Abstract
Retrotransposons like L1 are silenced in somatic cells by a variety of mechanisms acting at different levels. Protective mechanisms include DNA methylation and packaging into inactive chromatin to suppress transcription and prevent recombination, potentially supported by cytidine deaminase editing of RNA. Furthermore, DNA strand breaks arising during attempted retrotranspositions ought to activate cellular checkpoints, and L1 activation outside immunoprivileged sites may elicit immune responses. A number of observations indicate that L1 sequences nevertheless become reactivated in human cancer. Prominently, methylation of L1 sequences is diminished in many cancer types and full-length L1 RNAs become detectable, although strong expression is restricted to germ cell cancers. L1 elements have been found to be enriched at sites of illegitimate recombination in many cancers. In theory, lack of L1 repression in cancer might cause transcriptional deregulation, insertional mutations, DNA breaks, and an increased frequency of recombinations, contributing to genome disorganization, expression changes, and chromosomal instability. There is however little evidence that such effects occur at a gross scale in human cancers. Rather, as a rule, L1 repression is only partly alleviated. Unfortunately, many techniques commonly used to investigate genetic and epigenetic alterations in cancer cells are not well suited to detect subtle effects elicited by partial reactivation of retroelements like L1 which are present as abundant, but heterogeneous copies. Therefore, effects of L1 sequences exerted on the local chromatin structure, on the transcriptional regulation of individual genes, and on chromosome fragility need to be more closely investigated in normal and cancer cells.
INTRODUCTION
In normal somatic human cells, transcription of retrotransposon sequences like L1 and illegitimate recombination involving them are suppressed, restricting their activity to developing germ cells and placental tissues [1–3]. Suppression of retroelement activity prevents not only retrotransposition, but also various disturbances of transcription by retroelement promoters, interference by retroelement enhancers, the activity of retroelement-encoded enzymes, and illegitimate recombination between homologous elements. Moreover, while L1 sequences have the potential to create genomic instability, they probably exert certain beneficial, “symbiotic” effects. For instance, silencing of retrotransposons in somatic cells may help to organize the genome into macro- and microdomains with differential transcriptional activity. Failure to silence retroelements in cancer cells could therefore permit adverse activities of retroelements as well as perturb any beneficial effects.
The present paper summarizes current knowledge about L1 (LINE-1) retrotransposons in human cancer. For comparison, some observations on human endogenous retroviruses (HERV) are included [4]. Throughout the text, the emphasis will be on identifying open questions, of which there are plenty, as should become evident.
Since L1 general biology is treated in detail in recent reviews [1–3] and other contributions in this issue, only a short introduction will follow here. L1 sequences represent the major class of LTR-less retrotransposons in humans and constitute about 18% of the human genome. While they are interspersed throughout the genome, including euchromatic and heterochromatic regions, they are particularly frequent in gene-poor regions that correspond to chromosomal G-bands. Full-length elements are 6 kb in size and contain an internal promoter at the 5′-end that generates a genomic transcript which also serves as an mRNA. The RNA contains two open reading frames, ORF1 and ORF2. ORF1 encodes p40, an RNA-binding protein with cis preference for L1 RNA. ORF2 encodes the endonuclease and reverse transcriptase required for retrotransposition. Only a fraction of L1 elements in the human genome are intact. Most are truncated, usually at the 5′-end, and mutated, often at many sites. The up to 400, 000 elements that are still distinctly recognizable as L1 can be categorized into several subclasses. Most and perhaps all elements still capable of retrotransposition belong to a subclass named Ta. Normally, transcriptional activity of L1 is restricted to developing germ cells and to cells of the placenta. In somatic cells, L1 transcription and retrotransposition is prevented by a variety of control mechanisms, including methylation of L1 DNA and specifically L1 promoters. In the germline, these mechanisms are relaxed, and retrotransposition does occur occasionally.
Potential dangers
Dangers resulting from L1 reactivation in cancer cells comprise the direct adverse effects of retrotransposition, enhanced illegitimate recombination, and multiple ways of disturbance of transcriptional activity and gene regulation. In the human genome, fewer than 100 L1 elements are thought to be sufficiently intact for retrotransposition [3]. However, while the danger of retrotransposition is posed only by these intact L1s, other adverse effects can be exerted by a larger number of elements. In addition, reactivation may interfere with potential “symbiotic” effects of L1 sequences such as their contribution to the global and local organization of the genome and the provision of gene regulatory sequences. Activities on the immune system can also be envisioned. These would be expected to have ambiguous consequences. It seems therefore imprecise to consider alterations of L1 in cancer solely as “reactivation,” other effects may more appropriately be characterized as “dysregulation.”
Retrotransposition
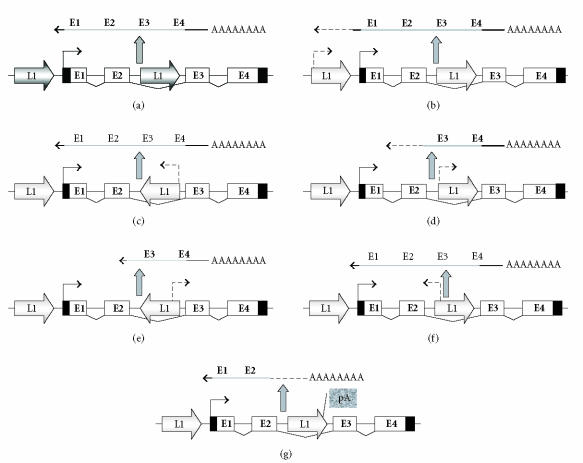
The mechanisms involved in L1 retrotransposition are now quite well understood [1–3]. The endonuclease encoded by L1 ORF2 induces single-strand breaks at AT-rich DNA target regions, preferably at consensus TTTT/A sites. Following L1 ORF2 endonuclease action, the L1 RNA poly-A sequence pairs with oligo-dT sequences in the target DNA, which serve as primers for reverse transcription by the L1 ORF2 encoded enzyme. Reverse transcription yields a branched DNA structure, which is presumably resolved by cellular DNA repair systems. The retrotransposition mechanism thus requires at least one recombination and creates two DNA single-strand breaks close to each other, which can in effect behave like a double-strand break. Therefore, attempted or successful retrotranspositions carry a high risk of eliciting chromosome breaks, deletions, translocations, and recombinations [5]. Moreover, successful retrotransposition events are likely to change the activity of genes at the insertion site. Diverse outcomes of insertions are conceivable, including increased or decreased transcriptional activity and the generation of novel, variant transcripts (Figure 1). On a genome-wide scale, the effects of retrotranspositions might be mitigated by the propensity of L1 elements to insert into AT-rich, gene-poor regions of the genome, and especially into or close to other elements [6]. However, even retrotranspositions outside transcriptional units can have catastrophic effects on a cell by inducing DNA strand breaks and initiating a breakage-fusion-bridge cycles [7].
Figure 1.
Potential effects of L1 sequences on transcriptional regulation. (a) schematic view of a human gene. One L1 element is located upstream of the gene and one within. Panels (b)–(g) show various disturbances that could be caused by partial or complete reactivation of L1 elements: (b) deregulation by upstream L1 promoter; (c) transcriptional interference by the promoter of an L1 in inverse direction to the gene; (d) generation of an alternative 5′-truncated transcript by an internal L1 promoter in sense direction; (e) generation of an alternative 5′-truncated transcript by the antisense promoter of an internal L1 element in inverse direction to the gene; (f) transcriptional interference by the antisense promoter of an internal L1 element oriented in sense direction; (g) generation of a truncated transcript by use of the poly-adenylation site of an internal L1 element. Note that most effects do not require intact retrotransposons.
