Abstract
The purpose of this paper is to present a general view of the current applications of fuzzy logic in medicine and bioinformatics. We particularly review the medical literature using fuzzy logic. We then recall the geometrical interpretation of fuzzy sets as points in a fuzzy hypercube and present two concrete illustrations in medicine (drug addictions) and in bioinformatics (comparison of genomes).
INTRODUCTION
The diagnosis of disease involves several levels of uncertainty and imprecision, and it is inherent to medicine.
A single disease may manifest itself quite differently, depending on the patient, and with different intensities. A single symptom may correspond to different diseases. On the other hand, several diseases present in a patient may interact and interfere with the usual description of any of the diseases.
The best and most precise description of disease entities uses linguistic terms that are also imprecise and vague. Moreover, the classical concepts of health and disease are mutually exclusive and opposite. However, some recent approaches consider both concepts as complementary processes in the same continuum [1–6]. According to the definition issued by the World Health Organization (WHO), health is a state of complete physical, mental, and social well-being, and not merely the absence of disease or infirmity. The loss of health can be seen in its three forms: disease, illness, and sickness.
To deal with imprecision and uncertainty, we have at our disposal fuzzy logic. Fuzzy logic introduces partial truth values, between true and false.
According to Aristotelian logic, for a given proposition or state we only have two logical values: true-false, black-white, 1-0. In real life, things are not either black or white, but most of the times are grey. Thus, in many practical situations, it is convenient to consider intermediate logical values. Let us show this with a very simple medical example. Consider the statement “you are healthy”. Is it true if you have only a broken nail? Is it false if you have a terminal cancer? Everybody is healthy to some degree h and ill to some degree i. If you are totally healthy, then of course h = 1, i = 0. Usually, everybody has some minor health problems and h < 1, but
| (1) |
In the other extreme situation, h = 0, and i = 1 so that you are not healthy at all (you are dead). In the case you have only a broken nail, we may write h = 0.999,i = 0.001; if you have a painful gastric ulcer, i = 0.6,h = 0.4, but in the case you have a terminal cancer, probably i = 0.95, h = 0.05. As we will see, this is a particular case of Kosko's hypercube: the one-dimensional case [4].
Uncertainty is now considered essential to science and fuzzy logic is a way to model and deal with it using natural language. We can say that fuzzy logic is a qualitative computational approach.
Since uncertainty is inherent in fields such as medicine and massive data in bioinformatics, and fuzzy logic takes into account such uncertainty, fuzzy set theory can be considered as a suitable formalism to deal with the imprecision intrinsic to many biomedical and bioinformatics problems. Fuzzy logic is a method to render precise what is imprecise in the world of medicine. Several examples and illustrations are mentioned below.
FUZZY LOGIC IN MEDICINE
The complexity of medical practice makes traditional quantitative approaches of analysis inappropriate. In medicine, the lack of information, and its imprecision, and, many times, contradictory nature are common facts. The sources of uncertainty can be classified as follows [7].
Information about the patient.
Medical history of the patient, which is usually supplied by the patient and/or his/her family. This is usually highly subjective and imprecise.
Physical examination. The physician usually obtains objective data, but in some cases the boundary between normal and pathological status is not sharp.
Results of laboratory and other diagnostic tests, but they are also subject to some mistakes, and even to improper behavior of the patient prior to the examination.
The patient may include simulated, exaggerated, understated symptoms, or may even fail to mention some of them.
We stress the paradox of the growing number of mental disorders versus the absence of a natural classification [8]. The classification in critical (ie, borderline) cases is difficult, particularly when a categorical system of diagnosis is considered.
Fuzzy logic plays an important role in medicine [7, 9–14]. Some examples showing that fuzzy logic crosses many disease groups are the following.
To predict the response to treatment with citalopram in alcohol dependence [15].
To analyze diabetic neuropathy [16] and to detect early diabetic retinopathy [17].
To calculate volumes of brain tissue from magnetic resonance imaging (MRI) [20], and to analyze functional MRI data [21].
To characterize stroke subtypes and coexisting causes of ischemic stroke [1, 3, 22, 23].
To improve decision-making in radiation therapy [24].
To control hypertension during anesthesia [25].
To determine flexor-tendon repair techniques [26].
To detect breast cancer [27, 28], lung cancer [28], or prostate cancer [29].
To assist the diagnosis of central nervous systems tumors (astrocytic tumors) [30].
To discriminate benign skin lesions from malignant melanomas [31].
