Since no quarternary amino groups are present, the cationic charges are generated by protonation of the amino groups and hence are dependent on the pH in the environment (eg, 20% at pH 7.4, see [50] for review). Due to its ability to condense and compact the DNA into complexes, which form small colloidal particles allowing efficient cellular uptake through endocytosis, PEI has been introduced as a potent DNA transfection reagent in a variety of cell lines and in animals for DNA delivery (for review, see [51, 52] and references therein). In fact, in several studies PEI has been shown to be able to deliver large DNA molecules such as 2.3 Mb yeast artificial chromosomes (YACs) [53] as well as plasmids or small oligonucleotides [48, 54–56] into mammalian cells in vitro and in vivo. The N/P ratio, which indicates the ratio of the nitrogen atoms of PEI to DNA phosphates in the complex and thus describes the amount of PEI used for complex formation independent of its molecular weight, influences the efficiency of DNA delivery. A positive net charge of the complexes, resulting from high N/P ratios, inhibits due to electrostatic repulsion their aggregation and improves their solubility in aqueous solutions as well as their interaction with the negatively charged extracellular matrix components and thus their cellular uptake [57]. Additionally, the strong buffer capacity, described by the “proton sponge hypothesis” which postulates enhanced transgene delivery by cationic polymer-DNA complexes (polyplexes) containing H+ buffering polyamines due to enhanced endosomal Cl− accumulation and osmotic swelling/lysis [48], seems to be responsible for the fact that PEI-based delivery does not require endosome disruptive agents for lysosomal escape. This tight condensation of the DNA molecules as well as the buffering capacity of PEI in certain cellular compartments like endosomes and lysosomes also protects DNA from degradation [48, 49, 58, 59]. PEIs have been successfully used for nonviral gene delivery in vitro and in vivo. While initial publications showed increased transfection efficacies when using high molecular weight PEIs [45], more recent studies demonstrated the advantages of certain low molecular weight PEIs [47, 60, 61]. The higher transfection efficacy of low molecular weight PEIs may be due to a more efficient uptake of the resulting PEI/DNA complexes, a better intracellular release of the DNA and/or lower in vitro cytotoxicity as compared to high molecular weight PEI [60–63] . In fact, a decrease in the molecular weight of the PEI leads to an increase in complex size which may be favourable at least for in vitro use [64, 65]. On the other hand, other PEIs with very low molecular weight (< 2 kd) display little or no transfection efficacy even at very high N/P ratios which may be attributed to the fact that a decrease in the molecular weight of PEI has been shown to translate into an increasingly lower ability to form small complexes [63]. Therefore, low molecular weight PEIs require higher N/P ratios for optimal transfection efficacies as compared to higher molecular weight PEIs since higher N/P ratios lead to an increase in compaction with reduced complex sizes and a reduced tendency of the complexes to aggregate due to hydrophobic interactions [61, 63, 64]. Nevertheless, while several parameters have been extensively studied, some precise determinants for transfection efficacy remain to be elucidated (see [50, 66] for review). Also, the mechanism of the cytotoxic effects of PEI complexes is only poorly understood. It may rely on the formation of large aggregates in the range of up to 2 μm which, when formed on the cell surface, impairs membrane functions finally leading to cell necrosis [60]. Clearly, there is a trend towards low molecular weight PEIs as rather nontoxic delivery reagents in vitro and in vivo, which combine high biocompatibility and reduced side-effects thus also allowing to employ larger PEI/DNA complex amounts without significant cytotoxicity.
Figure 2.
Polyethylenimine (PEI)-mediated siRNA transfer. Upper panel: PEIs are synthetic linear (a) or branched (b) polymers with an amino group in every third position. Dependent on the pH, some of these amino nitrogens are protonated giving PEI a high cationic charge density. Lower panel: proposed mechanism of PEI-mediated siRNA transfer. Due to electrostatic interactions, PEI is able to complex negatively charged siRNAs leading to a compaction and the formation of small colloidal particles which are endocytosed. The “proton sponge effect” exhibited by PEI complexes leads to osmotic swelling and ultimately to the disruption of the endosomes. siRNAs are protected from degradation due to their tight condensation in the complex and the buffering capacity of PEI. Upon their release from the PEI-based complex, intact siRNAs are incorporated into the RISC complex and induce RNAi (see Figure 1).
More recently, the use of polyethylenimines has been extended towards the complexation and delivery of RNA molecules, especially small RNA molecules like 37 nt all-RNA ribozymes [67–69] and siRNAs [70] (Figure 2). While chemically unmodified RNA molecules are very instable and prone to rapid degradation, the PEI complexation has been shown to lead to an almost complete protection against enzymatic or nonenzymatic degradation. In fact, PEI-complexed siRNAs, which are [32P]-labeled for better detection, remain intact in vitro for several hours even in the presence of RNase A or fetal calf serum at 37°C, while non-complexed siRNAs are rapidly degraded (Figure 3(a)). This indicates that siRNA molecules are efficiently condensed and thus fully covered and protected by PEI. Indeed, the analysis of PEI/siRNA complexes by atomic force microscopy showed the absence of free siRNAs or siRNA molecule ends and thus confirms these findings regarding an efficient complexation (Grzelinski et al, submitted). However, while the complex stability seems to be sufficient for siRNA protection with all PEIs tested (Werth et al, in press; Aigner et al, unpublished data), several of these complexes do not show any targeting efficacy at all. In fact, only when using certain polyethylenimines, PEI/siRNA complexes are efficiently delivered into target cells in vitro, where siRNAs are released and display bioactivity (Figures 1 and 2). In general and as seen before for PEI/DNA complexes (see above), the transfection efficacy is dependent on the PEI used, also indicating that the siRNA targeting efficiency mainly depends on the endocytotic uptake of the complex and/or its intracellular decomposition rather than on the in vitro complex stability. Good results were obtained with commercially available JetPEI [70] while the in vivo JetPEI from the same supplier showed only poor siRNA delivery efficacies [71]. Likewise, a novel low molecular weight PEI based on the fractionation of a commercially available polyethylenimine demonstrates high siRNA protection and delivery efficacies in vitro (Werth et al, in press). Under certain conditions, the PEI/RNA (siRNA or ribozyme) complexes retain their physical stability and biological activity also after lyophilization ([72] and Werth et al, in press). Although the PEI transfection is only transient, data from our lab show that PEI/siRNA effects are stable for at least 7 days (Urban-Klein and Aigner, unpublished results). Finally, another study has explored the use of siRNA nanoplexes comprising of PEI that is PEGylated with an RGD peptide ligand attached at the distal end of the PEI. Again, siRNA nanoplexes protect siRNAs against serum degradation and show in vitro activity [73].
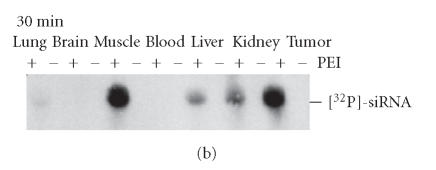
Figure 3.

