Abstract
Several signalling proteins involved in cell growth and differentiation represent attractive candidate targets for cancer diagnosis and/or therapy since they can act as oncogenes. Because of their high specificity and low immunogeneicity, using artificial small noncoding RNA (ncRNAs) as therapeutics has recently become a highly promising and rapidly expanding field of interest. Indeed, ncRNAs may either interfere with RNA transcription, stability, translation or directly hamper the function of the targets by binding to their surface. The recent finding that the expression of several genes is under the control of small single-stranded regulatory RNAs, including miRNAs, makes these genes as appropriate targets for ncRNA gene silencing. Furthermore, another class of small ncRNA, aptamers, act as high-affinity ligands and potential antagonists of disease-associated proteins. We will review here the recent and innovative methods that have been developed and the possible applications of ncRNAs as inhibitors or tracers in cancer medicine.
INTRODUCTION
The accumulation of multiple genetic alterations that affect the activity and/or expression of key proteins confers the proliferative and invasive characteristics of growth to cancer cells. Chromosomal deletions, rearrangements, and gene mutations are selected during cancer progression because these defect(s) lead to altered protein signalling networks and generate a survival advantage for the cancer cell [1].
The sequencing of the human genome, coupled to the availability of novel techniques as the high throughput screens and microarrays analysis, in less than a decade, has led to a vast accumulation of information about genes that are aberrantly regulated in cancers and has generated the realistic hope of identifying, at the molecular level, the fundamental processes that cause transformation from normal cell growth to malignancy.
The implications of this knowledge are profound because a detailed understanding of the complex interactions that occur at the genetic and protein levels provides attractive targets for rationally designing new drugs for new prevention and treatment approaches. Indeed, a major challenge of cancer research studies is to distinguish individuals at high risk of developing cancer thus to develop improved strategies for earlier diagnosis and more effective treatment with minimal side effects.
In the recent few years the increasing understanding on the function of small noncoding RNAs (ncRNAs) has as well generated a great enthusiasm because these molecules may provide an obvious potential use as powerful new tools in cancer medicine. Under the definition of ncRNAs falls a broad range of regulatory RNA molecules, such as ribozymes, antisense, interfering small RNAs or aptamers, that are either naturally found in several cell types or are artificially designed to target gene expression or protein function (Figure 1). The advantage of these biomolecules over other biochemical or chemical substances employed up to now include high potency and specificity for the target, use of in vitro techniques for their production, that considerably reduce production costs as well as the need for animal testing and that markedly increase specificities and quality assurance in diagnostic and therapeutic applications.
Figure 1.

Schematic representation of the mode of action of aptamers compared to other ncRNAs. Antisense, ribozymes, siRNAs, miRNAs recognise the target nucleic acid by complementary base pairing and, by activating an intracellular molecular machinery, impair the expression of the corresponding protein. Aptamers act by directly binding the target without interfering with its expression.
In this review, we will examine recent work in the possible applications of artificial small ncRNAs as versatile biomolecules to identify and validate cancer targets and as inhibitors or tracers in cancer medicine. The advantages and drawbacks of the competing methodologies will be discussed here.
A HETEROGENEOUS FAMILY OF RNA-BASED TOOLS
Small noncoding RNAs elicit at least four distinct types of responses that trigger specific gene inactivation, including destruction of homologous mRNA, inhibition of translation, de novo methylation of genomic regions that can block transcription of target genes, and chromosomal rearrangement.
Among small RNAs, the microRNAs (miRNAs) are 21-base-long RNA molecules that regulate gene expression. In mammalians, miRNAs act by imperfectly base-pairing predominantly within the 3′ untranslated region of target messenger RNAs and inhibiting protein translation [2]. Because of their importance in the regulation of gene expression miRNAs have been implicated in the modulation of several physiological and pathological cellular processes.
In RNA silencing mediated by siRNAs the sequence-specific gene inhibition is initiated by small RNA duplexes that cause the destruction of complementary target messenger RNA.
Aptamers are single-stranded oligonucleotides whose inhibiting function relies on a completely different mechanism with respect to miRNA and siRNA. These molecules are generated by an in vitro evolutionary selection-amplification scheme, named SELEX (systematic evolution of ligands by exponential enrichment) [3, 4]. Because they bind their target molecules at high affinity and specificity, displaying dissociation constants in the low picomolar to low nanomolar range, aptamers are considered as attractive therapeutic agents that rival antibodies.
