Abstract
The triumphant success of RNA interference (RNAi) in life sciences is based on its high potency to silence genes in a sequence-specific manner. Nevertheless, the first task for successful RNAi approaches is the identification of highly active small interfering RNAs (siRNAs). Early on, it has been found that the potency of siRNAs can vary drastically. Great progress was made when thermodynamic properties that influence siRNA activity were discovered. Design algorithms based on these parameters enhance the chance to generate potent siRNAs. Still, many siRNAs designed accordingly fail to silence their targeted gene, whereas others are highly efficient despite the fact that they do not fulfil the recommended criteria. Therefore, the accessibility of the siRNA-binding site on the target RNA has been investigated as an additional parameter which is important for RNAi-mediated silencing. These and other factors which are crucial for successful RNAi approaches will be discussed in the present review.
INTRODUCTION
RNA interference (RNAi) is a naturally occurring phenome-non of RNA-mediated gene silencing that is highly conserved among multicellular organisms (for recent reviews, see, eg, [1–4]). It is a post-transcriptional process initiated by double-stranded RNA molecules that induce degradation of a complementary target RNA. In the first step of the pathway, long double-stranded RNA molecules are chopped into shorter duplexes with 2 nucleotide overhangs at both 3′ ends by an endonuclease dubbed Dicer, the structure of which has been solved only recently [5]. The resulting 21 mer effector RNAs, named small or short interfering RNAs (siRNAs), are incorporated into a multimeric protein complex, the RNA-induced silencing complex (RISC). One of the two siRNA strands guides RISC to a complementary RNA. After hybridization the endonucleolytic “slicer” activity of RISC cleaves the target RNA, thus preventing its translation.
While long double-stranded RNA molecules can be employed to induce RNAi in lower eukaryotes, siRNAs being 21 nucleotides in length have to be used for gene silencing in mammalian cells in order to prevent the activation of an unspecific interferon response [6]. Due to the higher efficiency of siRNAs compared to traditional antisense oligonucleotides and ribozymes [7–9] and the relative ease of RNAi-mediated knockdown of target gene expression compared to knockout by homologous recombination, RNAi has rapidly become a standard technology in life sciences. Furthermore, siRNAs are not only new powerful research tools, but are also considered to be a promising new class of therapeutics [10–13].
In addition to siRNAs, endogenously expressed short double-stranded RNA molecules, referred to as microRNAs (miRNAs), entered the focus of current research (for a review, see [14]). These molecules are now believed to be important cellular gene regulators that play an important role in developmental processes and various diseases. At the beginning of the miRNA pathway, RNA polymerase II generates long primary RNAs that contain the miRNA sequences. These transcripts designated as pri-miRNAs are cleaved in the nucleus by an RNase III family enzyme, Drosha, to give the pre-miRNAs approximately 70–90 nucleotides with a 2 nucleotide 3′ overhang. After being exported to the cytoplasm, the pre-miRNA is recognized by Dicer and processed to generate the mature miRNA, which is incorporated into RISC. In contrast to siRNAs, however, miRNAs are capable of inhibiting translation of the targeted mRNA without degrading it (at least in mammalian cells). Still, the siRNA and miRNA pathways share many similarities. Elucidation of the mechanisms of miRNA activity therefore helps to understand the mode of action of siRNAs and vice versa.
Despite the great success of RNAi mediated approaches, the design of highly efficient siRNAs still remains a hurdle that has to be overcome. Initial expectations expressed on an antisense meeting in 2001 that there is no need to select for optimal siRNA target sequences [15] have soon been proven to be too optimistic, since a drastic variation of silencing efficiency was observed for different siRNAs directed against the same target RNA [16]. It thus became clear that either factors intrinsic to the siRNA or properties of the targeted mRNA are crucial for the success of an RNAi approach. In the present review our current knowledge about factors that influence the potency of siRNAs will be summarized and advice will be given that helps with the generation of efficient molecules. It will, however, become obvious that we do not yet know all relevant features so that even the sophisticated design algorithms available to date do not guarantee satisfactory activity of the proposed siRNAs.