Protection and in vivo delivery of siRNAs upon PEI complexation. In [70] (a) in vitro protection of siRNAs against nucleolytic degradation. [32P] end-labeled siRNAs, complexed (upper panel) or not complexed (lower panel) with PEI, were subjected to treatment with 1 % fetal calf serum at 37°C. At the time points indicated, the samples were analysed by agarose gel electrophoresis, blotting, and autoradiography. The bands represent full-length siRNA molecules indicating that PEI complexation leads to the efficient protection of siRNAs while noncomplexed siRNAs are rapidly degraded. (b,c) In vivo delivery of intact siRNAs upon PEI complexation. [32P]-labeled siRNAs, complexed (+) or not complexed (−) with PEI, were injected IP into mice bearing subcutaneous SKOV-3 ovarian carcinoma cell tumor xenografts, and after 30 min (b) or 4 h (b) total RNA from various organ and tissue homogenates was prepared and subjected to agarose gel electrophoresis prior to blotting and autoradiography. The bands represent intact [32P]-labeled siRNA molecules which for several hours are mainly found in tumor and muscle as well as in liver and, time-dependently, in kidney. Only little siRNA amounts are detected in the lung and traces in the brain.
The ultimate goal is the application of siRNAs in vivo which has been explored in some studies in different mouse models. Ge et al showed that PEI-complexed siRNAs targeting conserved regions of influenza virus genes are able to prevent and treat influenza virus infection in mice. Upon IV injection, PEI promoted the delivery of siRNAs into the lungs where, either given before or after virus infection, siRNA reduced influenza virus production in the lungs [74].
Most biological effects of the systemic application of PEI-complexed siRNAs, however, have been determined in different mouse tumor models and by targeting different proteins which have been shown previously to be tumor-relevant. This includes the epidermal growth factor receptor HER-2 (c-erbB-2/neu), the growth factor pleiotrophin (PTN), and vascular endothelial growth factor (VEGF) and its receptor (VEGF R2), and the fibroblast growth factor-binding protein FGF-BP.
The in vivo administration of PEI complexed, but not of naked siRNAs, through IP or subcutaneous injection resulted in the detection of intact siRNAs even hours after injection (Figure 3(b)). Radiolabeled siRNA molecules were found in several organs including subcutaneous tumors, muscle liver, kidney and, to a smaller extent, lung and brain. It is important to note that the siRNAs were actually internalized by the tissues as indicated by the fact that blood was negative for siRNAs (Figure 3(b)).
Overexpression of the HER-2 receptor has been observed in a wide variety of human cancers and cancer cell lines. Since HER-2 displays strong cell growth-stimulating and antiapoptotic effects especially through heterodimer formation with other members of the EGFR family, its overexpression has been established as a negative prognostic factor and linked to a more aggressive malignant behaviour of tumors (eg, [75]). Consequently, HER-2 qualifies as an attractive target molecule for antitumoral treatment strategies including anti-HER-2 antibodies, low molecular weight inhibitors, or HER-2-specific gene-targeting approaches. In fact, the relevance of HER-2 (over-)expression in tumor growth has been established in several in vitro HER-2 targeting studies including the use of ribozymes [76, 78, 79] or siRNAs [80, 81]. It was demonstrated that HER-2 reduction in vitro leads, among others, to the inhibition of cell proliferation and increased apoptosis.
The systemic treatment of athymic nude mice bearing subcutaneous SKOV-3 ovarian carcinoma tumor xenografts through IP injection of PEI-complexed HER-2-specific siRNAs led to marked antitumoral effects as seen by a significant reduction tumor growth (Figure 4) [70]. PEI-complexed nonspecific siRNAs or HER-2-specific, naked siRNAs had no effects. This was paralleled by the detection of intact HER-2-specific siRNAs in the tumors of the specific treatment group already 30 min after administration and for at least 4 h, and by the downregulation of HER-2 on mRNA and protein levels [70].
Figure 4.
Systemic treatment of mice with PEI-complexed HER-2-specific siRNAs leads to reduced growth of subcutaneous SKOV-3 tumor xenografts due to decreased HER-2 expression. In [70] athymic nude mice bearing subcutaneous tumor xenografts were injected IP with 0.6 nmoles HER-2-specific naked (open circles) or PEI-complexed (closed circles) siRNAs 2–3 times per week and tumor sizes were evaluated daily from the product of the perpendicular diameters of the tumors. Mean +/− standard error of the mean (SEM) is depicted and Student's unpaired t test was used for comparisons between data sets (**P < .03, ***P < .01). Differences in tumor growth reach significance at day 5 indicating the antitumoral effects of the PEI-complexed HER-2-specific siRNAs.
Another receptor, VEGF R2, was targeted in a study employing self-assembling nanoparticles based on siRNAs complexed PEI which is PEGylated with an RGD peptide ligand attached at the distal end of PEG. While the PEGylation allows steric stabilization and reduces nonspecific interactions of the complexes, the RGD motif provided tumor selectivity due to their ability to target integrins expressed on activated endothelial cells in the tumor vasculature. Upon IV administration into mice bearing subcutaneous N2A neuroblastoma tumor xenografts, a selective tumor uptake and a VEGF R2 downregulation were observed, resulting in decreased tumor growth and tumor angiogenesis [73].
The receptor ligand, VEGF, is a mitogenic and angiogenic growth factor stimulating tumor growth and angiogenesis in several tumors including prostate carcinoma. Thus, it may represent attractive target molecule for RNAi-based gene-targeting strategies also bearing in mind the double antitumoral effect due to reduction of tumor cell proliferation as well as tumor angiogenesis. The subcutaneous or intraperitoneal injection of VEGF-specific siRNAs complexed with a novel PEI obtained through fractionation of a commercially available PEI (Werth et al, in press) resulted in the reduction of tumor growth due to decreased VEGF expression levels (Höbel and Aigner, unpublished results). The same was true for PEI/siRNA-mediated targeting of FGF-BP (Dai and Aigner, unpublished results), which has been established previously as “rate-limiting” for tumor growth and angiogenesis in several tumors ([82, 83], see [84] for review).
Finally, PEI/siRNA-mediated targeting of pleiotrophin (PTN) exerted strong antitumoral effects. PTN is a secreted growth factor which shows mitogenic, chemotactic, angiogenic and transforming activity [85–93] and which is markedly upregulated in several human tumors including cancer of the breast, testis, prostate, pancreas, and lung as well as in melanomas, meningiomas, neuroblastomas, and glioblastomas. The in vivo treatment of nude mice through systemic subcutaneous or IP application of PEI-complexed PTN siRNAs led to the delivery of intact siRNAs into subcutaneous tumor xenografts and a significant inhibition of tumor growth. Likewise, in a clinically more relevant orthotopic mouse glioblastoma model with U87 cells growing intracranially, the injection of PEI-complexed PTN siRNAs into the CNS exerted antitumoral effects. This establishes, also in a complex and relevant orthotopic tumor model, the potential of PEI/siRNA-mediated PTN gene targeting as a novel therapeutic option in GBM, and further extends the modes of delivery of PEI/siRNA complexes intrathecal strategies as employed in the therapy of glioblastomas with antisense oligonucleotides.