To visualize nerve fibers in the human brain [32].
To represent quantitative estimates of drug use [33].
To study the auditory P50 component in schizophrenia [34].
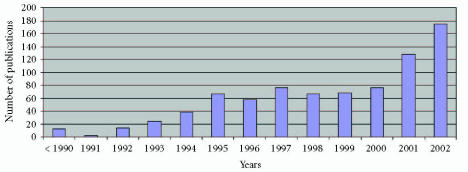
We used the database MEDLINE to identify the medical publications using fuzzy logic. We used as keywords fuzzy logic and grade of membership. The total number of articles per year appears in Table 1. The data is from 1991 to 2002 and includes also the number of those publications in 1990 and before. It results in a total of 804 articles and agrees essentially with the numbers indicated in [7, 13]. We plan to screen databases in the engineering literature that covers medicine-related articles since it is difficult to publish medical results using a fuzzy logic approach. In the future we will compare the figures obtained.
Table 1.
Number of papers per year in medicine using fuzzy logic.
| Year | Number |
|---|---|
| ≤1990 | 13 |
| 1991 | 2 |
| 1992 | 14 |
| 1993 | 24 |
| 1994 | 38 |
| 1995 | 66 |
| 1996 | 58 |
| 1997 | 76 |
| 1998 | 66 |
| 1999 | 68 |
| 2000 | 76 |
| 2001 | 128 |
| 2002 | 175 |
Figure 1 indicates an exponential growth in the number of articles in medicine making use of fuzzy technology. The preliminary data we have for 2003 and 2004 [38] supports this tendency.
Figure 1.
Number of publications per year indexed in MEDLINE using fuzzy logic.
FUZZY LOGIC IN BIOINFORMATICS
Bioinformatics derives knowledge from computer analysis of biological data. This data can consist of the information stored in the genetic code, and also experimental results (and hence imprecision) from various sources, patient statistics, and scientific literature. Bioinformatics combines computer science, biology, physical and chemical principles, and tools for analysis and modeling of large sets of biological data, the managing of chronic diseases, the study of molecular computing, cloning, and the development of training tools of bio-computing systems [39]. Bioinformatics is a very active and attractive research field with a high impact in new technological development [40].
Molecular biologists are currently engaged in some of the most impressive data collection projects. Recent genome-sequencing projects are generating an enormous amount of data related to the function and the structure of biological molecules and sequences. Other complementary high-throughput technologies, such as DNA microarrays, are rapidly generating large amounts of data that are too overwhelming for conventional approaches to biological data analysis. We have at our disposal a large number of genomes, protein structures, genes with their corresponding expressions monitored in experiments, and single-nucleotide polymorphisms (SNPs) [41]. For example, the EMBL Nucleotide Sequence Database (http://www.ebi.ac.uk/embl) has increased in 12 months from 18.3 million entries comprising 23 Gb (Release 71, September 2002) to 27.2 million entries comprising over 33 Gb (Release 76, September 2003) as indicated in [42].
Handling this massive amount of data, in many cases imprecise and fuzzy, requires powerful integrated bioinformatics systems and new technologies.
Fuzzy logic and fuzzy technology are now frequently used in bioinformatics. The following are some examples.
To increase the flexibility of protein motifs [43].
To study differences between polynucleotides [44].
To analyze experimental expression data [45] using fuzzy adaptive resonance theory.
To align sequences based on a fuzzy recast of a dynamic programming algorithm [46].
DNA sequencing using genetic fuzzy systems [47].
To cluster genes from microarray data [48].
To predict proteins subcellular locations from their dipeptide composition [49] using fuzzy k-nearest neighbors algorithm.
To simulate complex traits influenced by genes with fuzzy-valued effects in pedigreed populations [50].
To attribute cluster membership values to genes [51] applying a fuzzy partitioning method, fuzzy C-means.
To map specific sequence patterns to putative functional classes since evolutionary comparison leads to efficient functional characterization of hypothetical proteins [52]. The authors used a fuzzy alignment model.
To analyze gene expression data [53].
To unravel functional and ancestral relationships between proteins via fuzzy alignment methods [54], or using a generalized radial basis function neural network architecture that generates fuzzy classification rules [55].
To analyze the relationships between genes and decipher a genetic network [56].
To process complementary deoxyribonucleic acid (cDNA) microarray images [57]. The procedure should be automated due to the large number of spots and it is achieved using a fuzzy vector filtering framework.