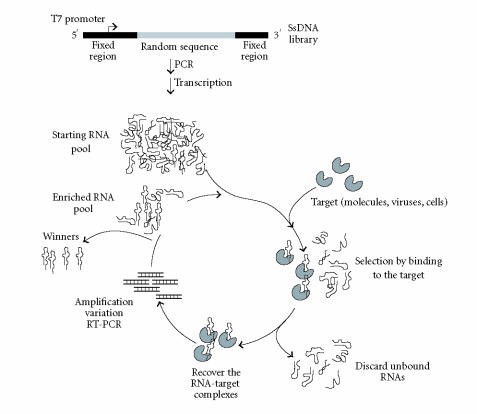
As shown in Figure 2, the starting point for the generation of an aptamer is the synthesis of a nucleic acid library (RNA, DNA, or modified RNA) of large sequence complexity followed by the selection for oligonucleotides able to bind with high affinity and specificity to a target molecule. Randomisation of a synthetic sequence stretch from 22 up to 100 nucleotides in length has been used to create an enormous diversity of possible sequences (4N different molecules) which in consequence generate a vast array of different conformations with different binding properties. The SELEX method includes the following steps: (i) incubating the library with the target molecule under conditions favourable for binding; (ii) partitioning: molecules that, under the conditions employed, adopt conformations that permit binding to a specific target are then partitioned from other sequences; (iii) dissociating the nucleic acid-protein complexes; and (iv) amplifying of the nucleic acids pool to generate a library of reduced complexity enriched in sequences that bind to the target. This library will be then used as starting pool for the next round of selection. After reiterating these steps for a variable number of cycles, the resulting oligonucleotides are subjected to DNA sequencing. The sequences corresponding to the initially variable region of the library are screened for conserved sequences and structural elements indicative of potential binding sites and subsequently tested for their ability to bind specifically to the target molecule. This selection scheme works since single-stranded nucleic acids fold up into unique 3D shapes in a similar manner to proteins, each structure being unique and dictated by the sequence of the nucleic acid.
Figure 2.
Schematic representation of the SELEX process. The single-stranded (ss) DNA library is amplified by polymerase chain reaction (PCR) in order to generate the double-stranded DNA pool that will be transcribed by T7 RNA polymerase. The pool of RNA molecules with different conformations will be used for the selection process (see text for details).
By starting with 1015 random DNA sequences (thus, to a first approximation, 1015 specific shapes), it is possible to select (through 10–15 rounds of selection-amplification) specific binding reagents for virtually each targeted human protein.
UNDERSTANDING THE MOLECULAR MECHANISMS OF CANCER
Determining gene function
Determining by reverse genetics the function and the biological relevance of a given protein for a particular cancer type is a critical step to validate the most promising molecular targets for drug development. The possible strategies that are usually used to understand the function of a specific gene in a cell are either based on techniques that impair the expression of the candidate target gene or rely on the use of products that act by specifically interfering or inhibiting the function but not the expression of the final product. In both cases the resulting phenotype turns out as a powerful source of information on the function of the target protein (Figure 1).
(a) Gene silencing: the generation of null mutants by homologous recombination of a given gene in a cell or in an entire organism has been extensively used to create models of several human diseases, including cancer. Using this technique (named gene knockout), in which the gene of interest is irreversibly disrupted and the synthesis of the encoded products abolished, allowed to make an incredible and rapid progress in our understanding of the function of several oncogenes and tumor-suppressor genes. However, triggering gene silencing by homozygous gene ablation is laborious and expensive and thus rather inappropriate for a large-scale screening. An alternative strategy has been recently developed to determine the roles of particular genes in cancer that is based on the use of ncRNAs for gene silencing. The RNA interference (RNAi) has proven to be a precious approach that permits loss-of-function phenotypic screens in mammalian somatic cells or in whole animals at high specificity. Indeed, in the last few years the sequencing of the entire human genome coupled to advancements in high-throughput oligonucleotide synthesis and better prediction of active sequences is allowing the design of RNAi constructs against virtually any transcript. Furthermore, in contrast to the knockout approach, the RNA interference-based strategies achieve loss of function phenotypes without the loss of genomic information of the targeted gene (recently reviewed in 5). This leaves the possibility to restore the exact expression of the endogenous gene once the RNAi vector is silenced or removed.
(b) Functional inhibitors: a major drawback to gain information on the function of a given protein by impairing its expression is that proteins and enzymes involved in crucial functions, such as cell growth and differentiation, frequently act in concert with various partners thus forming large stable complexes that dictate its function in the cell environment. Therefore, depleting a single key protein from the cell will change, or even disrupt, at the same time one or more of these multiprotein complexes. As a consequence, the resulting phenotype will be produced by the simultaneous impairment of several protein functions and the understanding stays frequently ambiguous. Furthermore, silencing a gene gives no information about which region of the putative target protein is important for its function.