Effects on transcription
Theoretically, a wide range of effects on the transcription of host genes can be exerted by L1 regulatory elements and transcriptional sequences that are located close to or within them (Figure 1). The L1 promoter is moderately strong [8, 9] and the polyadenylation signal is relatively weak permitting a substantial amount of read-through [10, 11]. Therefore, active L1 promoters located in sense orientation 5′ to a gene could override the normal transcriptional controls of a gene to deregulate expression. Active L1 promoters located in sense orientation within the transcriptional unit could generate alternative 5′-truncated transcripts. Indeed, many regulatory elements of human genes are derived from L1 or HERV sequences [12]. Even some protein-coding sequences are derived from retroelements. A prominent example is syncytin1, a crucial protein required for the formation of syncytic cells in the placenta which has evolved from an HERV env protein [13]. The gene encoding syncytin1 is now consequently named ERVWE1 for “endogenous retrovirus W envelope protein1.” As in this case, regulatory sequences derived from L1 or HERV sequences are often more active during germ cell or embryonic development than in somatic cells. In cancer cells, decreased methylation and a more open chromatin structure of such sequences could therefore allow the reexpression of genes or transcripts that are normally restricted to germ cells or the embryo, that is, oncofetal or cancer-testis gene expression.
Alternative transcripts may also be generated by the use of polyadenylation sites of intragenic L1 sequences, especially if these are 5′-truncated. As mentioned above, L1 polyadenylation signals are weak. It is not known which mechanism ensures that they are normally ignored in elements located within a transcriptional unit. Consequently, it is difficult to estimate how altered methylation and chromatin structure in cancer cells would affect their recognition.
Retroelements oriented in opposite direction inside a transcriptional unit might interfere with transcription by antisense effects, most prominently through formation of dsRNA. This mechanism is implicated in the generation of heterochromatin in some organisms [14, 15]. In mammalian cells, dsRNA ought to induce general cellular antiviral defense mechanisms, for example, by activating PKR, or leads to the production of siRNAs and gene-specific downregulation. Interestingly, transcripts containing Alu sequences in sense direction appear to be edited and consequently destabilized in human cells [16–18]. This mechanism provides an obvious means of posttranscriptional gene regulation. It is possible that a similar process acts on L1 sequences, but it is currently unknown to what extent intact or partial L1 sequences in pre-mRNA are edited and whether such sequences are employed for posttranscriptional gene regulation, in normal or in cancer cells. A recent study [19] suggests that L1 RNAs are not edited, at least not by the usual APOBEC3G cytidine deaminase.
A further possibility has been suggested by the recent discovery of an antisense promoter near the 5′-end of intact L1 sequences [20]. When active, this promoter could exert several effects on cellular genes, depending on its orientation. If located in sense direction, antisense transcripts could lead to downregulation; if located in antisense direction, it might lead to overexpression of normal transcripts or the emergence of novel transcripts. Accordingly, demethylation of L1 sequences in cancer cells may not only activate their canonical sense, but also their antisense promoters [21].
Effects of L1-encoded proteins
Intact L1 elements contain two open reading frames. ORF1 encodes a p40 RNA-binding protein supposed to act as a chaperone and transport factor for L1 RNA. ORF2 encodes an endonuclease and a reverse transcriptase. The properties of these enzymes have meanwhile been studied quite well in vitro [22–24], but their impact on normal and cancer cells remains difficult to estimate. One open question is how many L1 elements are actually capable of expressing active proteins, especially, whether only intact elements form the source. It is thought that less than 100 L1 sequences are capable of retrotransposition which all belong to the Ta family [25]. They are the most likely source of reverse transcriptase, endonuclease, and p40 protein in germ cells and the embryo as well as in cancer cells. However, many elements of other families are also intact, except for missense and stop mutations. They could still give rise to one or the other intact protein, as well as variant proteins. Alu retrotransposition uses the enzymatic machinery provided by L1 and is therefore dependent on expression of L1 proteins [26, 27]. Similarly, complementation of transposition in trans among L1 sequences is inefficient, but not impossible [28], that is, full-length L1s with mutated protein-coding sequences might still be capable of retrotransposition, if proteins are supplemented by other elements. The proteins provided by L1s are also most likely involved in the formation of pseudogenes. It is unknown, however, whether their endogenous expression levels in cancer cells are sufficient to support retrotransposition.
Importantly, the potential danger of proteins encoded by L1s depends critically on their ability to exert effects beyond aiding retrotransposition in cis or in trans. In the context of cancer, dangers posed by the endonuclease are most obvious. The endonuclease introduces single-strand breaks “(nicks)” into DNA with moderately stringent specificity [23]. Its activity is further restricted by chromatin structure [22]. The ultimate result of single-strand breaks introduced by the endonuclease in a cell depends on several factors. A first factor is the cell cycle phase. Nicks in S-phase are most problematic, because they can be converted into double-strand breaks by the replication complex. DNA repair competence and capacity constitute a second factor that may differ between normal and cancer cells. Thirdly, the presence of L1 RNA and other proteins at the nicked site would be thought to influence the type and efficiency of repair.
The potential impact of L1 reverse transcriptase and RNA-binding protein similarly depend on their specificity, actually in two respects. First, to which extent are they specific for L1 (and Alu) sequences? Second, are reverse transcription and RNA-chaperoning their sole activities? Drug inhibitors of reverse transcriptase and, more specifically, siRNA directed against L1 RT decrease the proliferation of cancer cell lines [29]. Such effects are difficult to explain by the known function of the enzyme in mediating L1 and Alu retrotranspositions.
Illegitimate recombination
Successful and abortive retrotransposition can create chromosomal instability and initiate illegitimate recombination by inducing DNA strand breaks and by generating a branched DNA structure. However, even in the absence of retrotransposition, the presence of thousands of intact, rather long (6 kb), and relatively homologous sequences in the genome plus the presence of ten thousands of truncated and mutated sequences carries a permanent risk of illegitimate recombination between elements located at different sites. In the germline, recombination between different L1 elements contributes to human evolution, but also elicits inherited diseases. In somatic cells, recombination ought to be restricted strictly to homologous recombination repair of DNA double-strand breaks using homologous sequences from sister chromatids or at most from the homologous chromosome. Any other recombination event involves deletions, insertions, or translocations. It is generally assumed that recombination between L1 sequences in somatic cells is suppressed by dense DNA methylation and tight packaging into chromatin. Decreased methylation and relaxed chromatin structure of L1 sequences in cancer cells might therefore facilitate illegitimate recombination contributing to chromosomal instability.
Disturbance of normal genome organization
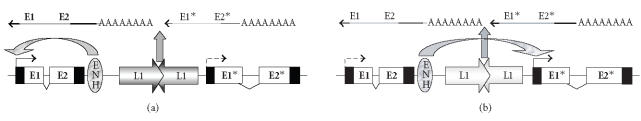
L1 sequences are thought to be involved in the organization of the human genome, their presence influencing short-range and long-range chromatin structures. L1 sequences are overrepresented in the late-replicating G-bands of human chromosomes [30]. It is plausible that their presence is responsible for their more heterochromatic character. L1s are also overrepresented on the X-chromosome [31] where they may act as “way stations” during X-chromosome inactivation [32, 33]. In a similar fashion, methylated L1 sequences on other chromosomes which are packaged into hypermethylated and deacetylated chromatin may constitute the cores of localized facultative heterochromatic regions. A fraction of centromeric heterochromatin also consists of retrotransposons, mostly of L1s [34]. Intriguingly, some L1 sequences are associated with nuclear matrix attachment regions [35] and may contribute to the organization of chromatin loops. L1 clusters located between genes may furthermore contribute to the segmentation of the genome into transcriptional units, helping to prevent interference by regulatory elements from neighboring genes (Figure 2). Such a “boundary” function would explain why HOX clusters, which require long-range interactions for their proper expression pattern, are largely devoid of retroelement sequences [30]. Importantly, the organization of the genome into subregions and loops pertains not only to transcription, but also to replication and imposes restrictions on the extent of DNA repair and recombination.