To classify amino acid sequences into different superfamilies [58].
THE FUZZY HYPERCUBE
In 1992, Kosko [4] introduced a geometrical interpretation of fuzzy sets as points in a hypercube. In 1998, Helgason and Jobe [1] used the unit hypercube to represent concomitant mechanisms in stroke. Indeed, for a given set
| (2) |
a fuzzy subset is just a mapping
| (3) |
and the value μ(x) expresses the grade of membership of the element x ∈ X to the fuzzy subset μ.
For example, let X be the set of persons of some population and let the fuzzy set μ be defined as healthy subjects. If John is a member of the population (the set X), then, μ (John) gives the grade of healthiness of John, or the grade of membership of John to the set of healthy subjects. If λ is the fuzzy set that describes the grade of depression, then λ (Mary) is the degree of depression of Mary.
Thus, the set of all fuzzy subsets (of X) is precisely the unit hypercube In = [0, 1]n, as any fuzzy subset μ determines a point P ∈ In given by . Reciprocally, any point generates a fuzzy subset μ defined by μ(xi) = ai, . Nonfuzzy or crisp subsets of x are given by mappings , and are located at the 2n corners of the n-dimensional unit hypercube In. For graphic representations of the two-dimensional and three-dimensional hypercube, we refer to [59].
Given,
| (4) |
, not both equal to the empty set not both equal to the empty , we define the difference between p and q as
| (5) |
Of course d( ∅, ∅) = 0. We know that d is indeed a metric [60]. Hypercubical calculus has been described in [61], while some biomedical applications of the fuzzy unit hypercube are given in [1, 6, 59]. Recently, the fuzzy hypercube has been utilized to study differences between polynucleotides [59] and to compare genomes [44, 62].
AN APPLICATION TO DRUG ADDICTIONS
We now present an example of the use of the fuzzy hypercube in a medical case of consumption of drugs.
Consider the following fuzzy variables: smoking and alcohol drinking. If you do not smoke, then your degree of being a smoker is evidently 0. If you smoke, for example, six cigarettes per day, we say that your degree of being a smoker is 0.8. If the consumption is ten or more, the degree is 1. See [63 Figure 3.8] for a geometrical representation of the fuzzy concept of being a smoker.
With respect to the other fuzzy variable, if you drink no alcohol, the degree of this variable is 0. If you drink more than 75 cc of alcohol per day, the degree of alcoholism is 1. For 25 cc/d, the degree could be 0.4 and for 50 cc/d, 0.8.
Thus, the fuzzy set μ = (0, 0) corresponds to a nonsmoker and teetotaler. Some further examples are the following: the set μ = (1, 0) represents a heavy smoker, but a teetotaler, and the set μ = (0.8, 1) is a person who smokes about six cigarettes a day and is a risk consumer of alcohol.
Suppose you correspond to the fuzzy set λ = (1, 1), have recently had some health problems, and your physician has advised you to reduce your consumption of cigarettes and alcohol by half. The ideal situation for your health is, of course, the point μ = (0, 0), but it is possibly difficult to achieve.
Cigarette smoking and alcohol drinking during adolescence have been shown to be associated with a greater possibility of concurrent and future substance-related disorders (Lewinsohn etal [64]; Nelson and Wittchen [65]). In order to report patterns of drug use and to describe factors associated with substance use in adolescents, a cross-sectional survey was carried out in a representative population sample of 2550 adolescents, aged 12 to 17 years, from Galicia (an autonomous region located in the Northwest of Spain). The original survey covered the use of alcohol, tobacco, illicit drugs, and other psychoactive substances. For tobacco smoking and alcohol drinking, each subject of the population sample was assigned a fuzzy degree of addiction (or risk use) and mapped into the two-dimensional hypercube I2 by an expert.
Several subjects occupy the same point in the two-dimensional hypercube. For example Figure 2 represents the number of subjects in the cross-sectional survey according to the two fuzzy degrees of addiction. The reader can see that there are 1278 subjects corresponding to the point (0,0), that is, nonsmoker and teetotaler. Also 7 adolescents are at the point (0.8,0.2). There are 121 subjects on the line of probability x1 + x2 = 1. Indeed (see Figure 2), 23 + 1 + 1 + 2 + 2 + 7 + 1 + 84 = 121.
Figure 2.