CONCLUSION
Only a few years after their discovery, siRNAs are catching up with ribozymes and antisense oligonucleotides as efficient tools for gene targeting in vitro and, more recently, also in vivo. This includes the exploration of their potential as therapeutics which will lead to the development of siRNA-based therapeutic strategies. Their ultimate success, however, will strongly depend on the development of powerful and feasible siRNA delivery strategies which need to address several issues including the stability/stabilization of siRNA molecules while preserving their efficacy and maintaining their gene-silencing activity, an efficient delivery into the target organ(s) as well as a sufficiently long siRNA half life in the organism and particularly in the target organ. Thus, siRNA delivery strategies must provide siRNA protection and transfection efficacy, the absence of toxic and nonspecific effects, they must be efficacious also when using small amounts of siRNAs and must be applicable in various treatment regimens and in various diseases even when this requires to overcome biological barriers after their administration to reach their target tissue or target organ. The research done on DNA-based gene delivery, ribozyme-targeting, and antisense technology will facilitate this process since it already provides a basis of established technologies. This is also true for the complexation of siRNAs with polyethylenimine, which may represent a promising avenue for siRNA applications in vivo. This may eventually lead to novel therapeutic strategies.
ACKNOWLEDGMENTS
The work of A. Aigner is supported by the Deutsche Forschungsgemeinschaft (AI 24/5-1) and by the Deutsche Krebshilfe. The author would like to apologize to the authors whose primary works have not been cited due to length considerations.
ABBREVIATIONS
ABBREVIATIONS
- dsRNA,
double-stranded RNA,
- FGF-BP,
fibroblast growth factor-binding protein,
- GFP,
green fluorescent protein,
- HER-2,
human epidermal growth factor receptor-2,
- IP,
intraperitoneal,
- ODN,
oligodeoxynucleotide,
- PEI,
polyethylenimine,
- PTN,
pleiotrophin,
- RISC,
RNA-induced silencing complex,
- RNAi,
RNA interference,
- siRNA,
small interfering RNA,
- shRNA,
short hairpin RNA.
References
- 1.Stephenson ML, Zamecnik PC. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proceedings of the National Academy of Sciences of the United States of America. 1978;75(1):285–288. doi: 10.1073/pnas.75.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zamecnik PC, Stephenson ML. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proceedings of the National Academy of Sciences of the United States of America. 1978;75(1):280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cech TR, Zaug AJ, Grabowski PJ. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell. 1981;27(3 pt 2):487–496. doi: 10.1016/0092-8674(81)90390-1. [DOI] [PubMed] [Google Scholar]
- 4.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 5.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35(3 pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 6.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 7.Jorgensen R. Altered gene expression in plants due to trans interactions between homologous genes. Trends in Biotechnology. 1990;8(12):340–344. doi: 10.1016/0167-7799(90)90220-r. [DOI] [PubMed] [Google Scholar]
- 8.Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81(4):611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286(5441):950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 10.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: doublestranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101(1):25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 12.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes and Development. 2001;15(2):188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNAdirected nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404(6775):293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 14.Collins RE, Cheng X. Structural domains in RNAi. FEBS Letters. 2005;579(26):5841–5849. doi: 10.1016/j.febslet.2005.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nykänen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107(3):309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 16.Elbashir SM, Harborth J, Weber K, Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26(2):199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- 17.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. The EMBO Journal. 2001;20(23):6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Research. 2002;30(8):1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim D-H, Behlke MA, Rose SD, Chang M-S, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nature Biotechnology. 2005;23(2):222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 20.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nature Biotechnology. 2004;22(3):326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 21.Donis-Keller H. Site specific enzymatic cleavage of RNA. Nucleic Acids Research. 1979;7(1):179–192. doi: 10.1093/nar/7.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bohula EA, Salisbury AJ, Sohail M, et al. The efficacy of small interfering RNAs targeted to the type 1 insulin-like growth factor receptor (IGF1R) is influenced by secondary structure in the IGF1R transcript. Journal of Biological Chemistry. 2003;278(18):15991–15997. doi: 10.1074/jbc.M300714200. [DOI] [PubMed] [Google Scholar]
- 23.Lee NS, Dohjima T, Bauer G, et al. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nature Biotechnology. 2002;20(5):500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- 24.Ding Y, Lawrence CE. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Research. 2003;31(24):7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vickers TA, Koo S, Bennett CF, Crooke ST, Dean NM, Baker BF. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. Journal of Biological Chemistry. 2003;278(9):7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 26.Xu Y, Zhang H-Y, Thormeyer D, et al. Effective small interfering RNAs and phosphorothioate antisense DNAs have different preferences for target sites in the luciferase mRNAs. Biochemical and Biophysical Research Communications. 2003;306(3):712–717. doi: 10.1016/s0006-291x(03)01024-6. [DOI] [PubMed] [Google Scholar]