THERMODYNAMIC PROPERTIES OF EFFICIENT siRNAs
Early on, recommendations have been given for the selection of siRNA target sites [17]: the selected region should preferably be located in the coding region, at least 50 nucleotides downstream of the start codon; the GC-content should be approximately 50%; and a sequence motive AA N19 TT was suggested to be advantageous. A blast search is necessary to ensure that the siRNA has no significant homologies with other genes than the intended target. Even though these selection criteria have been employed with great success in numerous RNAi experiments, a further increased hit rate for highly potent siRNAs was desirable for the generation of large libraries. Significant progress towards the design of active siRNAs was achieved when an unexpected asymmetry concerning the incorporation of the two strands of siRNAs and miRNAs was found in two independent studies [18, 19]. Analysis of the known miRNA sequences in the context of miRNA precursor hairpins revealed a low stability of the 5′ end of the antisense strand compared to the 5′ end of the sense strand [18]. Subsequently, the same feature was observed for siRNAs. Functional duplexes displayed a lower relative thermodynamic stability at the 5′ end of the antisense strand than nonfunctional duplexes. The finding that the relative stabilities of the base pairs at the termini of the two siRNA strands that determine the degree to which each strand is fed into the RNAi pathway led to the hypothesis that strand incorporation into RISC is determined by an RNA helicase that initiates dissociation of the miRNA or siRNA duplex at the end with the lower thermodynamic stability [19].
These findings were further refined in a systematic analysis of 180 siRNAs targeting the mRNAs of two genes [20]. In addition to the relative stability of both ends of the siRNA, base preferences at certain positions of the duplex were identified in functional siRNAs. A set of eight criteria was used in an algorithm intended to improve the selection of potent siRNAs (Table 1 and Figure 1). A total of 6 or more points according to this scoring system was proposed to significantly increase the probability for efficient gene silencing.
Table 1.
Design criteria for siRNAs according to Reynolds et al [20].
| Criterion | Score |
| GC content between 30–52% | 1 |
| (in the 19 mer siRNA duplex) | |
| A or U at positions 15–19 | 1 for each |
| A at position 19 | 1 |
| A at position 3 | 1 |
| U at position 10 | 1 |
| G or C at position 19 | −1 |
| G at position 13 | −1 |
Figure 1.
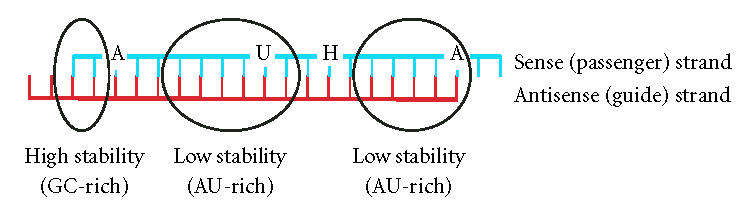
Features of efficient siRNAs according to Reynolds et al [20]. The relative stability of both ends of the siRNA as well as the bases in certain positions influences the activity of siRNAs (H = A, C or U).
Independent studies analysing the activities of siRNAs against different mRNAs confirmed the basic outcome of these studies [eg, [21, 22]]. Although some base preferences at certain positions of the siRNAs were either questioned or added to the list, the relative thermodynamic stability of the siRNA termini was verified to be a major determinant of the functionality of siRNAs. Somewhat different results were obtained, when a database was compiled consisting of 398 siRNAs against 92 genes from 30 different studies, in order to overcome a major shortcoming of earlier studies, the low number of genes being targeted [23]. Bioinformatic analysis of the data set led to a set of rules (termed “Stockholm rules”) that differs from the scoring systems described above.
Various academic groups and commercial vendors developed a software for designing siRNAs based on the identified features of active siRNAs. A list of freely available web tools is given in Table 2. Some additional prediction servers were introduced in a special web server issue of Nucleic Acids Research of July 2004.
Table 2.
Web sites for the design of effective siRNAs (based on [24] with modifications).
| Source | URL |
| Dharmacon | www.dharmacon.com/sidesign/ |
| Hannon Lab | katahdin.cshl.org:9331/homepage/siRNA/RNAi.cgi?type=siRNA |
| Integrated DNA Technologies | scitools.idtdna.com/RNAi |
| Sonnhammer Lab | sisearch.cgb.ki.se |
| Invitrogen | rnaidesigner.invitrogen.com/sirna |
| McManus Lab | |
| Qiagen | http://www1.qiagen.com/Products/GeneSilencing/CustomSiRna/SiRnaDesigner.aspx |
| Sfold Algorithm | http://sfold.wadsworth.org/index.pl |
| Whitehead Institute | http://Jura.wi.mit.edu/siRNAext |
| MWG-Biotech | http://www.mwg-biotech.com/html/s_synthetic_acids/s_sirna_design.shtml |
In a more recent study, a set of approximately 2200 randomly selected siRNAs targeting 24 mRNA species was used to train a neuronal network to predict the activity of siRNAs [25]. Statistic analysis of the large data set revealed some of the criteria discovered previously, but also identified new motives that are overrepresented in potent siRNAs. The approach to train an artificial neuronal network goes beyond earlier efforts like the above-mentioned scoring system, which uses a linear summation of parameters, in that it can handle complex sequence motifs and synergistic relations between two or more parameters. The neuronal network-based algorithm was finally employed to design a library of approximately 50.000 siRNAs that cover the human genome with a redundancy of two siRNAs per gene.