Figure 2.
Postulated boundary effect of intergenic L1 clusters. Being strongly methylated and tightly packed into chromatin, clustered L1 sequences might act as boundaries between genes, restricting the interaction of an enhancer (ENH) to one gene (a). In cancer cells, L1 hypomethylation could destroy this function and cause deregulation by allowing enhancer interaction with a neighboring gene (b).
Accordingly, alterations of DNA methylation and chromatin structure at L1 sequences in cancer cells could have effects not only on transcription, but also on DNA replication timing and on the extents of recombination and DNA repair. Deregulation of gene expression could not only be caused by activation of L1 elements, but also through altered chromatin structure at inactive L1s allowing transcriptional interference by neighboring enhancers or silencers. Not only in this particular situation DNA replication patterns could be disturbed, with normally late-replicating DNA shifting towards earlier periods within S-phase. Barrier functions of repeat DNA in the genome could be alleviated, allowing DNA processing during repair and Holiday junctions formed during recombination to pass through stretches of DNA that are less accessible in normal cells [36].
Effects on cell stress and immune responses
Endogenous retroelements have been implicated in the regulation of cell stress responses, of the immune system, and in the pathogenesis of several human autoimmune diseases. The strongest data on regulation of human retroelements by cell stress concern Alu sequences [37]. Likewise, the most convincing data on regulation of the immune system by retroelements and on the involvement in autoimmune diseases implicates HERVs [4]. There are, however, indications that L1 sequences too are induced during stress responses [37], during cytotoxic chemotherapy [38], and by UV exposure of skin cells [39]. Furthermore, L1 sequences may act in a similar fashion as HERVs in at least one autoimmune disease, rheumatoid arthritis [40]. In this disease, synovial fibroblasts become aberrantly activated in a fashion that resembles in many respects fibroblast activation in the stroma of malignant tumors, with enhanced proliferation, migration, and secretion of cytokines, chemokines, and proteases. The fibroblast genomes at large and L1 promoters in particular were found to become hypomethylated. Concurrently, full-length L1 RNA could be detected [41]. Overexpression of the p40 ORF1 protein in this disease has been suggested to activate stress-induced protein kinases [42]. It is thought that L1 hypomethylation and expression provide an amplification step in the pathogenesis of the disease by enhancing immune responses [40].
The function of the activation of retroelements during cellular stress responses is poorly understood. Conceivably, it forms part of a signaling system that alerts the immune system to the presence of infected or altered cells [43]. If that proposition is true, hypomethylation and activation of L1 sequences in cancer cells are likely to influence the immune response to malignant tumors. In support of this idea, some HERV proteins have been found to behave as tumor antigens [44, 45], but it is not known whether proteins encoded by L1 do so too. A similarly open question is to which extent hypomethylated repeat DNA liberated from tumor cells elicits danger signals in cells regulating the immune response. Interestingly, L1 activation is considered as a cause of increased plasma DNA levels in tumor patients [46].
Observations
Many of the effects that can be envisioned to be exerted by activated L1 retrotransposons have indeed been observed in the human germline and during fetal development [1–3, 47, 48]. In cancer cells, mainly three phenomena point towards a reactivation of retroelements. Retroelement DNA sequences become hypomethylated, transcripts as well as protein products can be detected, and L1 sequences are located at sites of breakage and recombination. For L1 retrotransposons, the most convincing data are available for hypomethylation. Data on L1 expression are scarce, in contrast to several reports on the expression of HERV gene products. L1 sequences have been found at or near deletion ends and translocation breakpoints, but the precise frequency and the mechanisms involved remain to be determined. Intriguingly, actual retrotransposition events are exceptional.
Altered methylation
In a large number of human cancers, decreased methylation of L1 sequences has been documented (Table 1). This decrease occurs in the context of general alterations in DNA methylation patterns that accompany carcinogenesis in many human tissues. These are regarded as part of an important epigenetic mechanism driving cancer development and progression [63]. Alterations of methylation in cancer cells comprise “hypermethylation” which occurs focally and in a largely specific fashion, typically at CpG islands surrounding the transcriptional start regions of individual genes. Somewhat paradoxically, in many, but not all cancers, increased methylation at specific sites is found alongside a decrease in methylation levels of the overall genome. The decrease in methylation appears to be relatively unspecific and is therefore commonly designated as “genome-wide” or “global” hypomethylation [64, 65]. In normal somatic cells, the bulk of methylcytosine is found in repetitive sequences such as L1, HERVs, and Alus, but also at CpG-rich satellites such as SAT2 and SAT3. The overall decrease in methylation found in cancer cells therefore reflects a largely parallel decrease in the methylation of retrotransposon sequences [61]. As a rule, however, L1 and HERV sequences seem to be more strongly affected than Alus [65].
Table 1.
Hypomethylation and expression of L1 in human cancers.
| Change reported | Cancer type | Remarks | References |
| Expression | Teratocarcinoma | Cell lines | [49, 50] |
| Hypomethylation | Various | Cell lines | [51] |
| Hypomethylation | Various | Cell lines | [46] |
| Hypomethylation | Many | Considerable differences between cancer types | [52] |
| Hypomethylation, expression | Bladder cancer | Expression weaker than in teratocarcinomas | [53, 54] |
| Hypomethylation | Renal carcinoma | Cell lines only | [53] |
| Hypomethylation | Prostate cancer | Increases with stage and metastasis | [55, 56, 57] |
| Hypomethylation | Liver carcinoma | — | [58] |
| Hypomethylation, expression | Liver carcinoma | Hypomethylation, but not cancer-specific expression | [59] |
| Hypomethylation | Various cancers | Differences between cancer types | [52] |
| Hypomethylation | Colon cancer | Begins in preneoplastic mucosa | [60] |
| Hypomethylation | Gastric cancer | Correlates with overall hypomethylation | [61] |
| Hypomethylation | Ovarian carcinoma | — | [62] |
Hypomethylation of L1 sequences has traditionally been investigated by Southern blot analysis following digestion of DNA with methylation-sensitive restriction enzymes [53]. Recently, methods employing PCR following bisulfite treatment of DNA have been developed for this purpose [52, 66]. These techniques are promising, since they can also be applied to small amounts of suboptimal quality DNA. However, because of the heterogeneity of L1 sequences, the extent of their demethylation is difficult to estimate precisely, especially by PCR-based methods. Southern blot analyses suggest that in cancer cell lines up to 70%–80% of CpG sites in L1 sequences become demethylated. Decreases in L1 methylation appear to parallel those in HERVs. Accordingly, individual HERV proviruses are essentially unmethylated in cancer cell lines with strong hypomethylation [53]. Nevertheless, L1 hypomethylation is anything but uniform in different cancers, in two respects. First, different extents of hypomethylation are found in cancers of the same type. These differences are also observed in cancer cell lines and are therefore not explained by differences in the proportion of tumor cells in tissue samples. Second, L1 hypomethylation appears to develop at different stages in the development of different cancers. For instance, it is found at early stages of colon and bladder cancers [53, 60], but only in higher-stage prostate carcinomas [55, 56] while primary renal carcinomas lack significant LINE-1 hypomethylation [52, 53]. Germ cell cancers are a special case since they have generally hypomethylated genomes, presumably due to their origin from cells with lower methylation levels [67, 68]. Accordingly, L1 [51, 69] and HERV [70] sequences are strongly hypomethylated in testicular cancers. Finally, note that very little is known on the methylation of individual L1 sequences [71], and accordingly, whether their hypomethylation in cancers is uniform [72].