- 27.Kretschmer-Kazemi Far R, Sczakiel G. The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucleic Acids Research. 2003;31(15):4417–4424. doi: 10.1093/nar/gkg649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandy P, Ventura A, Jacks T. Mammalian RNAi: a practical guide. BioTechniques. 2005;39(2):215–224. doi: 10.2144/05392RV01. [DOI] [PubMed] [Google Scholar]
- 29.Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi. Nature Biotechnology. 2003;21(6):635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 30.Hornung V, Guenthner-Biller M, Bourquin C, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nature Medicine. 2005;11(3):263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 31.Sledz CA, Holko M, De Veer MJ, Silverman RH, Williams BR-G. Activation of the interferon system by short-interfering RNAs. Nature Cell Biology. 2003;5(9):834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 32.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz A-L, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nature Genetics. 2003;34(3):263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 33.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 34.Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(17):9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nature Biotechnology. 2005;23(4):457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 36.Kim D-H, Longo M, Han Y, Lundberg P, Cantin E, Rossi JJ. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nature Biotechnology. 2004;22(3):321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 37.Lehrman S. Virus treatment questioned after gene therapy death. Nature. 1999;401(6753):517–518. doi: 10.1038/43977. [DOI] [PubMed] [Google Scholar]
- 38.Liu Q, Muruve DA. Molecular basis of the inflammatory response to adenovirus vectors. Gene Therapy. 2003;10(11):935–940. doi: 10.1038/sj.gt.3302036. [DOI] [PubMed] [Google Scholar]
- 39.Sun JY, Anand-Jawa V, Chatterjee S, Wong KK Jr. Immune responses to adeno-associated virus and its recombinant vectors. Gene Therapy. 2003;10(11):964–976. doi: 10.1038/sj.gt.3302039. [DOI] [PubMed] [Google Scholar]
- 40.Donahue RE, Kessler SW, Bodine D, et al. Helper virus induced T cell lymphoma in nonhuman primates after retroviral mediated gene transfer. Journal of Experimental Medicine. 1992;176(4):1125–1135. doi: 10.1084/jem.176.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merdan T, Kopeček J, Kissel T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Advanced Drug Delivery Reviews. 2002;54(5):715–758. doi: 10.1016/s0169-409x(02)00046-7. [DOI] [PubMed] [Google Scholar]
- 42.Duzgunes N, De Ilarduya CT, Simoes S, Zhdanov RI, Konopka K, Pedroso de Lima MC. Cationic liposomes for gene delivery: novel cationic lipids and enhancement by proteins and peptides. Current Medicinal Chemistry. 2003;10(14):1213–1220. doi: 10.2174/0929867033457403. [DOI] [PubMed] [Google Scholar]
- 43.Liu D, Ren T, Gao X. Cationic transfection lipids. Current Medicinal Chemistry. 2003;10(14):1307–1315. doi: 10.2174/0929867033457386. [DOI] [PubMed] [Google Scholar]
- 44.Zhao H-F, L'Abbé D, Jolicoeur N, et al. High-throughput screening of effective siRNAs from RNAi libraries delivered via bacterial invasion. Nature Methods. 2005;2(12):967–973. doi: 10.1038/nmeth812. [DOI] [PubMed] [Google Scholar]
- 45.Godbey WT, Wu KK, Mikos GA. Size matters: molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. Journal of Biomedical Materials Research. 1999;45(3):268–275. doi: 10.1002/(sici)1097-4636(19990605)45:3<268::aid-jbm15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 46.Tang MX, Szoka FC. The influence of polymer structure on the interactions of cationic polymers with DNA and morphology of the resulting complexes. Gene Therapy. 1997;4(8):823–832. doi: 10.1038/sj.gt.3300454. [DOI] [PubMed] [Google Scholar]
- 47.Bieber T, Elsässer H-P. Preparation of a low molecular weight polyethylenimine for efficient cell transfection. BioTechniques. 2001;30(1):74–77, 80–81. doi: 10.2144/01301st03. [DOI] [PubMed] [Google Scholar]
- 48.Boussif O, Lezoualc'h F, Zanta MA, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(16):7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Behr JP. The proton sponge: a trick to enter cells the viruses did not exploit. Chimia. 1997;51:34–36. [Google Scholar]
- 50.Neu M, Fischer D, Kissel T. Recent advances in rational gene transfer vector design based on poly(ethylene imine) and its derivatives. Journal of Gene Medicine. 2005;7(8):992–1009. doi: 10.1002/jgm.773. [DOI] [PubMed] [Google Scholar]
- 51.Kichler A. Gene transfer with modified polyethylenimines. Journal of Gene Medicine. 2004;6(suppl 1):S3–S10. doi: 10.1002/jgm.507. [DOI] [PubMed] [Google Scholar]
- 52.Wagner E, Kircheis R, Walker GF. Targeted nucleic acid delivery into tumors: new avenues for cancer therapy. Biomedicine and Pharmacotherapy. 2004;58(3):152–161. doi: 10.1016/j.biopha.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 53.Marschall P, Malik N, Larin Z. Transfer of YACs up to 2.3 Mb intact into human cells with polyethylenimine. Gene Therapy. 1999;6(9):1634–1637. doi: 10.1038/sj.gt.3300975. [DOI] [PubMed] [Google Scholar]
- 54.Abdallah B, Hassan A, Benoist C, Goula D, Behr JP, Demeneix BA. A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: polyethylenimine. Human Gene Therapy. 1996;7(16):1947–1954. doi: 10.1089/hum.1996.7.16-1947. [DOI] [PubMed] [Google Scholar]
- 55.Boletta A, Benigni A, Lutz J, Remuzzi G, Soria MR, Monaco L. Nonviral gene delivery to the rat kidney with polyethylenimine. Human Gene Therapy. 1997;8(10):1243–1251. doi: 10.1089/hum.1997.8.10-1243. [DOI] [PubMed] [Google Scholar]
- 56.Goula D, Benoist C, Mantero S, Merlo G, Levi G, Demeneix BA. Polyethylenimine-based intravenous delivery of transgenes to mouse lung. Gene Therapy. 1998;5(9):1291–1295. doi: 10.1038/sj.gt.3300717. [DOI] [PubMed] [Google Scholar]
- 57.Oh Y-K, Suh D, Kim JM, Choi H-G, Shin K, Ko JJ. Polyethylenimine-mediated cellular uptake, nucleus trafficking and expression of cytokine plasmid DNA. Gene Therapy. 2002;9(23):1627–1632. doi: 10.1038/sj.gt.3301735. [DOI] [PubMed] [Google Scholar]
- 58.Ferrari S, Pettenazzo A, Garbati N, Zacchello F, Behr J-P, Scarpa M. Polyethylenimine shows properties of interest for cystic fibrosis gene therapy. Biochimica et Biophysica Acta. 1999;1447(2-3):219–225. doi: 10.1016/s0167-4781(99)00153-0. [DOI] [PubMed] [Google Scholar]