Taken together, the analysis of the sequences of active and nonfunctional siRNAs clearly revealed that the two strands of an siRNA duplex are not equally eligible for assembly into RISC. Rather, the relative stability of both ends of the siRNA is widely considered to determine which of the strands will preferentially participate in the RNAi pathway. It is therefore advisable to take into account the proposed criteria for active siRNAs when designing siRNAs against a new target. It has to be mentioned, however, that following these algorithms does not guarantee for the success of an RNAi approach. On the contrary, numerous highly efficient siRNAs have been published that do not obey the rules. Before addressing further determinants of siRNA activity in more detail, a short summary of structural studies will be given that may account for the asymmetric incorporation of the two siRNA strands into RISC.
STRUCTURAL BASIS FOR STRAND ASYMMETRY
In recent years, significant progress has been made to elucidate the molecular basis of RNAi and to understand the asymmetric strand incorporation (for a review, see [26]). The catalytic activity of RISC, termed slicer, which leads to the cleavage of the target RNA, has been identified to be located in the Argonaute2 (Ago2) protein [27]. Ago2 contains two major domains referred to as PIWI and PAZ (acronym for PIWI/Argonaute/Zwille). Crystallographic analysis revealed the PIWI domain at the C-terminus of the protein to closely resemble the structure of RNase H [28]. This enzyme cleaves the RNA component of an RNA/DNA hybrid. The PIWI domain of Ago2 can thus be regarded as a variant of the RNase H structure motive specialized in cleavage of one strand of double-stranded RNAs.
Recombinant human Ago2 and an siRNA were found to form a minimal RISC that accurately cleaves substrate RNAs [29]. Interestingly, only single-stranded siRNA could be specifically incorporated into recombinant Ago2, whereas photoreactive double-stranded siRNA did not crosslink with Ago2. This finding indicates the importance of the RISC loading complex (RLC) for efficient incorporation of the siRNA into the Ago2 protein. In Drosophila melanogaster, a heterodimer consisting of Dicer-2 and the double-stranded RNA binding protein R2D2, which contains the siRNA, was found to be important for RISC assembly [30]. R2D2 binds the thermodynamically more stable end of the siRNA, that is, the 3′ end of the guide strand, and can thus determine which one of the strands will be associated with Ago2. It has therefore been described as the “protein sensor for siRNA thermodynamic asymmetry.”
In human cells, the HIV-1 trans-activating response RNA-binding protein (TRBP) has been found to recruit the Dicer complex to Ago2 [34]. Based on these findings a model has been proposed for RISC assembly and function [31] that is depicted in Figure 2. In cytoplasm, RISC containing Dicer, TRBP, and Ago2 recognizes hairpin RNAs like pre-miRNAs. The RNase III Dicer generates ∼22 nt long duplexes which remain associated with RISC as a ribonucleoprotein complex. In analogy to R2D2 from Drosophila, TRBP and Dicer are likely to sense the thermodynamic asymmetry between the two ends of the duplex. Two recent reports suggest that the passenger strand is cleaved, before being removed from the Ago2 protein [32, 33]. The guide strand remains bound to the active RISC and recognizes target RNAs by complementary base pairing. The PIWI domain of Ago2 cleaves the target RNA. After release of the cleavage products, RISC can undergo further rounds of target RNA destruction. Interestingly, none of these steps requires energy from ATP hydrolysis. Although RISC can utilize 21 mer siRNA duplexes, pre-miRNA-type Dicer substrates result in a 10-fold higher activity [31].
Figure 2.