Although genome-wide hypomethylation in human cancers has been known for more than twenty years, the mechanisms eliciting this alteration are still unknown. Hypothetical mechanisms include insufficient levels of methyl group donors, ultimately of S-adenosylmethionine, inadequate expression or regulation of DNA methyltransferases, reexpression of DNA demethylases, and altered expression of chromatin regulators directing DNA methyltransferases [64, 65].
The last mechanism is particularly interesting in the present context. Retroelements constitute approximately 45% of the human genome [30] and contain an at least proportionate amount of methylcytosine. Moreover, they appear to be preferentially recognized by the DNA methylation machinery and—at least in some circumstances—appear to act as “centers of methylation” from which methylation spreads into adjacent sequences [73]. Therefore, genome-wide hypomethylation could theoretically arise as a consequence of a defect in the recognition of retroelements as methylation targets.
Unfortunately, it is still not known how retroelements are distinguished for silencing in mammalian genomes. The L1 promoter is as active in somatic as in embryonic cells [9]. Therefore, L1 silencing in somatic cells cannot be simply a consequence of transcriptional inactivity. Instead, silencing must have been actively established during fetal development and is faithfully maintained through cell proliferation and differentiation in normal somatic cells. DNA methylation of retroelements is established first during germ cell development and then again during gastrulation, when the genome at large becomes de novo methylated, except for sequences that are actively protected, such as CpG islands and active imprinted genes [74]. De novo methylation in the mouse embryo requires DNA methyltransferases, specifically Dnmt3A and Dnmt3B as well as Dnmt1 for maintenance of the established methylation [74, 75]. In male germ cells, Dnmt3L is required for proper L1 methylation [76]. It is not entirely clear whether methylation of L1 during development requires specific chromatin regulators directing the methyltransferases. One candidate is SMARCA6, as its mouse orthologue Lsh has been found to be required for proper methylation of L1 sequences. Inactivation of Lsh in mice causes L1 hypomethylation, but only limited disturbances of the methylation of single-copy genes [77]. In comparison, inactivation of another chromatin protein ATRX causes hypomethylation of rDNA, but leaves L1 methylation intact [78]. This suggests that the specificity of DNA methylation may be regulated by specific “chromatin regulator” proteins.
A variety of chromatin regulator proteins have been reported to be aberrantly expressed or even mutated in human cancers [65, 79– 81]. However, many of these changes are rare or are specific to particular cancers. It is therefore difficult to envision a change in a single “master regulator” of L1 methylation as the cause of the widely distributed hypomethylation of these sequences. More likely, L1 hypomethylation could be associated with the general reorganization of chromatin structure in aneuploid cancer cells that disturbs the compartmentation of the genome [65, 79, 80]. Genome-wide alterations in histone modification have recently been described in cancer cells [82, 83]. Given the high proportion of L1 sequences in the human genome, these are likely to affect these retrotransposons and to interact with their methylation. Note that the relation between DNA methylation and histone modifications at L1 sequences is far from being understood [84].
L1 expression in cancers
The mechanisms underlying hypomethylation changes in human cancers are not understood, but even the description of these changes is fragmentary. For instance, methylation of HERVs has been studied in only a few cancers. Available data suggest that they are affected by genome-wide hypomethylation in parallel to LINE-1 sequences (Table 1). In selected cancers, endogenous retroviral sequences may be almost completely unmethylated. Expressed sequences derived from HERVs are found in germ cell cancers and antibodies directed against HERV-encoded proteins are found in the blood of patients [70]. In cancers of somatic cell origin, bona fide transcripts for envelope and auxiliary proteins have been reported, especially in breast cancer [44, 85], and recently in melanoma [86]. Some results suggest that HERV expression occurs in a wider range of cancers and even normal tissues [87, 88]. These data need further verification to exclude artifacts from genomic DNA and unspliced transcripts. Moreover, the somewhat surprising findings that different transcripts from different subfamilies may be expressed in a cancer-type-specific fashion call for a closer analysis of the mechanisms involved.
There are no sufficiently systematic studies of L1 expression in human cancers. The available data suggest that expression of full-length L1 sequences is by far the strongest in teratocarcinomas, while weaker expression is observed in a wider range of carcinomas exhibiting hypomethylation [53]. This expression pattern therefore resembles that of HERVs. Since HERVs also give rise to spliced transcripts, RNA analyses can provide a first indication of which protein products are expressed. For L1, this question needs to be addressed using antibodies. So far, no definite data have been published on the expression of the proteins encoded by the retrotransposons in human cancer. Their presence in germ cell cancers and teratocarcinoma cell lines, however, is very likely [89].
Involvement of L1 in chromosome breakage and recombination
Whereas retrotransposition events take place quite regularly in the germline, at an estimated rate of 1 event per 100 births [3, 4], very few have been reported in cancer cells [90, 91]. Similarly, although L1 sequences have been shown to become incorporated at sites of double-strand break DNA repair in model experiments [92], according sequence changes have only exceptionally been observed in human cancers [93]. In spite of the caveats discussed below, it is therefore probably safe to conclude that actual retrotransposition events are rare in human cancers and do not regularly contribute to genomic instability.
The evidence is better for indirect mechanisms by which retrotransposons could promote chromosomal instability in human cancer. L1 hypomethylation and chromosomal instability correlate well with each other in several cancer types [55, 58,94]. A similar relationship has been observed between the hypomethylation of tandem satellite sequences and alterations of the chromosomes that carry them as large juxtacentromeric region [95–97]. In this case, hypomethylation of the satellite sequences is thought to cause decondensation of pericentromeric chromatin and an increased propensity for chromosomal breaks and rearrangements in this region. In a similar fashion, hypomethylation of retroelement sequences dispersed in the genome could facilitate illegitimate recombination. In favor of this idea, L1 sequences are enriched at the ends of 3p14.1 and 9p21 deletions in carcinomas [36, 98, 99] and homozygous deletions arise preferentially in chromosomal regions with high LINE content [100]. It has also been suggested that L1 and HERV sequences are involved in the formation of double-minute circular chromosomes in cancer cells [101, 102].
The most straightforward hypothesis accounting for these findings is that decreased methylation and presumably more open chromatin structure at L1 sequences in cancer cells favors the illegitimate recombination between elements at different genomic locations, for example, during homologous recombination repair of DNA strand breaks. However, closer analyses of the deletion ends in solid tumors indicate that this hypothesis is probably incorrect. While deletion ends are indeed often located in or near L1 sequences and particularly L1 clusters, the breakpoints invariably show hallmarks of DNA double-strand break repair by nonhomologous end-joining (NHEJ). Typically, one end of the deletion is located in or close to an L1 sequence, while the other end is provided by an unrelated single copy or repeat sequence [36, 99, 103]. Such structures also appeared as occasional end products of repair of DNA double-strand breaks induced by a restriction endonuclease at a specific chromosomal site [104]. A plausible explanation for this structure is that processing by the NHE1 protein complex damaged DNA ends is slowed down at L1 sequences by denser chromatin, favoring reannealing and ligation there [36]. If this explanation is correct, retroelement hypomethylation in cancer could paradoxically diminish the tendency of breakpoints to be located at L1 sequences. It would instead tend to increase the size of deleted and recombinated sequences, because DNA processing and Holiday junctions arising during recombination repair could move further through more open chromatin.
Presently, either hypothesis remains speculative for several reasons. First of all, far too few chromosomal breakpoints have been investigated, especially in carcinomas. Secondly, it has not been established for any chromosomal alteration whether hypomethylation of repeat sequences at the affected site preceded it. Thirdly, L1 repeats are not randomly distributed in the genome. They might be associated with local structures that are particularly prone to breakage, such as fragile sites or the anchorage sites of chromatin loops.