- 59.Godbey WT, Barry MA, Saggau P, Wu KK, Mikos AG. Poly(ethylenimine)-mediated transfection: a new paradigm for gene delivery. Journal of Biomedical Materials Research. 2000;51(3):321–328. doi: 10.1002/1097-4636(20000905)51:3<321::aid-jbm5>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 60.Fischer D, Bieber T, Li Y, Elsässer H-P, Kissel T. A novel nonviral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharmaceutical Research. 1999;16(8):1273–1279. doi: 10.1023/a:1014861900478. [DOI] [PubMed] [Google Scholar]
- 61.Fischer D, Bhattacharya R, Osburg B, Bickel U. Inhibition of monocyte adhesion on brain-derived endothelial cells by NFkappaB decoy/polyethylenimine complexes. Journal of Gene Medicine. 2005;7(8):1063–1076. doi: 10.1002/jgm.747. [DOI] [PubMed] [Google Scholar]
- 62.Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24(7):1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 63.Kunath K, von Harpe A, Fischer D, et al. Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. Journal of Controlled Release. 2003;89(1):113–125. doi: 10.1016/s0168-3659(03)00076-2. [DOI] [PubMed] [Google Scholar]
- 64.Ogris M, Steinlein P, Kursa M, Mechtler K, Kircheis R, Wagner E. The size of DNA/transferrin-PEI complexes is an important factor for gene expression in cultured cells. Gene Therapy. 1998;5(10):1425–1433. doi: 10.1038/sj.gt.3300745. [DOI] [PubMed] [Google Scholar]
- 65.Wightman L, Kircheis R, Rössler V, et al. Different behavior of branched and linear polyethylenimine for gene delivery in vitro and in vivo. Journal of Gene Medicine. 2001;3(4):362–372. doi: 10.1002/jgm.187. [DOI] [PubMed] [Google Scholar]
- 66.Lungwitz U, Breunig M, Blunk T, Göpferich A. Polyethylenimine-based non-viral gene delivery systems. European Journal of Pharmaceutics and Biopharmaceutics. 2005;60(2):247–266. doi: 10.1016/j.ejpb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 67.Eigner A, Fischer D, Merdan T, Brus C, Kissel T, Czubayko F. Delivery of unmodified bioactive ribozymes by an RNA stabilizing polyethylenimine LMW PEI efficiently down regulates gene expression. Gene Therapy. 2002;9(24):1700–1707. doi: 10.1038/sj.gt.3301839. [DOI] [PubMed] [Google Scholar]
- 68.Brus C, Petersen H, Aigner A, Czubayco F, Kissel T. Physicochemical and biological characterization of polyethylenimine-graft- poly(ethylene glycol) block copolymers as a delivery system for oligonucleotides and ribozymes. Bioconjugate Chemistry. 2004;15(4):677–684. doi: 10.1021/bc034160m. [DOI] [PubMed] [Google Scholar]
- 69.Brus C, Petersen H, Aigner A, Czubayko F, Kissel T. Efficiency of polyethylenimines and polyethylenimine-graft-poly (ethylene glycol) block copolymers to protect oligonucleotides against enzymatic degradation. European Journal of Pharmaceutics and Biopharmaceutics. 2004;57(3):427–430. doi: 10.1016/j.ejpb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 70.Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Therapy. 2005;12(5):461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- 71.Hassani Z, Lemkine GF, Erbacher P, et al. Lipid-mediated siRNA delivery down-regulates exogenous gene expression in the mouse brain at picomolar levels. Journal of Gene Medicine. 2005;7(2):198–207. doi: 10.1002/jgm.659. [DOI] [PubMed] [Google Scholar]
- 72.Brus C, Kleemann E, Aigner A, Czubayko F, Kissel T. Stabilization of oligonucleotide-polyethylenimine complexes by freeze-drying: physicochemical and biological characterization. Journal of Controlled Release. 2004;95(1):119–131. doi: 10.1016/j.jconrel.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 73.Schiffelers RM, Ansari A, Xu J, et al. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Research. 2004;32(19):e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ge Q, Filip L, Bai A, Nguyen T, Eisen HN, Chen J. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(23):8676–8681. doi: 10.1073/pnas.0402486101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244(4905):707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 76.Juhl H, Downing SG, Wellstein A, Czubayko F. HER-2/neu is rate-limiting for ovarian cancer growth. Conditional depletion of HER-2/neu by ribozyme targeting. Journal of Biological Chemistry. 1997;272(47):29482–29486. doi: 10.1074/jbc.272.47.29482. [DOI] [PubMed] [Google Scholar]
- 77.Thybusch-Bernhardt A, Aigner A, Beckmann S, Czubayko F, Juhl H. Ribozyme targeting of HER-2 inhibits pancreatic cancer cell growth in vivo. European Journal of Cancer. 2001;37(13):1688–1694. doi: 10.1016/s0959-8049(01)00181-2. [DOI] [PubMed] [Google Scholar]
- 78.Suzuki T, Anderegg B, Ohkawa T, et al. Adenovirus-mediated ribozyme targeting of HER-2/neu inhibits in vivo growth of breast cancer cells. Gene Therapy. 2000;7(3):241–248. doi: 10.1038/sj.gt.3301065. [DOI] [PubMed] [Google Scholar]
- 79.Czubayko F, Downing SG, Hsieh SS, et al. Adenovirus-mediated transduction of ribozymes abrogates HER-2/neu and pleiotrophin expression and inhibits tumor cell proliferation. Gene Therapy. 1997;4(9):943–949. doi: 10.1038/sj.gt.3300483. [DOI] [PubMed] [Google Scholar]
- 80.Choudhury A, Charo J, Parapuram SK, et al. Small interfering RNA (siRNA) inhibits the expression of the Her2/Neu gene, upregulates HLA class I and induces apoptosis of Her2/Neu positive tumor cell lines. International Journal of Cancer. 2004;108(1):71–77. doi: 10.1002/ijc.11497. [DOI] [PubMed] [Google Scholar]
- 81.Yang G, Cai KQ, Thompson-Lanza JA, Bast RC Jr, Liu J. Inhibition of breast and ovarian tumor growth through multiple signaling pathways by using retrovirus-mediated small interfering RNA against Her-2/neu gene expression. Journal of Biological Chemistry. 2004;279(6):4339–4345. doi: 10.1074/jbc.M311153200. [DOI] [PubMed] [Google Scholar]
- 82.Czubayko F, Liaudet-Coopman EDE, Aigner A, Tuveson AT, Berchem G, Wellstein A. A secreted FGF-binding protein can serve as the angiogenic switch in human cancer. Nature Medicine. 1997;3(10):1137–1140. doi: 10.1038/nm1097-1137. [DOI] [PubMed] [Google Scholar]
- 83.Aigner A, Renneberg H, Bojunga J, Apel J, Nelson PS, Czubayko F. Ribozyme-targeting of a secreted FGF-binding protein (FGF-BP) inhibits proliferation of prostate cancer cells in vitro and in vivo. Oncogene. 2002;21(37):5733–5742. doi: 10.1038/sj.onc.1205560. [DOI] [PubMed] [Google Scholar]
- 84.Abuharbeid S, Czubayko F, Aigner A. The fibroblast growth factor-binding protein FGF-BP. to appear in The International Journal of Biochemistry & Cell Biology. doi: 10.1016/j.biocel.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 85.Fang WJ, Hartmann N, Chow D, Riegel AT, Wellstein A. Pleiotrophin stimulates fibroblasts and endothelial and epithelial cells and is expressed in human cancer. Journal of Biological Chemistry. 1992;267(36):25889–25897. [PubMed] [Google Scholar]