To overcome these disadvantages, additional approaches have been developed that enable to interfere with a given protein function. Indeed, using monoclonal antibodies, peptides, and small molecules to directly target the protein in a drug-like manner has the advantage to interfere with the protein activity without depleting the protein itself and thus with low destabilisation of the proteomic status of the cell. During the past decade, as excellent alternatives to these inhibitors, the RNA-based aptamers have proven to be highly selective ligands and efficient inhibitors of a wide variety of proteins implicated in cancer. Aptamers have a larger surface area as compared to small interfering compounds thus presenting more points of contact with the target protein. Furthermore, these molecules have been shown either to inhibit their target by competitive mechanisms or to interfere with its conformation by noncompetitive mechanisms [6]. The ability to select aptamers directed against purified soluble targets has recently incredibly progressed thanks to the automation of part of the in vitro selection processes so that several targets can be isolated in parallel strongly reducing the time required for the selection. Aptamers for protein targets of biomedical interest have been reported and many of them are actually under clinical trials for cancer treatment (Table 1). In particular, Food and Drug Administration has recently approved one aptamer developed by Eyetech (Macugen™) that inhibits the human vascular endothelial growth factor 165 (VEGF165), for the treatment of age-related macular degeneration [7]. Since an obvious potential therapeutic use for aptamers to VEGF is in cancer, this aptamer was tested in a mouse model of nephroblastoma [8]. Renal histopathology revealed an 84% reduction in tumor weight in the aptamer-treated kidneys compared to the controls. Furthermore, lung metastases were seen in 20% of the aptamer-treated mice compared to 60% of control animals. This same aptamer was also tested in a murine model of neuroblastoma, where it resulted in 53% reduction in tumor growth compared to control [9]. Another aptamer that inhibits receptor tyrosine kinase activation by binding to the corresponding ligand is the aptamer against platelet-derived growth factor (PDGF). It has been successfully used in vivo in animal models of cancer due to its high specificity in the fact that it suppresses PDGF B-chain (PDGF-BB) but not the epidermal- or fibroblast-growth-factor-2-induced proliferation [10]. Furthermore, the SELEX methodology has been used to identify high-affinity 2'-aminopyrimidine RNA ligands to the potent angiogenic factor, the basic fibroblast growth factor (bFGF). In cell culture, these aptamers inhibited bFGF binding to both low-affinity sites and high-affinity sites on FGF receptor-1 [11].
Table 1.
Therapeutic aptamers in cancer treatment.
| Aptamer | Aptamer activity | The rapeutic application | |
| in vitro | in vivo | ||
| Macugen | Inhibition of VEGF165 | Inhibition of the VEGF-induced | Approved by FDA for treatment of age-related |
| vascular permeability | macular degeneration | ||
| ProMune | Agonist for toll-like receptor 9 (TLR 9) | Activate the immune system through TLR 9 against cancer | Phase 2: melanoma |
| Phase 1: renal cell carcinoma; | |||
| non- Hodgkin's lymphoma; cutaneous T-cell | |||
| lymphoma, non-small-cell lung cancer | |||
| Agro 100 | Binding to nucleolin | Antiproliferative activity in a broad | Phase 1 |
| array of tumor cell types; enhancement | Phase 2 launched in 2005 | ||
| of chemotherapeutic agents effects | for advanced solid malignancies | ||
| HYB2055 | Agonist for TLR 9 | Antitumor activity in nude mouse | Phase 2 for advanced solid malignancies |
| xenografts with colon, breast | |||
| lung cancer, and glioma cell lines | |||
| VaxImmune | Agonist for TLR 9 | Elicits a powerful immune response | Phase 2 for several different cancer indications |
| adjuvant | against infectious disease and cancers | ||
Even though several aptamers that inhibit receptor tyrosine kinases by binding to their soluble ligands have now been selected, targeting the receptor itself that is a large insoluble glycosylated protein has only been recently addressed [12]. To this aim we developed a general protocol of differential whole-cell SELEX to target cell surface bound proteins in their natural physiological environment. We have evolved aptamers able to inhibit an active mutant of the receptor tyrosine kinase, Ret, by targeting its extracellular domain in which such activating mutation is located. By this method, the selection procedure was performed by using as target the RETC634Y mutant expressed on PC12 cells. A library of 2′-fluoro pyrimidine RNAs was incubated with parental PC12 cells to remove aptamers that bind nonspecifically to the cell surface. To select for aptamers that specifically bound the mutant receptor, the supernatant was incubated with PC12–RETC634Y cells. Unbound sequences were washed off, the whole process reiterated 16 times, and the bound winning sequences cloned. The resulting aptamers did not bind to a recombinant EC C634Y RET fragment highlighting the strength of the whole-cell approach. Among the selected aptamers, the best inhibitor (D4) binds specifically to the Ret receptor tyrosine kinase and blocks its downstream signalling effects on cell differentiation and transformation [12]. The results suggest that the differential whole-cell SELEX approach will be useful in the isolation of other lead therapeutic compounds and diagnostic cell-surface markers. Aptamers that have high affinity and specificity for tissues have also been produced, demonstrating that complex targets, including tumour tissue, are compatible with the SELEX process. “Tissue SELEX” methodology could be favourable when the precise molecular target is unknown but the target is, for example, a specific type of cells. A fluorescence based SELEX-procedure was applied against transformed endothelial cells as a complex target to detect microvessels of rat experimental glioma, a fatal brain tumour which is highly vascularized. A secondary selection scheme, named deconvolution-SELEX, was carried out to facilitate the isolation of ligands for components of interest within the targeted mixture. Other examples of proteins that have been reported as targets for development of aptamers as promising therapeutics include the tenascin-C, the Ras binding domain of Raf-1, the prostate specific membrane antigen (PMSA), the protein kinase CβII, the epidermal growth factor receptor-3 (ErbB3/HER3), and the CXCL10 chemokine [13].