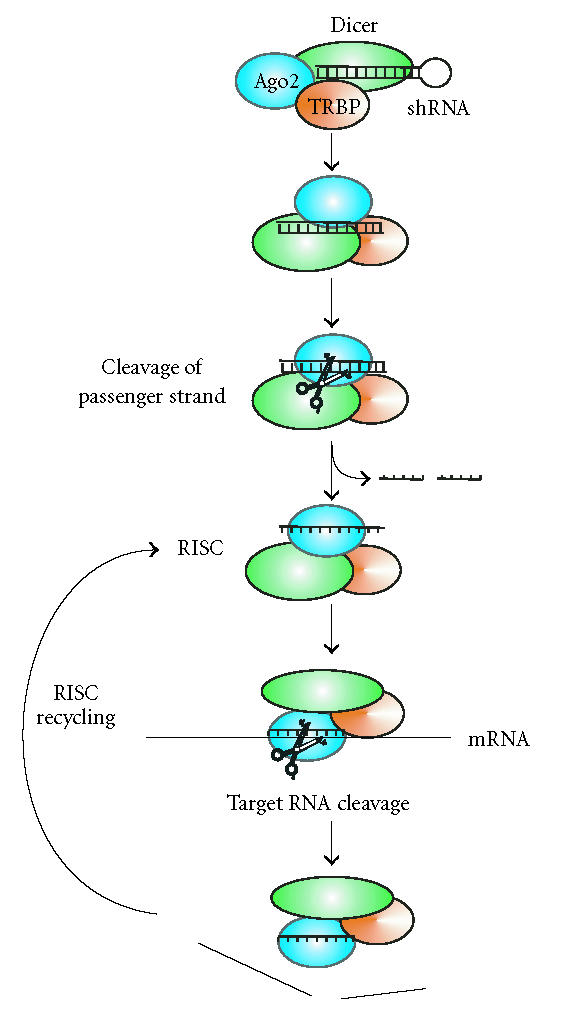
Model for assembly and function of RISC according to [31] under consideration of [32, 33].
TARGET SITE ACCESSIBILITY
Although there is no doubt that the design criteria described above increase the success rate to generate active siRNAs, a survey of published RNAi experiments readily reveals that many siRNAs are highly potent although they do not fulfil the recommendations. Even more intriguing is the fact that siRNAs may be unsuitable to silence their target although they comply with these rules. It is thus obvious that additional features have to be considered to optimize the efficiency of RNAi. Some earlier studies had already suggested that the structure of the target RNA may influence siRNA activity [35–37]. When it became clear that the design algorithms based solely on thermodynamic parameters of the siRNA are helpful tools, but do not guarantee success of RNAi approaches, target-site accessibility came back into the focus.
Luo and Chang [38] described the local mRNA structure at the target site as the main cause for the positional effect of different siRNAs. As a reliable parameter for target site accessibility, they introduced the “hydrogen bond index” representing the average number of hydrogen bonds formed between nucleotides in the target region and the rest of the mRNA. This index, which has to be determined by bioinformatic secondary structure prediction, was found to correlate inversely with the gene-silencing effect. Further experiments revealed that the tight stem-loop structure of the HIV-1 transactivation response element (TAR) is detrimental to silencing by RNAi [39]. In contrast, the location of the siRNA-binding site within a translated or noncoding region of the mRNA had only marginal effects.
A systematic global analysis was performed with a set of siRNAs directed against two target RNAs, for which the accessibility of the siRNA target sites was determined by an iterative computational approach and by experimental RNase H mapping [40]. IC50-values as well as the maximal extent of target suppression were significantly improved for siRNAs against accessible local target sites compared to those siRNAs which targeted inaccessible regions of the mRNAs. In contrast, the relative thermodynamic stability of both ends of the siRNA was not found to be a suitable marker for siRNA activity. This finding was further strengthened by a kinetic analysis of isolated human RISC [41]. An siRNA directed against the highly structured RNA of the HIV-1 TAR was found to be incapable of target RNA cleavage. When the tight structure was disrupted by the addition of an oligonucleotide consisting of 2′-O-methyl RNA, target-site accessibility increased leading to enhanced cleavage of the TAR RNA.
In a recent study, we aimed at deciphering the contributions of both factors, that is, the thermodynamic properties of the siRNA and the target RNA structure, to the efficiency of an RNAi approach by constructing a set of intentionally designed target sites [42]. A highly active siRNA, which is capable of silencing its full-length target RNA in the subnanomolar range, maintained its potency when directed against the isolated target site fused to the green fluorescent protein (GFP). Interestingly, a fusion construct with the siRNA-binding site in reverse orientation was found to be silenced to a much lower extent, confirming the existence of a strand bias. However, incorporation of the original target site into a tight hairpin structure was detrimental to silencing as well. Further experimental and bioinformatic analysis of a set of target RNAs with varying degrees of target-site accessibility revealed a linear correlation between the local free energy in the siRNA-binding region and the extent of gene knockdown. These findings demonstrate that the thermodynamic properties of the siRNA itself as well as the structure of the target RNA both influence the efficiency of an siRNA. We therefore proposed a model, according to which the outcome of an RNAi approach is determined at two points of the multistep process (Figure 3). Firstly, asymmetric strand incorporation into RISC is controlled by thermodynamic properties of the siRNA; secondly, accessibility of the target site may further modulate the efficiency of silencing. Even siRNAs with favorable thermodynamic properties may thus be incapable of inhibiting gene expression in cases in which the binding region is inaccessible due to tight secondary structures.