- 86.Bowden ET, Stoica GE, Wellstein A. Anti-apoptotic signaling of pleiotrophin through its receptor, anaplastic lymphoma kinase. Journal of Biological Chemistry. 2002;277(39):35862–35868. doi: 10.1074/jbc.M203963200. [DOI] [PubMed] [Google Scholar]
- 87.Wellstein A, Fang WJ, Khatri A, et al. A heparin-binding growth factor secreted from breast cancer cells homologous to a developmentally regulated cytokine. Journal of Biological Chemistry. 1992;267(4):2582–2587. [PubMed] [Google Scholar]
- 88.Milner PG, Li YS, Hoffman RM, Kodner CM, Siegel NR, Deuel TF. A novel 17 kD heparin-binding growth factor (HBGF-8) in bovine uterus: purification and N-terminal amino acid sequence. Biochemical and Biophysical Research Communications. 1989;165(3):1096–1103. doi: 10.1016/0006-291x(89)92715-0. [DOI] [PubMed] [Google Scholar]
- 89.Zhang N, Zhong R, Wang ZY, Deuel TF. Human breast cancer growth inhibited in vivo by a dominant negative pleiotrophin mutant. Journal of Biological Chemistry. 1997;272(27):16733–16736. doi: 10.1074/jbc.272.27.16733. [DOI] [PubMed] [Google Scholar]
- 90.Czubayko F, Schulte AM, Berchem GJ, Wellstein A. Melanoma angiogenesis and metastasis modulated by ribozyme targeting of the secreted growth factor pleiotrophin. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(25):14753–14758. doi: 10.1073/pnas.93.25.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Czubayko F, Riegel AT, Wellstein A. Ribozyme-targeting elucidates a direct role of pleiotrophin in tumor growth. Journal of Biological Chemistry. 1994;269(33):21358–21363. [PubMed] [Google Scholar]
- 92.Schulte AM, Lai S, Kurtz A, Czubayko F, Riegel AT, Wellstein A. Human trophoblast and choriocarcinoma expression of the growth factor pleiotrophin attributable to germ line insertion of an endogenous retrovirus. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(25):14759–14764. doi: 10.1073/pnas.93.25.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Choudhuri R, Zhang H-T, Donnini S, Ziche M, Bicknell R. An angiogenic role for the neurokines midkine and pleiotrophin in tumorigenesis. Cancer Research. 1997;57(9):1814–1819. [PubMed] [Google Scholar]
- 94.Zender L, Hütker S, Liedtke C, et al. Caspase 8 small interfering RNA prevents acute liver failure in mice. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(13):7797–7802. doi: 10.1073/pnas.1330920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Giladi H, Ketzinel-Gilad M, Rivkin L, Felig Y, Nussbaum O, Galun E. Small interfering RNA inhibits hepatitis B virus replication in mice. Molecular Therapy. 2003;8(5):769–776. doi: 10.1016/s1525-0016(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 96.Klein C, Bock CT, Wedemeyer H, et al. Inhibition of hepatitis B virus replication in vivo by nucleoside analogues and siRNA. Gastroenterology. 2003;125(1):9–18. doi: 10.1016/s0016-5085(03)00720-0. [DOI] [PubMed] [Google Scholar]
- 97.Lewis DL, Hagstrom JE, Loomis AG, Wolff JA, Herweijer H. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nature Genetics. 2002;32(1):107–108. doi: 10.1038/ng944. [DOI] [PubMed] [Google Scholar]
- 98.Song E, Lee S-K, Wang J, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nature Medicine. 2003;9(3):347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 99.Heidel JD, Hu S, Liu XF, Triche TJ, Davis ME. Lack of interferon response in animals to naked siRNAs. Nature Biotechnology. 2004;22(12):1579–1582. doi: 10.1038/nbt1038. [DOI] [PubMed] [Google Scholar]
- 100.Matsui Y, Kobayashi N, Nishikawa M, Takakura Y. Sequence-specific suppression of mdr1a/1b expression in mice via RNA interference. Pharmaceutical Research. 2005;22(12):2091–2098. doi: 10.1007/s11095-005-8178-8. [DOI] [PubMed] [Google Scholar]
- 101.Contreras JL, Vilatoba M, Eckstein C, Bilbao G, Anthony Thompson J, Eckhoff DE. Caspase-8 and caspase-3 small interfering RNA decreases ischemia/reperfusion injury to the liver in mice. Surgery. 2004;136(2):390–400. doi: 10.1016/j.surg.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 102.Sato Y, Ajiki T, Inoue S, et al. Gene silencing in rat-liver and limb grafts by rapid injection of small interference RNA. Transplantation. 2005;79(2):240–243. doi: 10.1097/01.tp.0000147786.52502.2f. [DOI] [PubMed] [Google Scholar]
- 103.Liang Z, Yoon Y, Votaw J, Goodman MM, Williams L, Shim H. Silencing of CXCR4 blocks breast cancer metastasis. Cancer Research. 2005;65(3):967–971. [PMC free article] [PubMed] [Google Scholar]
- 104.Merl S, Michaelis C, Jaschke B, Vorpahl M, Seidl S, Wessely R. Targeting 2A protease by RNA interference attenuates coxsackieviral cytopathogenicity and promotes survival in highly susceptible mice. Circulation. 2005;111(13):1583–1592. doi: 10.1161/01.CIR.0000160360.02040.AB. [DOI] [PubMed] [Google Scholar]
- 105.Duxbury MS, Matros E, Ito H, Zinner MJ, Ashley SW, Whang EE. Systemic siRNA-mediated gene silencing: a new approach to targeted therapy of cancer. Annals of Surgery. 2004;240(4):667–676. doi: 10.1097/01.sla.0000140755.97224.9a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. EphA2: a determinant of malignant cellular behavior and a potential therapeutic target in pancreatic adenocarcinoma. Oncogene. 2004;23(7):1448–1456. doi: 10.1038/sj.onc.1207247. [DOI] [PubMed] [Google Scholar]
- 107.Duxbury MS, Ito H, Benoit E, Zinner MJ, Ashley SW, Whang EE. RNA interference targeting focal adhesion kinase enhances pancreatic adenocarcinoma gemcitabine chemosensitivity. Biochemical and Biophysical Research Communications. 2003;311(3):786–792. doi: 10.1016/j.bbrc.2003.10.060. [DOI] [PubMed] [Google Scholar]
- 108.Hamar P, Song E, Kökeny G, Chen A, Ouyang N, Lieberman J. Small interfering RNA targeting Fas protects mice against renal ischemia-reperfusion injury. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(41):14883–14888. doi: 10.1073/pnas.0406421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tompkins SM, Lo C-Y, Tumpey TM, Epstein SL. Protection against lethal influenza virus challenge by RNA interference in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(23):8682–8686. doi: 10.1073/pnas.0402630101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bradley SP, Rastellini C, Da Costa MA, et al. Gene silencing in the endocrine pancreas mediated by short-interfering RNA. Pancreas. 2005;31(4):373–379. doi: 10.1097/01.mpa.0000179730.69081.64. [DOI] [PubMed] [Google Scholar]