Perspectives
Consequences of L1 activity in the human germline are well documented. Retrotranspositions in the germline take place at a significant rate of approximately 1 event per 100 births [3]. In addition, a substantial number of recombination events involving L1 elements have been detected, typically because they elicited translocations or rearrangements causing inherited diseases [2, 47, 48]. Specifically, L1 retrotransposition and illegitimate recombination in the germline are causes of inherited and congenital cancers. For instance, a germline deletion in the MLH1 gene carries hallmarks of recombination initiated by a failed L1 retrotransposition event [105].
In contrast, in spite of considerable evidence hinting at a reactivation of L1 retrotransposons in a variety of human cancers, there is limiting evidence for major consequences of this process. This may be due to two very different reasons. One is technical: even typical effects expected from L1 reactivation are difficult to detect by the techniques commonly used to investigate genetic alterations in human cancers. The other is biological: reactivation may be partial and the mechanisms ensuring silencing of L1 DNA sequences and limiting the effects of transcribed sequences may remain functional to some degree. Perhaps, limited reactivation of L1 sequences may exert effects more through the loss of symbiotic functions than through direct adverse effects on genomic stability. This possibility is even more difficult to ascertain.
In general, investigations of genetic and epigenetic changes in human cancers avoid dealing with repeat sequences and focus on single-copy protein-coding genes. Mutation analysis of genes is typically restricted to coding sequences and employs PCR techniques to analyze individual exons or mRNA. Insertions or recombinations caused by L1 or other retroelements would often not be detected by this approach, unless they occur within exons. Therefore, it might not be coincidental that reports describing oncogene activation and tumor suppressor inactivation by L1 insertion date from a period when Southern blot analysis was more en vogue.
Similarly, recombination and deletions in cancers are well documented at the level accessible by cytogenetic techniques, but are not well investigated at the molecular level, with the important exception of translocations in hematological cancers. In these, retroelements have indeed been found at many translocation sites, although their role in the generation of the translocations is not clear. In contrast, very few studies have addressed the precise structure of chromosomal breakpoints in solid tumors. A recent genome-wide study of 505 cancer cell lines yielded a strong association between LINE content and the presence of homozygous deletions, but no breakpoints were characterized in detail [100]. Detailed analyses of deletion endpoints at the FRA3B fragile site [98] and around CDKN2A at 9p21 [99] revealed a preponderance of L1 sequences at or close to the deletion endpoints. Such analyses remain tedious even with the finished human genome sequence having become available. Therefore, we know little on the structure of amplicons, another category of unstable sequence in cancer cells, and next to nothing on the sites of illegitimate recombination in cancer cells. L1 sequences have been detected in double minutes, an important intermediate in one amplification mechanism, and have been proposed, but not proven to be involved in their formation [101]. By a comparison of loss of heterozygosity analysis and cytogenetic techniques of chromosome 8p in bladder cancer cell lines, recombination events were recently shown to be much more frequent and were shown to take place across much smaller regions than hitherto assumed [106]. However, it is not known and difficult to determine what initiated the recombination events and which sequences precisely were involved. In summary, therefore, whereas it seems unlikely that retrotransposition is common in human cancer cells, the role of L1s in recombination and chromosome breakage is probably underestimated due to a lack of studies with appropriate methodology.
A similar argument can be made for epigenetic effects of L1 sequences in cancer. In genome-wide screens for altered methylation in cancer, repeat sequences are often and understandably considered a nuisance and typically removed by prehybridization. Overall changes in L1 methylation are therefore well documented, but data on the behavior of individual sequences is lacking. Bisulfite sequencing is restricted to a few hundred bp per PCR and is prone to artifacts from template switching and target priming when applied to repeat sequences. An elegant solution may be hairpin PCR. This method has revealed that in fetal fibroblasts, the promoters of most full-length L1 sequences are densely and symmetrically methylated, while selected elements are unmethylated [71]. The obvious question is which elements are these. Accordingly, it is not clear whether the number of completely unmethylated elements increases in cancer cells or whether the decrease in methylation is distributed across all L1 sequences. These questions extend of course to the issue of chromatin structure at L1 elements.
The contribution of L1s to altered gene expression in cancer is still more difficult to ascertain. There are many unexplained instances of altered gene expression in cancers. Perhaps most striking are reports on frequent downregulation of genes (usually tumor suppressor candidates) without detectable genetic alterations in their vicinity and altered DNA methylation in regulatory sequences. Recently, increased expression of miRNA has been introduced as a potential cause of such enigmatic observations [107]. In the light of potential effects of L1 sequences, perhaps effects exerted by L1 elements in or near affected genes should also be considered. This suggestion likewise applies the mechanisms generating aberrant transcripts in cancer cells [21]. Again, this is a highly difficult issue, especially if genes that are investigated are alternatively spliced even in normal cells. The argument can be broadened further to encompass the potential boundary function of repeat elements. Its disturbance by altered chromatin structure in cancer cells may result in more or less subtle up- and downregulation. More than two decades of intense work have been spent on a small number of selected loci to understand the mechanisms of action of long-range regulatory elements and boundary elements at all. It is understandable that very little is known on how they are altered in cancer cells. Still, it may be advisable to consider such effects and others mentioned in this paragraph when encountering instances of altered gene regulation in cancer that cannot be straightforwardly explained by mutations or altered DNA methylation of gene regulatory sequences.
The difficulties in determining the impact of L1 sequences in cancer cells resulting from methodological limitations are compounded by biological factors. Several layers of mechanisms control L1 expression and activity. DNA methylation inactivates the L1 promoter [9]. It is likely that this restriction of transcriptional activity is aided by an inactive chromatin structure [108], although this cannot be considered proven [86]. A second set of L1 controls appears to act at the RNA level, perhaps exerted by cytidine deaminases, leading to RNA instability [11]. Alu-containing RNAs are subject to editing [16–18], but the evidence for L1 RNA is scanty. A third level of control is enacted at the retrotransposition step. Active TP53 prevents retrotransposition [109], but this is very likely not the only barrier at this step. Last and perhaps not least, there is some evidence that retroelement activation might attract immune responses. As discussed elsewhere in detail [43], such responses are better documented for HERVs, but they might additionally or concurrently select against cells with strongly activated L1 retrotransposons outside immunoprivileged organs.
There is good evidence for three of these protective mechanisms to be impeded in cancer cells: DNA methylation is decreased, TP53 and checkpoints are often defective, and immune responses to advanced cancers are muted. We know very little about another mechanism, control of retroelements at the RNA level. In summary, therefore, the regulation of L1 genomic structure, expression, and retrotransposition is clearly perturbed in many human cancers, but inactivation of all tiers of control may be rare. Some cancer types exhibit few changes, for example, renal cell carcinoma, where even L1 DNA methylation appears to be maintained, while germ cell cancers appear to represent the other end of the spectrum [49, 50, 52, 53]. Even in these, however, retrotransposition events appear to be rare and the evidence for major contributions of retrotransposon activity to the cancer phenotype is limited. Presumably, at least one of the multiple safeguards against retrotransposition holds up. A likely candidate is TP53 [109], since germ cell tumors are among the few cancer types in which mutations of this tumor suppressor are rare [68].
CONCLUSIONS
Activation of L1 retrotransposons in cancer cells is expected to exert a variety of effects on the tumor phenotype, if it occurs. Of course, this statement hinges on the “if,” and our present knowledge does not allow firm conclusions. Considering that L1 retrotransposons make up almost a fifth of our genome, there are astonishingly large gaps in our knowledge on their general biology, and consequentially in our knowledge on their behavior in cancer. As argued above, there is an obvious need for more systematic investigations of DNA methylation and chromatin structure of L1 DNA, of the expression of full-length transcripts and L1-encoded proteins on one hand and for exemplary studies of individual elements and their influence on adjacent genes in cancer cells on the other hand. At this stage, it is probably safe to conclude that L1 retrotransposons do become reactivated to various degrees in different cancers, but that some of the many safeguards that prevent retrotransposition and their adverse effects in somatic cells hold up in most cancers. Perhaps, even cancer cells cannot survive with fully active retrotransposons. It follows that more subtle effects of L1 dysregulation in cancer cells, which may include adverse actions as well as loss of symbiotic functions, should be a focus of investigation.