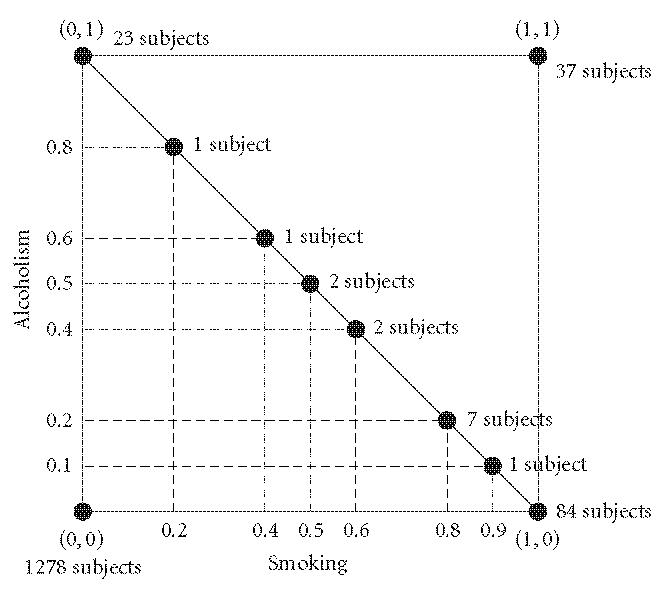
Number of subjects in the two-dimensional fuzzy hypercube I2.
Most subjects were inside the hypercube but outside the line of probability. This means that the vast majority of subjects () are outside the line of probability. This is in agreement with the fundamental limitation of probability theory with respect to clinical science in general [1] and agrees with its results ().
We refer to [59] for details on the general theory of fuzzy midpoints and their applications. It has been used recently to average biopolymers [66].
AN APPLICATION TO THE COMPARISON OF GENOMES
Whole genome sequence comparison is important in bioinformatics [44, 67].
The complete genome sequence of Mycobacterium tuberculosis H37Rv is available at http://www.ncbi.nlm.nih.gov with accession number NC–000962.
The genome comprises 4 411 529 base pairs, contains around 4000 genes, and has a very high guanine+cytosine content [68]. Computing [44] the number of the nucleotides at the three base sites of a codon in the coding sequences of M tuberculosis (Table 2), and then calculating the corresponding fractions, we have the fuzzy set of frequencies of the genome sequence of M tuberculosis (Table 3). This set can be considered as a point in the hypercube I12. Indeed, the point
| (6) |
Table 2.
Number of nucleotides at the three base sites of a codon in the coding sequence of Mycobacterium tuberculosis.
| T | C | A | G | |
|---|---|---|---|---|
| First base | 216 051 | 409 011 | 228 244 | 470 868 |
| Second base | 269 638 | 416 457 | 233 472 | 404 607 |
| Third base | 217 803 | 458 256 | 210 892 | 437 223 |
Table 3.
Fractions of nucleotides at the three base sites of a codon in the coding sequence of Mycobacterium tuberculosis.
| T | C | A | G | |
|---|---|---|---|---|
| First base | 0.1632 | 0.3089 | 0.1724 | 0.3556 |
| Second base | 0.2036 | 0.3145 | 0.1763 | 0.3056 |
| Third base | 0.1645 | 0.3461 | 0.1593 | 0.3302 |
Aquifex aeolicus was one of the earliest diverging, and is one of the most thermophilic, bacteria known [69]. It can grow on hydrogen, oxygen, carbon dioxide, and mineral salts. The complex metabolic machinery needed for A aeolicus to function as a chemolithoautotroph (an organism which uses an inorganic carbon source for biosynthesis and an inorganic chemical energy source) is encoded within a genome that is only one-third the size of the E coli genome.
The corresponding data for A aeolicus was obtained from http://www.ncbi.nlm.nih.gov with accession number NC_000918, and is presented in Tables 4 and 5, respectively. The complete genome sequence has 1 551 335 base pairs. The fuzzy set of frequencies of the genome of A aeolicus is
| (7) |
Table 4.
Number of nucleotides at the three base sites of a codon in the coding sequence of Aquifex aeolicus.
| T | C | A | G | |
|---|---|---|---|---|
| First base | 82 722 | 77 800 | 157 096 | 167 050 |
| Second base | 159 068 | 84 092 | 168 591 | 72 917 |
| Third base | 103 692 | 119 016 | 147 956 | 114 004 |
Table 5.