- 111.Hino T, Yokota T, Ito S, et al. In vivo delivery of small interfering RNA targeting brain capillary endothelial cells. Biochemical and Biophysical Research Communications. 2006;340(1):263–267. doi: 10.1016/j.bbrc.2005.11.173. [DOI] [PubMed] [Google Scholar]
- 112.Filleur S, Courtin A, Ait-Si-Ali S, et al. SiRNA-mediated inhibition of vascular endothelial growth factor severely limits tumor resistance to antiangiogenic thrombospondin-1 and slows tumor vascularization and growth. Cancer Research. 2003;63(14):3919–3922. [PubMed] [Google Scholar]
- 113.Ocker M, Neureiter D, Lueders M, et al. Variants of bcl-2 specific siRNA for silencing antiapoptotic bcl-2 in pancreatic cancer. Gut. 2005;54(9):1298–1308. doi: 10.1136/gut.2004.056192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lingor P, Koeberle P, Kügler S, Bähr M. Down-regulation of apoptosis mediators by RNAi inhibits axotomy-induced retinal ganglion cell death in vivo. Brain. 2005;128(3):550–558. doi: 10.1093/brain/awh382. [DOI] [PubMed] [Google Scholar]
- 115.Lomas-Neira JL, Chung C-S, Wesche DE, Perl M, Ayala A. In vivo gene silencing (with siRNA) of pulmonary expression of MIP-2 versus KC results in divergent effects on hemorrhage-induced, neutrophil-mediated septic acute lung injury. Journal of Leukocyte Biology. 2005;77(6):846–853. doi: 10.1189/jlb.1004617. [DOI] [PubMed] [Google Scholar]
- 116.McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature. 2002;418(6893):38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 117.Reich SJ, Fosnot J, Kuroki A, et al. Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in mouse model. Molecular Vision. 2003;9:210–216. [PubMed] [Google Scholar]
- 118.Schiffelers RM, Xu J, Storm G, Woodle MC, Scaria PV. Effects of treatment with small interfering RNA on joint inflammation in mice with collagen-induced arthritis. Arthritis and Rheumatism. 2005;52(4):1314–1318. doi: 10.1002/art.20975. [DOI] [PubMed] [Google Scholar]
- 119.Kim TW, Lee J-H, He L, et al. Modification of professional antigen-presenting cells with small interfering RNA in vivo to enhance cancer vaccine potency. Cancer Research. 2005;65(1):309–316. [PubMed] [Google Scholar]
- 120.Bitko V, Musiyenko A, Shulyayeva O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nature Medicine. 2005;11(1):50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 121.Zhang X, Shan P, Jiang D, et al. Small interfering RNA targeting heme oxygenase-1 enhances ischemia-reperfusion-induced lung apoptosis. Journal of Biological Chemistry. 2004;279(11):10677–10684. doi: 10.1074/jbc.M312941200. [DOI] [PubMed] [Google Scholar]
- 122.Li B-J, Tang Q, Cheng D, et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nature Medicine. 2005;11(9):944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bradley SP, Kowalik TF, Rastellini C, et al. Successful incorporation of short-interfering RNA into islet cells by in situ perfusion. Transplantation Proceedings. 2005;37(1):233–236. doi: 10.1016/j.transproceed.2004.12.181. [DOI] [PubMed] [Google Scholar]
- 124.Pillé J-Y, Denoyelle C, Varet J, et al. Anti-RhoA and anti-RhoC siRNAs inhibit the proliferation and invasiveness of MDA-MB-231 breast cancer cells in vitro and in vivo. Molecular Therapy. 2005;11(2):267–274. doi: 10.1016/j.ymthe.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 125.Aharinejad S, Paulus P, Sioud M, et al. Colony-stimulating factor-1 blockade by antisense oligonucleotides and small interfering RNAs suppresses growth of human mammary tumor xenografts in mice. Cancer Research. 2004;64(15):5378–5384. doi: 10.1158/0008-5472.CAN-04-0961. [DOI] [PubMed] [Google Scholar]
- 126.Dorn G, Patel S, Wotherspoon G, et al. siRNA relieves chronic neuropathic pain. Nucleic Acids Research. 2004;32(5):e49. doi: 10.1093/nar/gnh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Takabatake Y, Isaka Y, Mizui M, et al. Exploring RNA inteference as a therapeutic strategy for renal disease. Gene Therapy. 2005;12(12):965–973. doi: 10.1038/sj.gt.3302480. [DOI] [PubMed] [Google Scholar]
- 128.Perl M, Chung C-S, Lomas-Neira J, et al. Silencing of Fas, but not caspase-8, in lung epithelial cells ameliorates pulmonary apoptosis, inflammation, and neutrophil influx after hemorrhagic shock and sepsis. American Journal of Pathology. 2005;167(6):1545–1559. doi: 10.1016/S0002-9440(10)61240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Makimura H, Mizuno TM, Mastaitis JW, Agami R, Mobbs CV. Reducing hypothalamic AGRP by RNA interference increases metabolic rate and decreases body weight without influencing food intake. BMC Neuroscience. 2002;3(1):18. doi: 10.1186/1471-2202-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Thakker DR, Natt F, Hüsken D, et al. Neurochemical and behavioral consequences of widespread gene knockdown in the adult mouse brain by using nonviral RNA interference. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(49):17270–17275. doi: 10.1073/pnas.0406214101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Thakker DR, Natt F, Hüsken D, et al. siRNA-mediated knockdown of the serotonin transporter in the adult mouse brain. Molecular Psychiatry. 2005;10(8):782–789. doi: 10.1038/sj.mp.4001687. [DOI] [PubMed] [Google Scholar]
- 132.Herard AS, Besret L, Dubois A, et al. siRNA targeted against amyloid precursor protein impairs synaptic activity in vivo. to appear in Neurobiology of Aging. doi: 10.1016/j.neurobiolaging.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 133.Kim B, Tang Q, Biswas PS, et al. Inhibition of ocular angiogenesis by siRNA targeting vascular endothelial growth factor pathway genes: Therapeutic strategy for herpetic stromal keratitis. American Journal of Pathology. 2004;165(6):2177–2185. doi: 10.1016/S0002-9440(10)63267-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nakamura H, Siddiqui SS, Shen X, et al. RNA interference targeting transforming growth factor-beta type II receptor suppresses ocular inflammation and fibrosis. Molecular Vision. 2004;10:703–711. [PubMed] [Google Scholar]
- 135.Yano J, Hirabayashi K, Nakagawa S-I, et al. Antitumor activity of small interfering RNA/cationic liposome complex in mouse models of cancer. Clinical Cancer Research. 2004;10(22):7721–7726. doi: 10.1158/1078-0432.CCR-04-1049. [DOI] [PubMed] [Google Scholar]