Identifying “cancer genes”
Two paradigmatic and elegant recent papers well demonstrate the power of siRNAs in identifying new cancer targets involved in cell survival. Apoptosis-based anticancer therapies are designed to achieve tumor eradication through the use of death-inducing molecules. Because of its specific toxicity against transformed cells, the tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) receptors are judged to be amongst the most promising apoptosis-based antitumor targets. To better understand the molecular mechanisms of TRAIL-induced apoptosis, Aza-Blanc et al [14] carried out a large RNAi-based screen to identify genes that modulate TRAIL-induced apoptosis. They used an siRNA library to individually target 510 human genes (corresponding to 380 predicted kinases and 130 other predicted proteins) transfected in the cervical carcinoma derived HeLa cell line. By using an assay that could detect both sensitization and resistance to TRAIL-induced apoptosis the authors identified a variety of genes that modulate both positively and negatively TRAIL activity. Furthermore, besides genes that encode well-characterised mediators of TRAIL, including the DR4 receptor for TRAIL and the caspase 8 or known modulators of apoptosis (Myc and JNK3), in the RNAi screen they detected and functionally characterised two previously unknown genes, which were found to modulate TRAIL-induced apoptosis. One of the genes, DOBI (Downstream of Bid), is required for progression of the apoptotic signal through the intrinsic mitochondrial cell death pathway indicating that it may function to mediate cytochrome c release induced by BID cleavage, the other, named MIRSA (Mina53- related suppressor of apoptosis), is a gene that acts to prevent TRAIL-induced death.
Changes in the expression and/or activity of kinases and phosphatases, key proteins of the cellular signalling pathways, are the most frequent molecular causes of cancer progression. MacKeigan et al [15] reported a large-scale RNAi approach to identify kinases and phosphatases that regulate cell survival and apoptosis. Authors transfected in HeLa cells two large siRNA library sets against human kinase and phosphatase, containing two siRNAs for each target (corresponding to 650 kinases and to 222 phosphatases). By the data obtained they predicted that 11% of kinases and 32% of phosphatases are constituted of genes whose expression is critical for cell survival. In addition to those for previously known “survival kinases” (Akt2, KD2, SGK, PKCdelta) they identified several genes with still unknown function, the silencing of two of which resulted in a strong increase in apoptosis. They also identified phosphatases that act as tumor suppressors to sensitize or promote apoptosis. Silencing of these “cell death phosphatases” resulted in marked cell protection to chemotherapeutic-induced cell death. Furthermore, the authors developed a further screen in the presence of low doses of chemotherapeutics by which they identified kinases whose silencing increases the rate of apoptosis. The kinases identified are thus promising targets for silencing in order to sensitize the cancer cell to low doses of chemotherapeutic agents, thus reducing unwanted side effects in chemotherapy.