ACKNOWLEDGMENTS
I am grateful to Dr. Andrea R. Florl for critical reading of the manuscript and to Sandy Fritzsche for help in compiling the reference list. Work in our lab is supported by the Deutsche Forschungsgemeinschaft (LI 1038/3-1) and the Deutsche Krebshilfe (70-3193 Schu 1).
References
- 1.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends in Genetics. 1997;13(8):335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 2.Ostertag EM, Kazazian HH Jr. Biology of mammalian L1 retrotransposons. Annual Review of Genetics. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 3.Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303(5664):1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 4.Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(suppl 2):14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Symer DE, Connelly C, Szak ST, et al. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110(3):327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 6.Boissinot S, Entezam A, Young L, Munson PJ, Furano AV. The insertional history of an active family of L1 retrotransposons in humans. Genome Research. 2004;14(7):1221–1231. doi: 10.1101/gr.2326704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gisselsson D. Chromosome instability in cancer: how, when, and why? Advances in Cancer Research. 2003;87:1–29. doi: 10.1016/s0065-230x(03)87164-6. [DOI] [PubMed] [Google Scholar]
- 8.Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Molecular and Cellular Biology. 1990;10(12):6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinhoff C, Schulz WA. Transcriptional regulation of the human LINE-1 retrotransposon L1.2B. Molecular Genetics and Genomics. 2003;270(5):394–402. doi: 10.1007/s00438-003-0931-2. [DOI] [PubMed] [Google Scholar]
- 10.Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH Jr. A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nature Genetics. 1994;7(2):143–148. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- 11.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429(6989):268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 12.van de Lagemaat LN, Landry JR, Mager DL, Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends in Genetics. 2003;19(10):530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Mi S, Lee X, Li X-P, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403(6771):785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 14.Schramke V, Allshire R. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science. 2003;301(5636):1069–1074. doi: 10.1126/science.1086870. [DOI] [PubMed] [Google Scholar]
- 15.Lippman Z, Gendrel A-V, Black M, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430(6998):471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 16.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biology. 2004;2(12):e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim DD, Kim TT, Walsh T, et al. Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Research. 2004;14(9):1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levanon EY, Eisenberg E, Yelin R, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nature Biotechnology. 2004;22(8):1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 19.Turelli P, Vianin S, Trono D. The innate antiretroviral factor APOBEC3G does not affect human LINE-1 retrotransposition in a cell culture assay. The Journal of Biological Chemistry. 2004;279(42):43371–43373. doi: 10.1074/jbc.C400334200. [DOI] [PubMed] [Google Scholar]
- 20.Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Molecular and Cellular Biology. 2001;21(6):1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nigumann P, Redik K, Mätlik K, Speek M. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics. 2002;79(5):628–634. doi: 10.1006/geno.2002.6758. [DOI] [PubMed] [Google Scholar]
- 22.Cost GJ, Golding A, Schlissel MS, Boeke JD. Target DNA chromatinization modulates nicking by L1 endonuclease. Nucleic Acids Research. 2001;29(2):573–577. doi: 10.1093/nar/29.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. The EMBO Journal. 2002;21(21):5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weichenrieder O, Repanas K, Perrakis A. Crystal structure of the targeting endonuclease of the human LINE-1 retrotransposon. Structure. 2004;12(6):975–986. doi: 10.1016/j.str.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Myers JS, Vincent BJ, Udall H, et al. A comprehensive analysis of recently integrated human Ta L1 elements. American Journal of Human Genetics. 2002;71(2):312–326. doi: 10.1086/341718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kajikawa M, Okada N. LINEs mobilize SINEs in the eel through a shared 3' sequence. Cell. 2002;111(3):433–444. doi: 10.1016/s0092-8674(02)01041-3. [DOI] [PubMed] [Google Scholar]
- 27.Hagan CR, Sheffield RF, Rudin CM. Human Alu element retrotransposition induced by genotoxic stress. Nature Genetics. 2003;35(3):219–220. doi: 10.1038/ng1259. [DOI] [PubMed] [Google Scholar]
- 28.Wei W, Gilbert N, Ooi SL, et al. Human L1 retrotransposition: cis preference versus trans complementation. Molecular and Cellular Biology. 2001;21(4):1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sciamanna I, Landriscina M, Pittoggi C, et al. Inhibition of endogenous reverse transcriptase antagonizes human tumor growth. Oncogene. 2005;24(24):3923–3931. doi: 10.1038/sj.onc.1208562. [DOI] [PubMed] [Google Scholar]
- 30.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 31.Ross MT, Grafham DV, Coffey AJ, et al. The DNA sequence of the human X chromosome. Nature. 2005;434(7031):325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey JA, Carrel L, Chakravarti A, Eichler EE. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(12):6634–6639. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen RS. X inactivation-specific methylation of LINE-1 elements by DNMT3B: implications for the Lyon repeat hypothesis. Human Molecular Genetics. 2003;12(19):2559–2567. doi: 10.1093/hmg/ddg268. [DOI] [PubMed] [Google Scholar]
- 34.Laurent AM, Puechberty J, Prades C, Gimenez S, Roizès G. Site-specific retrotransposition of L1 elements within human alphoid satellite sequences. Genomics. 1997;46(1):127–132. doi: 10.1006/geno.1997.4987. [DOI] [PubMed] [Google Scholar]
- 35.Khodarev NN, Bennett T, Shearing N, et al. LINE L1 retrotransposable element is targeted during the initial stages of apoptotic DNA fragmentation. Journal of Cellular Biochemistry. 2000;79(3):486–495. [PubMed] [Google Scholar]
- 36.Raschke S, Balz V, Efferth T, Schulz WA, Florl AR. Homozygous deletions of CDKN2A caused by alternative mechanisms in various human cancer cell lines. Genes, Chromosomes and Cancer. 2005;42(1):58–67. doi: 10.1002/gcc.20119. [DOI] [PubMed] [Google Scholar]
- 37.Li TH, Schmid CW. Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene. 2001;276(1-2):135–141. doi: 10.1016/s0378-1119(01)00637-0. [DOI] [PubMed] [Google Scholar]
- 38.Hagan CR, Rudin CM. Mobile genetic element activation and genotoxic cancer therapy: potential clinical implications. American Journal of PharmacoGenomics. 2002;2(1):25–35. doi: 10.2165/00129785-200202010-00003. [DOI] [PubMed] [Google Scholar]
- 39.Banerjee G, Gupta N, Tiwari J, Raman G. Ultraviolet-induced transformation of keratinocytes: possible involvement of long interspersed element-1 reverse transcriptase. Photodermatology, Photoimmunology & Photomedicine. 2005;21(1):32–39. doi: 10.1111/j.1600-0781.2005.00136.x. [DOI] [PubMed] [Google Scholar]