Figure 3.
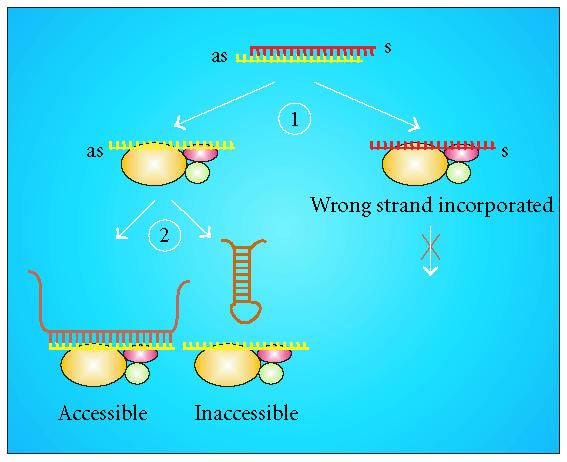
Efficiency of an siRNA is determined at two points of the RNAi pathway. (1) A strand bias exists that is defined by the intrinsic thermodynamic properties of the siRNA duplex, that is, by the relative stability of both ends. (2) A highly ordered structure may have a detrimental influence on the hybridisation of the siRNA/RISC to its target site and may therefore reduce the efficiency of the silencing process, even in cases in which the intended antisense strand is favored for incorporation into RISC. (Reprinted with slight modifications from the Journal of Molecular Biology; see [42], with kind permission from Elsevier.)
Design of siRNAs according to the criteria recommended by Reynolds et al [20] frequently results in satisfactory inhibition of gene expression. Some targets, however, are refractory to RNAi-mediated silencing, most likely due to the existence of stable secondary structures. For example, we and others failed to identify efficient siRNAs against the highly structured 5′ untranslated region of plus-stranded RNA viruses and were more successful when targeting less tightly arranged parts of the coding region [43–47]. In some cases, it might be advisable to take the target RNA structure into account as well. Several freely available design algorithms, for example, the Sfold web server (http://sfold.wadsworth.org [48]) and the siRNA design tool offered by MWG-biotech (http://www.mwg-biotech.com [49]) allow the design of siRNAs based on thermodynamic properties of the duplexes with consideration of the predicted secondary structure of the binding region of a potential siRNA.
STRATEGIES TO IMPROVE siRNA EFFICIENCY
Detailed bioinformatic analysis of the large set of sequence-activity relationships reported by Huesken et al [25] confirmed that the score according to Reynolds et al [20] as well as the target-site accessibility correlate with the extent of siRNA-mediated gene silencing. However, this investigation clearly revealed that both parameters are insufficient to fully explain or predict the potency of siRNAs (G. Schramm, personal communication). Thus, further factors can be expected to influence the functionality of siRNA molecules. Recently, Patzel et al [50] suggested that the structure of the guide strand could be another feature, which is crucial for the efficiency. Employing a series of siRNAs with different structures, guide strands that do not form defined structures or possess freely available terminal nucleotides, mainly at the 3′ end of the guide strand, were found to increase the efficiency of siRNAs (Figure 4). In contrast, structures with base-paired ends were virtually inactive. Interestingly, in this study neither the thermodynamic duplex profiles nor target mRNA structure were found to be of major importance for the silencing process.
Figure 4.
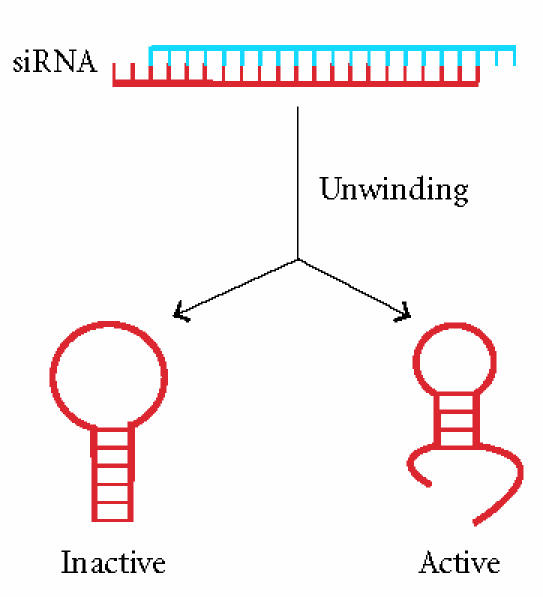
Influence of guide RNA structure on siRNA efficiency [50]. siRNA guide strands with base-paired termini were found to be inactive, whereas guide RNAs with freely accessible ends (mainly 3′ ends) were highly efficient.