On the other hand, as alternative to transfection, siRNAs can be also expressed within the cells as a short hairpin RNA (shRNA) that is then processed by the cell machinery to produce the small interfering double strand RNA. To facilitate the use of RNAi as a genetic tool in mammals, Paddison et al [16] and Berns et al [17] used a shRNA retrovirus-based library approach to develop strategies that allow a high throughput RNAi-based screen of mammalian genes. Berns et al screened a library targeting around 8.000 human genes for those that affect the function of the tumour suppressor, p53. Genes were subsequently identified by microarray detection of the shRNA sequence [17]. Paddison and colleagues developed a shRNA library targeting around 10.000 human and more than 5.000 mouse genes. To facilitate the use in virtually any cell types, their shRNA expression library was constructed in a vector that permits moving the shRNA encoding inserts to different vectors by bacterial mating and designed to function for both genetic selections and screens. Indeed, for facilitating the screening, in addition to the selection pressure both groups adopted a unique DNA “bar-code” sequence present in the vector which can be identified using microarrays containing oligonucleotides corresponding to the bar-code sequences.
The generation of large siRNA libraries has been further improved by using, instead of shRNAs that are transcribed by the RNA polymerase III, pri-miRNA based large transcripts that permit to generate siRNAs driven by the RNA polymerase II promoters that can be thus tightly regulated both in culture and in vivo, in animal models [18, 19].
Two recent reports [20, 21] address the use of large viral-based RNAi libraries to identify novel potential tumor-suppressor genes thus further underscoring the power of RNAi screening to understand the molecular mechanisms of neoplastic transformation.
Kolfschoten et al [20] used a shRNA retrovirus-based library [17] in combination with in vitro neoplastic transformation assays to screen for novel tumor-suppressor genes. To this aim they used a Ras-dependent transformation model of genetically modified human primary BJ fibroblast cells expressing the catalytic subunit of telomerase (hTERT), and SV40 small T antigen (ST) in combination with the inhibition of the expression of p53 and p16INK4A. In this cell line the expression of the oncogenic H-RasV12 is sufficient to confer anchorage-independence for survival and proliferation [22]. By this approach the authors identified few genes whose silencing substitutes for the activity of the oncogenic Ras one of which, the homeodomain transcription factor PITX1, was not previously implicated as possible tumor suppressor allowing anchorage-independent growth of fibroblasts. The authors showed that PITX1 regulates the Ras pathway and thereby tumorigenesis. The mechanism appears to involve constitutive activation of the Ras signalling pathways at the level of GTP loading onto Ras itself. Indeed, PITX1 directly controls the expression of RASAL1, one of the negative regulators of Ras belonging to the GTPase-activating protein (GAP) family. The evidence that PITX1 is a tumor suppressor was supported by the strong correlation between the low PITX1 levels present in colon cancer cell lines and wt Ras expression and the low PITX1 expression levels in prostate and bladder tumor tissues compared with normal tissues.
To identify genes that suppress oncogenic transformation, Wetsbrook et al [21] used genetically modified human epithelial mammary cells that have been immortalized by the expression of hTERT and SV40 large T antigen and also naturally express high levels of Myc (TLM-HMECs). Forced expression of an active mutant of PI3K confers to these cells the ability to grow in an anchorage-independence manner [23]. Based on the assumption that inactivation of a single tumor-suppressor gene may be sufficient to shift TLM-HMECs cells into a frankly transformed phenotype that can be selected for, the authors used a shRNA retrovirus-based library to infect TLM-HMECs [16]. In the screen they identified several potential suppressors of epithelial cell transformation genes that represent potential tumor suppressors. Most of them were associated with genes known to be involved in key intracellular signalling pathways including Ras, PI3K, and TGF-β signalling. In addition, they provided evidence that one of these candidates, the transcriptional repressor REST/NRSF, plays a previously unknown role in tumor suppression. REST encodes a transcriptional factor involved in repressing neuronal genes in non-neuronal cells. Even though the mechanism by which REST silencing releases the transformed state should be still elucidated, it likely involves regulation of PI3K. Indeed, impaired REST function promoted epithelial cell transformation, enhanced the intensity and duration of PI3K activity, eventually acting via the transcriptional control of neurotrophins.
Despite the expanding potentiality of siRNAs the key challenges for their development for gene silencing is largely dependent on the improvement of siRNA specificity. Not every sequence works, and a success rate of about one of three should be expected. In addition, although the effects are generally thought to be highly sequence specific, one potential concern in using siRNAs for phenotypic screens is that the observed effects could be due to inhibition of either the intended target or of an off-target mRNA [24]. Indeed, siRNAs are not perfectly selective and results should be confirmed with an independent siRNA targeting of the same transcript to understand whether or not some of the effects see result from an off-target transcript. Generation of libraries of multiple siRNAs for each gene has been therefore the most frequent approach used to avoid this drawback and be safe to conclude that the effects are specific to the targeted gene.