- 136.Hassan A, Tian Y, Zheng W, Ji H, Sandberg K, Verbalis JG. Small interfering RNA-mediated functional silencing of vasopressin V 2 receptors in the mouse kidney. Physiological Genomics. 2005;21(3):382–388. doi: 10.1152/physiolgenomics.00147.2004. [DOI] [PubMed] [Google Scholar]
- 137.Landen CN Jr, Chavez-Reyes A, Bucana C, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Research. 2005;65(15):6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 138.Miyawaki-Shimizu K, Predescu D, Shimizu J, Broman M, Predescu S, Malik AB. siRNA-induced caveolin-1 knockdown in mice increases lung vascular permeability via the junctional pathway. American Journal of Physiology. Lung Cellular and Molecular Physiology. 2006;290(2):L405–L413. doi: 10.1152/ajplung.00292.2005. [DOI] [PubMed] [Google Scholar]
- 139.Sioud M, Sørensen DR. Cationic liposome-mediated delivery of siRNAs in adult mice. Biochemical and Biophysical Research Communications. 2003;312(4):1220–1225. doi: 10.1016/j.bbrc.2003.11.057. [DOI] [PubMed] [Google Scholar]
- 140.Flynn MA, Casey DG, Todryk SM, Mahon BP. Efficient delivery of small interfering RNA for inhibition of IL-12p40 expression in vivo. Journal of Inflammation. 2004;1(1):4. doi: 10.1186/1476-9255-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Verma UN, Surabhi RM, Schmaltieg A, Becerra C, Gaynor RB. Small interfering RNAs directed against beta-catenin inhibit the in vitro and in vivo growth of colon cancer cells. Clinical Cancer Research. 2003;9(4):1291–1300. [PubMed] [Google Scholar]
- 142.Sørensen DR, Leirdal M, Sioud M. Gene silencing by systemic delivery of synthetic siRNAs in adult mice. Journal of Molecular Biology. 2003;327(4):761–766. doi: 10.1016/s0022-2836(03)00181-5. [DOI] [PubMed] [Google Scholar]
- 143.Nogawa M, Yuasa T, Kimura S, et al. Intravesical administration of small interfering RNA targeting PLK-1 successfully prevents the growth of bladder cancer. Journal of Clinical Investigation. 2005;115(4):978–985. doi: 10.1172/JCI23043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Maeda Y, Fukushima K, Nishizaki K, Smith RJH. In vitro and in vivo suppression of GJB2 expression by RNA interference. Human Molecular Genetics. 2005;14(12):1641–1650. doi: 10.1093/hmg/ddi172. [DOI] [PubMed] [Google Scholar]
- 145.Fukuyama S, Yoshino I, Yamaguchi M, et al. Blockage of the macrophage migration inhibitory factor expression by short interference RNA inhibited the rejection of an allogeneic tracheal graft. Transplant International. 2005;18(10):1203–1209. doi: 10.1111/j.1432-2277.2005.00197.x. [DOI] [PubMed] [Google Scholar]
- 146.Bollerot K, Sugiyama D, Escriou V, et al. Widespread lipoplex-mediated gene transfer to vascular endothelial cells and hemangioblasts in the vertebrate embryo. Developmental Dynamics. 2006;235(1):105–114. doi: 10.1002/dvdy.20579. [DOI] [PubMed] [Google Scholar]
- 147.Pal A, Ahmad A, Khan S, et al. Systemic delivery of RafsiRNA using cationic cardiolipin liposomes silences Raf-1 expression and inhibits tumor growth in xenograft model of human prostate cancer. International Journal of Oncology. 2005;26(4):1087–1091. [PubMed] [Google Scholar]
- 148.Chien P-Y, Wang J, Carbonaro D, et al. Novel cationic cardiolipin analogue-based liposome for efficient DNA and small interfering RNA delivery in vitro and in vivo. Cancer Gene Therapy. 2005;12(3):321–328. doi: 10.1038/sj.cgt.7700793. [DOI] [PubMed] [Google Scholar]
- 149.Luo MC, Zhang DQ, Ma SW, et al. An efficient intrathecal delivery of small interfering RNA to the spinal cord and peripheral neurons. Molecular Pain. 2005;1:29. doi: 10.1186/1744-8069-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bertrand J-R, Pottier M, Vekris A, Opolon P, Maksimenko A, Malvya C. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochemical and Biophysical Research Communications. 2002;296(4):1000–1004. doi: 10.1016/s0006-291x(02)02013-2. [DOI] [PubMed] [Google Scholar]
- 151.Palliser D, Chowdhury D, Wang Q-Y, et al. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature. 2006;439(7072):89–94. doi: 10.1038/nature04263. [DOI] [PubMed] [Google Scholar]
- 152.Soutschek J, Akinc A, Bramlage B, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 153.Morrissey DV, Lockridge JA, Shaw L, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nature Biotechnology. 2005;23(8):1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 154.Golzio M, Mazzolini L, Moller P, Rols MP, Teissié J. Inhibition of gene expression in mice muscle by in vivo electrically mediated siRNA delivery. Gene Therapy. 2005;12(3):246–251. doi: 10.1038/sj.gt.3302405. [DOI] [PubMed] [Google Scholar]
- 155.Leng Q, Mixson AJ. Small interfering RNA targeting Raf-1 inhibits tumor growth in vitro and in vivo. Cancer Gene Therapy. 2005;12(8):682–690. doi: 10.1038/sj.cgt.7700831. [DOI] [PubMed] [Google Scholar]
- 156.Takei Y, Kadomatsu K, Yuzawa Y, Matsuo S, Muramatsu T. A small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer Research. 2004;64(10):3365–3370. doi: 10.1158/0008-5472.CAN-03-2682. [DOI] [PubMed] [Google Scholar]
- 157.Minakuchi Y, Takeshita F, Kosaka N, et al. Atelocollagenmediated synthetic small interfering RNA delivery for effective gene silencing in vitro and in vivo. Nucleic Acids Research. 2004;32(13):e109. doi: 10.1093/nar/gnh093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Takeshita F, Minakuchi Y, Nagahara S, et al. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(34):12177–12182. doi: 10.1073/pnas.0501753102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ito M, Yamamoto S, Nimura K, Hiraoka K, Tamai K, Kaneda Y. Rad51 siRNA delivered by HVJ envelope vector enhances the anti-cancer effect of cisplatin. Journal of Gene Medicine. 2005;7(8):1044–1052. doi: 10.1002/jgm.753. [DOI] [PubMed] [Google Scholar]
- 160.Song E, Zhu P, Lee S-K, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nature Biotechnology. 2005;23(6):709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 161.Yin C, Xi L, Wang X, Eapen M, Kukreja RC. Silencing heat shock factor 1 by small interfering RNA abrogates heat shock-induced cardioprotection against ischemiareperfusion injury in mice. Journal of Molecular and Cellular Cardiology. 2005;39(4):681–689. doi: 10.1016/j.yjmcc.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 162.de Jonge J, Holtrop M, Wilschut J, Huckriede A. Reconstituted influenza virus envelopes as an efficient carrier system for cellular delivery of small-interfering RNAs. Gene Therapy. 2006;13(5):400–411. doi: 10.1038/sj.gt.3302673. [DOI] [PubMed] [Google Scholar]