A strategy to circumvent the need to identify suitable individual siRNAs is to use mixtures of siRNAs. To this end, long double-stranded RNA molecules have been processed in vitro by Escherichia coli RNase III [51]. The resulting pool of siRNAs, dubbed endoribonuclease-prepared siRNAs (esiRNA), can subsequently be transfected into cells to silence the corresponding gene. This efficient and cost effective method allowed the rapid generation of a large library consisting of more than 5000 esiRNAs [51]. It is still under debate whether this approach will elicit severe off-target effects due to the large number of sequences contained in the pool. It has, however, also been argued that pooling of siRNAs might decrease unspecific effects, since this strategy dilutes out the off-target effects of each individual siRNA, while retaining the total target-specific silencing capacity.
Two independent studies described additional approaches to enhance the efficiency of a single siRNA. Conventional siRNAs consist of a 19 mer double-stranded region and two nucleotide overhangs at the 3′ ends of each strand. Accordingly, short hairpin RNAs used for vector expression are designed with a 19 mer duplex, a loop connecting both strands, and two to four uridines at the 3′ end of the antisense strand. The two more recent publications now report that longer siRNA duplexes are up to 100-fold more potent than the corresponding conventional 21 mer siRNAs [52, 53]. In one of these studies a set of chemically synthesised siRNAs of varying length was used [52]. The optimum of silencing efficiency was found for siRNAs being 27 nucleotides in length. These 27 mers were even suitable to target sites that are refractory to silencing by 21 mer siRNAs. Importantly, the 27 mer duplexes did not activate the interferon response or protein kinase R. The authors of the second publication found 29 mer short hairpin RNAs to be particularly potent inducers of RNAi [53]. The higher efficiency of longer double-stranded RNA duplexes might be due to the fact that these siRNAs and shRNAs, respectively, are initially processed by Dicer to give 21 mers. As described above, mechanistic models based on copurification experiments [31] indicate that Dicer is involved in the loading process of siRNAs into RISC, thus explaining the improved potency of Dicer substrates compared to traditional 21 mer siRNAs. In a follow-up study, 27 mer duplexes with 2-base 3′-overhangs were found to be superior compared to blunt-end duplexes [54]. Interestingly, asymmetric strand utilization was found with the strand carrying the overhang being preferred for silencing. The authors conclude that Dicer processing confers functional polarity within the RNAi pathway for longer double-stranded RNAs.
Recently developed strategies to generate siRNAs from a miRNA environment went along the same lines to employ Dicer substrates for silencing. Stegmeier et al [55] generated an siRNA by replacing a naturally occurring miRNA by a target-specific siRNA sequence flanked by ∼125 bases of 5′ and 3′ sequence derived from the primary miRNA transcript. This construct can be expressed from both Pol III and Pol II promotors, thus opening the road to use tissue-specific promotors. The microRNA-type expression of shRNAs has been found to be superior compared to conventionally expressed isolated shRNAs and has been used to generate large libraries covering a substantial fraction of the predicted genes in the human and mouse genomes [56].
SUMMARY
Various factors have been identified that contribute to the efficacy of small interfering RNAs. Thermodynamic properties of a given siRNA itself influence its asymmetric incorporation into the RNA-induced silencing complex. Furthermore, the local structure of the targeted RNA might render the siRNA-binding region inaccessible, thus preventing efficient silencing. Additional factors like the availability of free ends of the siRNA antisense strand have been described to be relevant to the induction of RNAi. It is, however, clear that all of these features still do not provide an exhaustive description of the determinants of siRNA potency. We can therefore expect additional factors to be identified that contribute to the activity of siRNAs. Additional research is needed to further increase the success rate when designing siRNAs against a new target RNA.
ACKNOWLEDGMENTS
I wish to thank Prof. Dr. Volker A. Erdmann for his continuous support and all the members of my research team for their excellent work. Furthermore, I want to thank Diana Rothe, Steffen Schubert, Thomas Röhl, Harry Kurreck, and Gerrit Schramm for critical reading of the manuscript. The author's work is funded by the Deutsche Forschungsgemeinschaft (SFB/TR19), by the Fonds der Chemischen Industrie, and by the RiNA Network for RNA Technologies which is funded by the city of Berlin, the Federal Ministry of Education and Research, and the European Regional Development Fund.