DEFINING THE SIGNATURES OF CANCER CELLS: MICRORNAs PROFILING
Early detection together with the accurate description of the tumor type is crucial for a better diagnosis of cancer and a more effective therapy. Therefore, what is required to gain an increased survival rate of the patient is the identification of multiple biomarkers that can be measured simultaneously as a biological signature of the disease state. Using high-throughput technologies allows the identification of these signatures and their validation by the rapid comparison of samples from many different patients with the realistic hope of finding molecules that are informative of the type of cancer and with high predictive value for the patient.
The coordinated expression of specific miRNAs is believed to have a central role in diverse cellular processes, including cell proliferation and apoptosis, and their altered expression is involved in tumorigenesis. Indeed, expression profiling of differential miRNA has been shown to represent highly informative signatures for human cancers. Lu et al [25] developed a bead-based technique coupled to flow cytometry to determine the expression profiles of 217 human miRNAs in 332 cancer samples. They found that the expression pattern of a small set of miRNA dramatically varies across tumour types, reflecting the lineage and differentiation status of the tumor. In contrast, profiling expression data obtained on the same samples using 16,000 mRNAs was ineffective, thus implicating miRNA profiling as highly informative to classify human cancer types [25]. Furthermore, their results show that the expression pattern of a small set of miRNAs is highly informative to classify human cancer types.
Furthermore, as well illustrated by the three following recent papers, the development of microarrays containing all known miRNAs permitted to perform large-scale analysis of miRNA expression profiling in human cancers. In the first report the authors [26] evaluated the activity of miRNA genes in chronic lymphocytic leukemia (CLL) cells from 94 patients. Of the miRNA genes analysed, the researchers found that the activity pattern of 13 of them accurately predicted whether a person had the slow- or fast-progressing form of CLL. They also identified a germ-line mutation in the miR-16-1-miR-15 precursors, which caused low levels of miRNA expression and thus may be considered as cancer susceptibility genes for CLL (see below). In a second paper, by applying a similar approach, the same group examined miRNA expression profile in 76 breast tumors compared to normal breast tissue [27]. In this study 29 miRNAs were significantly deregulated in breast cancer (either over-expressed or downregulated). They found that miRNA expression was correlated with breast tumors' hormone status as well as its metastatic, invasive, and proliferative potential. Most important, their work demonstrated that the expression pattern of as few as five miRNAs (miR-10b, miR-125b, miR-145, miR-21, and miR-155) was sufficiently informative to successfully distinguish normal tissue from cancerous tissue. Finally, in the paper by Huiling He et al [28] the researchers examined samples of malignant tissue from 15 patients diagnosed with papillary thyroid carcinoma and compared them with the normal tissue adjacent to the tumors. They found 23 miRNAs that were significantly altered in the cancerous tissue when compared with the normal counterparts, with three of them (miR-146, miR-221, and miR-222) dramatically overexpressed, reaching 11-to-19-fold higher levels of expression in the tumors. Further investigation revealed that the expression pattern of miR-146, miR-221, and miR-222 if combined with that of two additional miRNAs (miR-21 and miR-181a), formed a “signature” that clearly predicted the presence of malignant tissue.
In conclusion, in several cancers the miRNA expression profiles seem to be sufficient to provide a “signature” that is directly associated to the clinical status of the disease. Indeed, the utility of single markers in diagnosis and monitoring of cancer is limited by the poor association of any single protein with a specific disease or stage of disease. Thus identifying the distinctive signature of a network of these regulatory signals would enable us by a more precise diagnosis to detect tumors earlier, at times when treatments are more effective.
CANCER SIGNATURE MEASUREMENT
Developing methods that allow clinicians and researchers to translate signature discoveries to routine clinical use by looking simultaneously at a large number of biomarkers has now become a major challenge in cancer diagnosis. Indeed, because they are readily accessible without any need of invasive intervention measuring molecules expressed in serum or plasma is highly preferable. However, many potential cancer biomarkers in biological fluids are present at low concentrations, presumably in the low nanomolar range. Therefore, the capability to measure multiple protein markers simultaneously depends on methods having not only low limits of detection with elevated signals, but also coupled to very low noise, thus capable to distinguish specific protein signalling in the presence of a huge excess of unrelated proteins.