- 40.Seemayer CA, Distler O, Kuchen S, et al. Rheumatoide Arthritis: Neue Entwicklungen in der Pathogeneses unter besonderer Berücksichtigung der synovialen Fibroblasten [Rheumatoid arthritis: new developments in the pathogenesis with special reference to synovial fibroblasts] Zeitschrift für Rheumatologie. 2001;60(5):309–318. doi: 10.1007/s003930170030. [DOI] [PubMed] [Google Scholar]
- 41.Neidhart M, Rethage J, Kuchen S, et al. Retrotransposable L1 elements expressed in rheumatoid arthritis synovial tissue: association with genomic DNA hypomethylation and influence on gene expression. Arthritis & Rheumatism. 2000;43(12):2634–2647. doi: 10.1002/1529-0131(200012)43:12<2634::AID-ANR3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 42.Kuchen S, Seemayer CA, Rethage J, et al. The L1 retroele- ment-related p40 protein induces p38delta MAP kinase. Autoimmunity. 2004;37(1):57–65. doi: 10.1080/08916930310001637977. [DOI] [PubMed] [Google Scholar]
- 43.Schulz WA, Steinhoff C, Florl AR. Methylation of endogenous human retroelements in health and disease. Current Topics in Microbiology and Immunology. 2006;310, in press. [DOI] [PubMed]
- 44.Armbruester V, Sauter M, Krautkraemer E, et al. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clinical Cancer Research. 2002;8(6):1800–1807. [PubMed] [Google Scholar]
- 45.Schiavetti F, Thonnard J, Colau D, Boon T, Coulie PG. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Research. 2002;62(19):5510–5516. [PubMed] [Google Scholar]
- 46.Alves G, Kawamura MT, Nascimento P, et al. DNA release by line-1 (L1) retrotransposon. Could it be possible? Annals of the New York Academy of Sciences. 2000;906:129–133. doi: 10.1111/j.1749-6632.2000.tb06602.x. [DOI] [PubMed] [Google Scholar]
- 47.Shaffer LG, Lupski JR. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annual Review of Genetics. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- 48.Ovchinnikov I, Rubin A, Swergold GD. Tracing the LINEs of human evolution. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(16):10522–10527. doi: 10.1073/pnas.152346799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bratthauer GL, Fanning TG. Active LINE-1 retrotransposons in human testicular cancer. Oncogene. 1992;7(3):507–510. [PubMed] [Google Scholar]
- 50.Skowronski J, Fanning TG, Singer MF. Unit-length line-1 transcripts in human teratocarcinoma cells. Molecular and Cellular Biology. 1988;8(4):1385–1397. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dante R, Dante-Paire J, Rigal D, Roizes G. Methylation patterns of long interspersed repeated DNA and alphoid repetitive DNA from human cell lines and tumors. Anticancer Research. 1992;12(2):559–563. [PubMed] [Google Scholar]
- 52.Chalitchagorn K, Shuangshoti S, Hourpai N, et al. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23(54):8841–8846. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 53.Florl AR, Löwer R, Schmitz-Dräger BJ, Schulz WA. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. British Journal of Cancer. 1999;80(9):1312–1321. doi: 10.1038/sj.bjc.6690524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jurgens B, Schmitz-Drager BJ, Schulz WA. Hypomethylation of L1 LINE sequences prevailing in human urothelial carcinoma. Cancer Research. 1996;56(24):5698–5703. [PubMed] [Google Scholar]
- 55.Schulz WA, Elo JP, Florl AR, et al. Genomewide DNA hypomethylation is associated with alterations on chromosome 8 in prostate carcinoma. Genes, Chromosomes and Cancer. 2002;35(1):58–65. doi: 10.1002/gcc.10092. [DOI] [PubMed] [Google Scholar]
- 56.Florl AR, Steinhoff C, Müller M, et al. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. British Journal of Cancer. 2004;91(5):985–994. doi: 10.1038/sj.bjc.6602030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santourlidis S, Florl AR, Ackermann R, Wirtz HC, Schulz WA. High frequency of alterations in DNA methylation in adenocarcinoma of the prostate. The Prostate. 1999;39(3):166–174. doi: 10.1002/(sici)1097-0045(19990515)39:3<166::aid-pros4>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 58.Takai D, Yagi Y, Habib N, Sugimura T, Ushijima T. Hypomethylation of LINE1 retrotransposon in human hepatocellular carcinomas, but not in surrounding liver cirrhosis. Japanese Journal of Clinical Oncology. 2000;30(7):306–309. doi: 10.1093/jjco/hyd079. [DOI] [PubMed] [Google Scholar]
- 59.Lin C-H, Hsieh S-Y, Sheen I-S, et al. Genome-wide hypomethylation in hepatocellular carcinogenesis. Cancer Research. 2001;61(10):4238–4243. [PubMed] [Google Scholar]
- 60.Suter CM, Martin DI, Ward RL. Hypomethylation of L1 retrotransposons in colorectal cancer and adjacent normal tissue. International Journal of Colorectal Disease. 2004;19(2):95–101. doi: 10.1007/s00384-003-0539-3. [DOI] [PubMed] [Google Scholar]
- 61.Kaneda A, Tsukamoto T, Takamura-Enya T, et al. Frequent hypomethylation in multiple promoter CpG islands is associated with global hypomethylation, but not with frequent promoter hypermethylation. Cancer Science. 2004;95(1):58–64. doi: 10.1111/j.1349-7006.2004.tb03171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Menendez L, Benigno BB, McDonald JF. L1 and HERV-W retrotransposons are hypomethylated in human ovarian carcinomas. Molecular Cancer. 2004;3(1):12. doi: 10.1186/1476-4598-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature Reviews. Genetics. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 64.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21(35):5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 65.Hoffmann MJ, Schulz WA. Causes and consequences of DNA hypomethylation in human cancer. Biochemistry and Cell Biology. 2005;83(3):296–321. doi: 10.1139/o05-036. [DOI] [PubMed] [Google Scholar]
- 66.Yang AS, Estécio MR, Doshi K, Kondo Y, Tajara EH, Issa J-PJ. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Research. 2004;32(3):e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smiraglia DJ, Szymanska J, Kraggerud SM, Lothe RA, Peltomäki P, Plass C. Distinct epigenetic phenotypes in seminomatous and nonseminomatous testicular germ cell tumors. Oncogene. 2002;21(24):3909–3916. doi: 10.1038/sj.onc.1205488. [DOI] [PubMed] [Google Scholar]
- 68.Oosterhuis JW, Looijenga LH. Testicular germ-cell tumours in a broader perspective. Nature Reviews. Cancer. 2005;5(3):210–222. doi: 10.1038/nrc1568. [DOI] [PubMed] [Google Scholar]
- 69.Alves G, Tatro A, Fanning TG. Differential methylation of human LINE-1 retrotransposons in malignant cells. Gene. 1996;176(1-2):39–44. doi: 10.1016/0378-1119(96)00205-3. [DOI] [PubMed] [Google Scholar]
- 70.Gotzinger N, Sauter M, Roemer K, Mueller-Lantzsch N. Regulation of human endogenous retrovirus-K Gag expression in teratocarcinoma cell lines and human tumours. Journal of General Virology. 1996;77(pt 12):2983–2990. doi: 10.1099/0022-1317-77-12-2983. [DOI] [PubMed] [Google Scholar]
- 71.Burden AF, Manley NC, Clark AD, Gartler SM, Laird CD, Hansen RS. Hemimethylation and non-CpG methylation levels in a promoter region of human LINE-1 (L1) repeated elements. The Journal of Biological Chemistry. 2005;280(15):14413–14419. doi: 10.1074/jbc.M413836200. [DOI] [PubMed] [Google Scholar]
- 72.Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nature Genetics. 2005;37(8):853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 73.Turker MS. Gene silencing in mammalian cells and the spread of DNA methylation. Oncogene. 2002;21(35):5388–5393. doi: 10.1038/sj.onc.1205599. [DOI] [PubMed] [Google Scholar]