References
- 1.Eckstein F. Small non-coding RNAs as magic bullets. Trends in Biochemical Sciences. 2005;30(8):445–452. doi: 10.1016/j.tibs.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Ryther RCC, Flynt AS, Phillips JA III, Patton JG. siRNA therapeutics: big potential from small RNAs. Gene Therapy. 2005;12(1):5–11. doi: 10.1038/sj.gt.3302356. [DOI] [PubMed] [Google Scholar]
- 3.Grünweller A, Hartmann RK. RNA interference as a gene-specific approach for molecular medicine. Current Medicinal Chemistry. 2005;12(26):3143–3161. doi: 10.2174/092986705774933489. [DOI] [PubMed] [Google Scholar]
- 4.Jayasena SD. Designer siRNAs to overcome the challenges in a stolen natural process. Journal of RNAi and Gene Silencing. 2006;2(1):109–117. [PMC free article] [PubMed] [Google Scholar]
- 5.MacRae IJ, Zhou K, Li F, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311(5758):195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 6.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 7.Bertrand J-R, Pottier M, Vekris A, Opolon P, Maksimenko A, Malvya C. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochemical and Biophysical Research Communications. 2002;296(4):1000–1004. doi: 10.1016/s0006-291x(02)02013-2. [DOI] [PubMed] [Google Scholar]
- 8.Grünweller A, Wyszko E, Bieber B, Jahnel R, Erdmann VA, Kurreck J. Comparison of different antisense strategies in mammalian cells using locked nucleic acids, 2'O-methyl RNA, phosphorothioates and small interfering RNA. Nucleic Acids Research. 2003;31(12):3185–3193. doi: 10.1093/nar/gkg409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyagishi M, Hayashi M, Taira K. Comparison of the suppressive effects of antisense oligonucleotides and siRNAs directed against the same targets in mammalian cells. Antisense and Nucleic Acid Drug Development. 2003;13(1):1–7. doi: 10.1089/108729003764097296. [DOI] [PubMed] [Google Scholar]
- 10.Leung RKM, Whittaker PA. RNA interference: from gene silencing to gene-specific therapeutics. Pharmacology and Therapeutics. 2005;107(2):222–239. doi: 10.1016/j.pharmthera.2005.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sioud M, Iversen PO. Ribozymes, DNAzymes and small interfering RNAs as therapeutics. Current Drug Targets. 2005;6(6):647–653. doi: 10.2174/1389450054863653. [DOI] [PubMed] [Google Scholar]
- 12.Shankar P, Manjunath N, Lieberman J. The prospect of silencing disease using RNA interference. Journal of the American Medical Association. 2005;293(11):1367–1373. doi: 10.1001/jama.293.11.1367. [DOI] [PubMed] [Google Scholar]
- 13.Uprichard SL. The therapeutic potential of RNA interference. FEBS Letters. 2005;579(26):5996–6007. doi: 10.1016/j.febslet.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 15.Stein CA. Antisense that comes naturally. Nature Biotechnology. 2001;19(8):737–738. doi: 10.1038/90783. [DOI] [PubMed] [Google Scholar]
- 16.Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Research. 2002;30(8):1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elbashir SM, Harborth J, Weber K, Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26(2):199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- 18.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115(2):209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 19.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 20.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nature Biotechnology. 2004;22(3):326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 21.Ui-Tei K, Naito Y, Takahashi F, et al. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Research. 2004;32(3):936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amarzguioui M, Prydz H. An algorithm for selection of functional siRNA sequences. Biochemical and Biophysical Research Communications. 2004;316(4):1050–1058. doi: 10.1016/j.bbrc.2004.02.157. [DOI] [PubMed] [Google Scholar]
- 23.Chalk AM, Wahlestedt C, Sonnhammer ELL. Improved and automated prediction of effective siRNA. Biochemical and Biophysical Research Communications. 2004;319(1):264–274. doi: 10.1016/j.bbrc.2004.04.181. [DOI] [PubMed] [Google Scholar]
- 24.Sandy P, Ventura A, Jacks T. Mammalian RNAi: a practical guide. BioTechniques. 2005;39(2):215–224. doi: 10.2144/05392RV01. [DOI] [PubMed] [Google Scholar]
- 25.Huesken D, Lange J, Mickanin C, et al. Design of a genome-wide siRNA library using an artificial neural network. Nature Biotechnology. 2005;23(8):995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- 26.Hutvagner G. Small RNA asymmetry in RNAi: function in RISC assembly and gene regulation. FEBS Letters. 2005;579(26):5850–5857. doi: 10.1016/j.febslet.2005.08.071. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 28.Song J-J, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of argonaute and its implications for RISC slicer activity. Science. 2004;305(5689):1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 29.Rivas FV, Tolia NH, Song JJ, et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nature Structural & Molecular Biology. 2005;12(4):340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 30.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306(5700):1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 31.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 32.