The use of aptamers as biorecognition element for the development of biosensors to detect protein targets offers over classical methods mainly based on antibodies, a lot of advantages, such as the possibility of easily regenerate the immobilised aptamers, their homogeneous preparation, and the possibility of using different detection methods due to easy labelling [29]. Moreover, the enormous diversity of random oligonucleotide libraries can exceed the diversity of antibodies in the mammalian genome by several orders of magnitude. Since aptamers are nucleic acids, experience with DNA, as in the production of DNA arrays, should be applicable to the development of aptamer-based biosensors. On the other side, the aptamer arrays can potentially expand the scope of DNA microarrays to recognise expressed proteins as well as expressed mRNAs. In this regard, numerous aptamers have already been selected against a wide array of proteins, and the possibility of acquiring aptamers against proteomes has been advanced by automation of the in vitro selection procedure. These considerations explain why now the aptamer-based technology for protein detection is in advanced stages of development as useful tools in clinical diagnosis and therapy. Furthermore, this technology has been improved by the use of modified aptamer molecules, named photoaptamers by Petach et al at SomaLogic, Inc [30–33]. These modified aptamers (either DNA or RNA) at specific locations include, in place of thymidine residues, the photoreactive 5-bromodeoxyuridine (BrdU) that can form a specific covalent crosslink with the target proteins. Indeed, short pulses of ultraviolet light at 308 nm induce a chemical crosslink between the BrdU residue and the electron rich amino acid on the target protein that is in a specific location in proximity and in the correct juxtaposition of the BrdU. Since this cross-linking event is dependent on the correct juxtaposition of the BrdU and the target amino acid, it conveys specificity to the photoaptamer-protein complex. This gives rise to multiplicative specificity by a photochemical cross-link that follows the initial affinity binding event. Proteins captured on the array are then measured either by universal protein staining or by using specific antibodies.
In order to measure simultaneously large numbers of proteins, even thousands, in biological fluids multiple capture photoaptamers can be deposited and covalently linked to the appropriate chip surface. Therefore, since photoaptamers covalently bind to their targets before staining, the photoaptamer arrays can be vigorously washed to remove background proteins, thus providing the needed potential for elevated signal-to-noise ratios.
The sensitivity and specificity of photoaptamers, combined with the ability to automate and scale up their selection and the ability to use them on solid surfaces, indicate that they could become an important factor in the development of proteomic technology.
IN VIVO DELIVERING
Whether being used as experimental tools and/or pharmaceutical drugs, small ncRNAs need to be able to cross cell membranes but negatively charged oligonucleotides will not pass through a lipid layer such as cell membranes. Similar to antisense oligonucleotides and ribozymes, the delivery of synthetic siRNAs and aptamers can be improved via the use of various delivery systems, which include synthetic carriers, composed primary of lipids. Larger carriers such as liposomes localise the drugs mainly to the blood compartment. However, angiogenic blood vessels in most tissues have gaps between adjacent endothelial cells large enough to cause the extravagation of liposome/siRNA complexes into the tumor. In addition, most solid tumors possess an enhanced vascular permeability and impaired lymphatic drainage, which leads to the accumulation of most liposomes within the tumor tissues. For these reasons, a big effort has recently been devoted to develop delivery vehicles that can efficiently deliver these RNAs molecules in vivo for the success of these molecules as therapeutics. Furthermore, the possibility to chemically modify and easily engineer small RNAs permits to express these molecules inside the target cell thus coupling the advantages of drug-based to those of gene-based therapy.
CONCLUDING REMARKS
Over the past few years the unexpected progress gathered in the knowledge of the mode of action of small noncoding RNAs is changing our point-of-view on the possible approaches to identify and to target tumor-associated genes. These flexible molecules have proven their enormous potential both as diagnostic and therapeutic tools in several fields of cancer medicine. Paradigmatic examples in the use of these small RNA molecules include that of siRNAs for high selective gene silencing, aptamers as high affinity inhibitory ligands, the miRNA profiling for a more accurate description of the tumor state, and the use of high sensitive aptamer-based biosensors to readily measure the proteomic status in biological fluids. Therefore, developing ncRNAs as the new generation of molecular tools for human health is an urgent challenge that in the next future should provide us with the ability to tailor therapies to the cancer patient more effectively.
ACKNOWLEDGMENTS
This work was supported by the European Molecular Imaging Laboratory (EMIL) Network (LSHC-2004-503569), and by the MIUR-FIRB Grant (#RBIN04J4J7).