- 74.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nature Reviews. Genetics. 2002;3(9):662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 75.Bestor TH. The DNA methyltransferases of mammals. Human Molecular Genetics. 2000;9(16):2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 76.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431(7004):96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 77.Huang J, Fan T, Yan Q, et al. Lsh, an epigenetic guardian of repetitive elements. Nucleic Acids Research. 2004;32(17):5019–5028. doi: 10.1093/nar/gkh821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gibbons RJ, McDowell TL, Raman S, et al. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nature Genetics. 2000;24(4):368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- 79.Ferreira R, Naguibneva I, Pritchard LL, Ait-Si-Ali S, Harel-Bellan A. The Rb/chromatin connection and epigenetic control: opinion. Oncogene. 2001;20(24):3128–3133. doi: 10.1038/sj.onc.1204337. [DOI] [PubMed] [Google Scholar]
- 80.Geiman TM, Robertson KD. Chromatin remodeling, histone modifications, and DNA methylation-how does it all fit together? Journal of Cellular Biochemistry. 2002;87(2):117–125. doi: 10.1002/jcb.10286. [DOI] [PubMed] [Google Scholar]
- 81.Lund AH, van Lohuizen M. Epigenetics and cancer. Genes & Development. 2004;18(19):2315–2335. doi: 10.1101/gad.1232504. [DOI] [PubMed] [Google Scholar]
- 82.Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nature Genetics. 2005;37(4):391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 83.Seligson DB, Horvath S, Shi T, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435(7046):1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 84.Martens JH, O'Sullivan RJ, Braunschweig U, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. The EMBO Journal. 2005;24(4):800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang-Johanning F, Frost AR, Jian B, et al. Detecting the expression of human endogenous retrovirus E envelope transcripts in human prostate adenocarcinoma. Cancer. 2003;98(1):187–197. doi: 10.1002/cncr.11451. [DOI] [PubMed] [Google Scholar]
- 86.Büscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Research. 2005;65(10):4172–4180. doi: 10.1158/0008-5472.CAN-04-2983. [DOI] [PubMed] [Google Scholar]
- 87.Sugimoto J, Matsuura N, Kinjo Y, Takasu N, Oda T, Jinno Y. Transcriptionally active HERV-K genes: identification, isolation, and chromosomal mapping. Genomics. 2001;72(2):137–144. doi: 10.1006/geno.2001.6473. [DOI] [PubMed] [Google Scholar]
- 88.Yi J-M, Kim H-M, Kim H-S. Expression of the human endogenous retrovirus HERV-W family in various human tissues and cancer cells. Journal of General Virology. 2004;85(pt 5):1203–1210. doi: 10.1099/vir.0.79791-0. [DOI] [PubMed] [Google Scholar]
- 89.Ergün S, Buschmann C, Heukeshoven J, et al. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. The Journal of Biological Chemistry. 2004;279(26):27753–27763. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- 90.Morse B, Rotherg PG, South VJ, Spandorfer JM, Astrin SM. Insertional mutagenesis of the myc locus by a LINE-1 sequence in a human breast carcinoma. Nature. 1988;333(6168):87–90. doi: 10.1038/333087a0. [DOI] [PubMed] [Google Scholar]
- 91.Miki Y, Nishisho I, Horii A, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Research. 1992;52(3):643–645. [PubMed] [Google Scholar]
- 92.Morrish TA, Gilbert N, Myers JS, et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nature Genetics. 2002;31(2):159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 93.Liu J, Nau MM, Zucman-Rossi J, Powell JI, Allegra CJ, Wright JJ. LINE-I element insertion at the t(11;22) translocation breakpoint of a desmoplastic small round cell tumor. Genes, Chromosomes and Cancer. 1997;18(3):232–239. doi: 10.1002/(sici)1098-2264(199703)18:3<232::aid-gcc10>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 94.Florl AR, Franke KH, Niederacher D, Gerharz CD, Seifert HH, Schulz WA. DNA methylation and the mechanisms of CDKN2A inactivation in transitional cell carcinoma of the urinary bladder. Laboratory Investigation. 2000;80(10):1513–1522. doi: 10.1038/labinvest.3780161. [DOI] [PubMed] [Google Scholar]
- 95.Qu GZ, Grundy PE, Narayan A, Ehrlich M. Frequent hypomethylation in Wilms tumors of pericentromeric DNA in chromosomes 1 and 16. Cancer Genetics and Cytogenetics. 1999;109(1):34–39. doi: 10.1016/s0165-4608(98)00143-5. [DOI] [PubMed] [Google Scholar]
- 96.Wong N, Lam WC, Lai PB, Pang E, Lau WY, Johnson PJ. Hypomethylation of chromosome 1 heterochromatin DNA correlates with q-arm copy gain in human hepatocellular carcinoma. The American Journal of Pathology. 2001;159(2):465–471. doi: 10.1016/S0002-9440(10)61718-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Widschwendter M, Jiang G, Woods C, et al. DNA hypomethylation and ovarian cancer biology. Cancer Research. 2004;64(13):4472–4480. doi: 10.1158/0008-5472.CAN-04-0238. [DOI] [PubMed] [Google Scholar]
- 98.Mimori K, Druck T, Inoue H, et al. Cancer-specific chromosome alterations in the constitutive fragile region FRA3B. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(13):7456–7461. doi: 10.1073/pnas.96.13.7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Florl AR, Schulz WA. Peculiar structure and location of 9p21 homozygous deletion breakpoints in human cancer cells. Genes, Chromosomes and Cancer. 2003;37(2):141–148. doi: 10.1002/gcc.10192. [DOI] [PubMed] [Google Scholar]
- 100.Cox C, Bignell G, Greenman C, et al. A survey of homozygous deletions in human cancer genomes. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(12):4542–4547. doi: 10.1073/pnas.0408593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jones RS, Potter SS. L1 sequences in HeLa extrachromosomal circular DNA: evidence for circularization by homologous recombination. Proceedings of the National Academy of Sciences of the United States of America. 1985;82(7):1989–1993. doi: 10.1073/pnas.82.7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang H, Qian J, Proffit J, Wilber K, Jenkins R, Smith DI. FRA7G extends over a broad region: coincidence of human endogenous retroviral sequences (HERV-H) and small polydispersed circular DNAs (spcDNA) and fragile sites. Oncogene. 1998;16(18):2311–2319. doi: 10.1038/sj.onc.1200202. [DOI] [PubMed] [Google Scholar]
- 103.Sasaki S, Kitagawa Y, Sekido Y, et al. Molecular processes of chromosome 9p21 deletions in human cancers. Oncogene. 2003;22(24):3792–3798. doi: 10.1038/sj.onc.1206589. [DOI] [PubMed] [Google Scholar]
- 104.Varga T, Aplan PD. Chromosomal aberrations induced by double strand DNA breaks. DNA Repair. 2005;4(9):1038–1046. doi: 10.1016/j.dnarep.2005.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Viel A, Petronzelli F, Della Puppa P, et al. Different molecular mechanisms underlie genomic deletions in the MLH1 Gene. Human Mutation. 2002;20(5):368–374. doi: 10.1002/humu.10138. [DOI] [PubMed] [Google Scholar]
- 106.Adams J, Williams SV, Aveyard JS, Knowles MA. Loss of heterozygosity analysis and DNA copy number measurement on 8p in bladder cancer reveals two mechanisms of allelic loss. Cancer Research. 2005;65(1):66–75. [PubMed] [Google Scholar]
- 107.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kondo Y, Issa JP. Enrichment for histone H3 lysine 9 methylation at Alu repeats in human cells. The Journal of Biological Chemistry. 2003;278(30):27658–27662. doi: 10.1074/jbc.M304072200. [DOI] [PubMed] [Google Scholar]
- 109.Haoudi A, Semmes OJ, Mason JM, Cannon RE. Retrotrans-position-competent human LINE-1 induces apoptosis in cancer cells with intact p53. Journal of Biomedicine and Biotechnology. 2004;2004(4):185–194. doi: 10.1155/S1110724304403131. [DOI] [PMC free article] [PubMed] [Google Scholar]