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123(4):607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 33.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123(4):621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 34.Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436(7051):740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kretschmer-Kazemi Far R, Sczakiel G. The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucleic Acids Research. 2003;31(15):4417–4424. doi: 10.1093/nar/gkg649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bohula EA, Salisbury AJ, Sohail M, et al. The efficacy of small interfering RNAs targeted to the type 1 insulin-like growth factor receptor (IGF1R) is influenced by secondary structure in the IGF1R transcript. Journal of Biological Chemistry. 2003;278(18):15991–15997. doi: 10.1074/jbc.M300714200. [DOI] [PubMed] [Google Scholar]
- 37.Vickers TA, Koo S, Bennett CF, Crooke ST, Dean NM, Baker BF. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. Journal of Biological Chemistry. 2003;278(9):7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 38.Luo KQ, Chang DC. The gene-silencing efficiency of siRNA is strongly dependent on the local structure of mRNA at the targeted region. Biochemical and Biophysical Research Communications. 2004;318(1):303–310. doi: 10.1016/j.bbrc.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 39.Yoshinari K, Miyagishi M, Taira K. Effects on RNAi of the tight structure, sequence and position of the targeted region. Nucleic Acids Research. 2004;32(2):691–699. doi: 10.1093/nar/gkh221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overhoff M, Alken M, Kretschmer-Kazemi Far R, et al. Local RNA target structure influences siRNA efficacy: a systematic global analysis. Journal of Molecular Biology. 2005;348(4):871–881. doi: 10.1016/j.jmb.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 41.Brown KM, Chu C-Y, Rana TM. Target accessibility dictates the potency of human RISC. Nature Structural and Molecular Biology. 2005;12(5):469–470. doi: 10.1038/nsmb931. [DOI] [PubMed] [Google Scholar]
- 42.Schubert S, Grünweller A, Erdmann VA, Kurreck J. Local RNA target structure influences siRNA efficacy: systematic analysis of intentionally designed binding regions. Journal of Molecular Biology. 2005;348(4):883–893. doi: 10.1016/j.jmb.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 43.Wilson JA, Jayasena S, Khvorova A, et al. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(5):2783–2788. doi: 10.1073/pnas.252758799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phipps KM, Martinez A, Lu J, Heinz BA, Zhao G. Small interfering RNA molecules as potential anti-human rhinovirus agents: in vitro potency, specificity, and mechanism. Antiviral Research. 2004;61(1):49–55. doi: 10.1016/j.antiviral.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 45.Werk D, Schubert S, Lindig V, et al. Developing an effective RNA interference strategy against a plus-strand RNA virus: silencing of coxsackievirus B3 and its cognate coxsackievirus-adenovirus receptor. Biological Chemistry. 2005;386(9):857–863. doi: 10.1515/BC.2005.100. [DOI] [PubMed] [Google Scholar]
- 46.Schubert S, Grunert H-P, Zeichhardt H, Werk D, Erdmann VA, Kurreck J. Maintaining inhibition: siRNA double expression vectors against coxsackieviral RNAs. Journal of Molecular Biology. 2005;346(2):457–465. doi: 10.1016/j.jmb.2004.11.074. [DOI] [PubMed] [Google Scholar]
- 47.Yuan J, Cheung PKM, Zhang HM, Chau D, Yang D. Inhibition of Coxsackievirus B3 replication by small interfering RNAs requires perfect sequence match in the central region of the viral positive strand. Journal of Virology. 2005;79(4):2151–2159. doi: 10.1128/JVI.79.4.2151-2159.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding Y, Chan CY, Lawrence CE. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Research. 2004;32(web server issue):W135–W141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schramm G, Ramey R. siRNA design including secondary structure target site prediction. Nature Methods. 2005;2(8):1–2. application note. [Google Scholar]
- 50.Patzel V, Rutz S, Dietrich I, Köberle C, Scheffold A, Kaufmann SHS. Design of siRNAs producing unstructured guide-RNAs results in improved RNA interference efficiency. Nature Biotechnology. 2005;23(11):1440–1444. doi: 10.1038/nbt1151. [DOI] [PubMed] [Google Scholar]
- 51.Kittler R, Putz G, Pelletier L, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432(7020):1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 52.Kim D-H, Behlke MA, Rose SD, Chang M-S, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nature Biotechnology. 2005;23(2):222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 53.Siolas D, Lerner C, Burchard J, et al. Synthetic shRNAs as potent RNAi triggers. Nature Biotechnology. 2005;23(2):227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- 54.Rose SD, Kim D-H, Amarzguioui M, et al. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Research. 2005;33(13):4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(37):13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silva JM, Li MZ, Chang K, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nature Genetics. 2005;37(11):1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]