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 4.Tuerk C, Gold L. Systemic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 5.Dykxhoorn DM, Lieberman J. The silent revolution: RNA interference as basic biology, research tool, and therapeutic. Annual Review of Medicine. 2005;56:401–423. doi: 10.1146/annurev.med.56.082103.104606. [DOI] [PubMed] [Google Scholar]
- 6.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Research. 2002;62(14):4029–4033. [PubMed] [Google Scholar]
- 7.Ruckman J, Green LS, Beeson J, et al. 2′-fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165): inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. Journal of Biological Chemistry. 1998;273(32):20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 8.Huang J, Moore J, Soffer S, et al. Highly specific antiangiogenic therapy is effective in suppressing growth of experimental Wilms tumors. Journal of Pediatric Surgery. 2001;36(2):357–361. doi: 10.1053/jpsu.2001.20716. [DOI] [PubMed] [Google Scholar]
- 9.Kim ES, Serur A, Huang J, et al. Potent VEGF blockade causes regression of coopted vessels in a model of neuroblastoma. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(17):11399–11404. doi: 10.1073/pnas.172398399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Floege J, Ostendorf T, Janssen U, et al. Novel approach to specific growth factor inhibition in vivo: antagonism of platelet-derived growth factor in glomerulonephritis by aptamers. American Journal of Pathology. 1999;154(1):169–179. doi: 10.1016/S0002-9440(10)65263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jellinek D, Green LS, Bell C, et al. Potent 2′-amino-2′-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry. 1995;34(36):11363–11372. doi: 10.1021/bi00036a009. [DOI] [PubMed] [Google Scholar]
- 12.Cerchia L, Ducongé F, Pestourie C, et al. Neutralizing aptamers from whole-cell SELEX inhibit the RET receptor tyrosine kinase. PLoS Biology. 2005;3(4):697–704. doi: 10.1371/journal.pbio.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Blank M, Weinschenk T, Priemer M, Schluesener H. Systematic evolution of a DNA aptamer binding to rat brain tumor microvessels: selective targeting of endothelial regulatory protein pigpen. Journal of Biological Chemistry. 2001;276(19):16464–16468. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- 14.Aza-Blanc P, Cooper CL, Wagner K, Batalov S, Deveraux QL, Cooke MP. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Molecular Cell. 2003;12(3):627–637. doi: 10.1016/s1097-2765(03)00348-4. [DOI] [PubMed] [Google Scholar]
- 15.MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nature Cell Biology. 2005;7(6):591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 16.Paddison PJ, Silva JM, Conklin DS, et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428(6981):427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 17.Berns K, Hijmans EM, Mullenders J, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428(6981):431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 18.Silva JM, Li MZ, Chang K, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nature Genetics. 2005;37(11):1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 19.Dickins RA, Hemann MT, Zilfou JT, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nature Genetics. 2005;37(11):1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 20.Kolfschoten IGM, van Leeuwen B, Berns K, et al. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell. 2005;121(6):849–858. doi: 10.1016/j.cell.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 21.Westbrook TF, Martin ES, Schlabach MR, et al. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121(6):837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 22.Voorhoeve PM, Agami R. The tumor-suppressive functions of the human INK4A locus. Cancer Cell. 2003;4(4):311–319. doi: 10.1016/s1535-6108(03)00223-x. [DOI] [PubMed] [Google Scholar]
- 23.Zhao JJ, Gjoerup OV, Subramanian RR, et al. Human mammary epithelial cell transformation through the activation of phosphatidylinositol 3-kinase. Cancer Cell. 2003;3(5):483–495. doi: 10.1016/s1535-6108(03)00088-6. [DOI] [PubMed] [Google Scholar]
- 24.Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi. Nature Biotechnology. 2003;21(6):635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 25.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 26.Calin GA, Ferracin M, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. New England Journal of Medicine. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 27.Iorio MV, Ferracin M, Liu C-G, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Research. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 28.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(52):19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Franciscis V, Cerchia L. Aptamer-based technologies as new tools for proteomics in diagnosis and therapy. In: Caldwell GW, Rahman AU, D'Andrea M, Choudhary MI, editors. Frontiers in Drug Design & Discovery. Vol 2. Ewing Township, NJ: Bentham Science; 2006. pp. 103–119. [Google Scholar]
- 30.Petach H, Ostroff R, Greef C, Husar GM. Processing of photoaptamer microarrays. Methods in Molecular Biology. 2004;264:101–110. doi: 10.1385/1-59259-759-9:101. [DOI] [PubMed] [Google Scholar]
- 31.Gander TR, Brody EN. Photoaptamer chips for clinical diagnostics. Expert Review of Molecular Diagnostics. 2005;5(1):1–3. doi: 10.1586/14737159.5.1.1. [DOI] [PubMed] [Google Scholar]
- 32.Golden MC, Collins BD, Willis MC, Koch TH. Diagnostic potential of PhotoSELEX-evolved ssDNA aptamers. Journal of Biotechnology. 2000;81(2-3):167–178. doi: 10.1016/s0168-1656(00)00290-x. [DOI] [PubMed] [Google Scholar]
- 33.Bock C, Coleman M, Collins B, et al. Photoaptamer arrays applied to multiplexed proteomic analysis. Proteomics. 2004;4(3):609–618. doi: 10.1002/pmic.200300631. [DOI] [PubMed] [Google Scholar]