Fractions of nucleotides at the three base sites of a codon in the coding sequence of Aquifex aeolicus.
| T | C | A | G | |
|---|---|---|---|---|
| First base | 0.1706 | 0.1605 | 0.3241 | 0.3446 |
| Second base | 0.3282 | 0.1735 | 0.3478 | 0.1504 |
| Third base | 0.2139 | 0.2455 | 0.3052 | 0.2352 |
Using the distance given in (5), it is possible to compute the distance between these two fuzzy sets representing the frequencies of the nucleotides of A aeolicus and M tuberculosis:
| (8) |
In [44] we calculate the difference between M tuberculosis and E coli K-12 obtaining
| (9) |
Using the corresponding data for E coli (see [44 Tables 3 and 3]), we get
| (10) |
ACKNOWLEDGMENTS
This research is partially supported by Ministerio de Educación y Ciencia and FEDER, Projects MTM2004–06652–C03–01 and MTM2004–06652–C03–01, and by Xunta de Galicia and FEDER, Project PGIDIT05PXIC20702PN.
References
- 1.Jobe TH, Helgason CM. The fuzzy cube and causal efficacy: representation of concomitant mechanisms in stroke. Neural Networks. 1998;11(3):549–555. doi: 10.1016/s0893-6080(97)00149-4. [DOI] [PubMed] [Google Scholar]
- 2.Helgason CM, Jobe TH. Perception-based reasoning and fuzzy cardinality provide direct measures of causality sensitive to initial conditions in the individual patient (Invited paper) International Journal of Computational Cognition. 2003;1(2):70–104. [Google Scholar]
- 3.Helgason CM, Malik DS, Cheng S-C, Jobe TH, Mordeson JN. Statistical versus fuzzy measures of variable interaction in patients with stroke. Neuroepidemiology. 2001;20(2):77–84. doi: 10.1159/000054764. [DOI] [PubMed] [Google Scholar]
- 4.Kosko B. Neural Networks and Fuzzy Systems. Englewood Cliffs, NJ: Prentice-Hall; 1992. [Google Scholar]
- 5.Kosko B. Fuzzy Thinking: The New Science of Fuzzy Logic. New York, NY: Hyperion Press; 1993. [Google Scholar]
- 6.Sadegh-Zadeh K. Fundamentals of clinical methodology: 3. Nosology. Artificial Intelligence in Medicine. 1999;17(1):87–108. doi: 10.1016/s0933-3657(99)00015-9. [DOI] [PubMed] [Google Scholar]
- 7.Abbod MF, von Keyserlingk DG, Linkens DA, Mahfouf M. Survey of utilisation of fuzzy technology in Medicine and Healthcare. Fuzzy Sets and Systems. 2001;120(2):331–349. [Google Scholar]
- 8.Marchais P. De l'esprit et des modes de classification en psychiatrie [Classification in psychiatry: principles, modes and ways of thinking] Annales Medico-Psychologiques. 2002;160(3):247–252. [Google Scholar]
- 9.Barro S, Marín R. Fuzzy Logic in Medicine. Heidelberg, Germany: Physica; 2002. [Google Scholar]
- 10.Boegl K, Adlassnig KP, Hayashi Y, Rothenfluh TE, Leitich H. Knowledge acquisition in the fuzzy knowledge representation framework of a medical consultation system. Artificial Intelligence in Medicine. 2004;30(1):1–26. doi: 10.1016/s0933-3657(02)00073-8. [DOI] [PubMed] [Google Scholar]
- 11.Mahfouf M, Abbod MF, Linkens DA. A survey of fuzzy logic monitoring and control utilisation in medicine. Artificial Intelligence in Medicine. 2001;21(1–3):27–42. doi: 10.1016/s0933-3657(00)00072-5. [DOI] [PubMed] [Google Scholar]
- 12.Mordeson JN, Malik DS, Cheng S-C. Fuzzy Mathematics in Medicine. Heidelberg, Germany: Physica; 2000. [Google Scholar]
- 13.Steimann F. On the use and usefulness of fuzzy sets in medical AI. Artificial Intelligence in Medicine. 2001;21(1–3):131–137. doi: 10.1016/s0933-3657(00)00077-4. [DOI] [PubMed] [Google Scholar]
- 14.Szczepaniak PS, Lisoba PJG, Kacprzyk J. Fuzzy Systems in Medicine. Heidelberg, Germany: Physica; 2000. [Google Scholar]
- 15.Naranjo CA, Bremner KE, Bazoon M, Turksen IB. Using fuzzy logic to predict response to citalopram in alcohol dependence. Clinical Pharmacology and Therapeutics. 1997;62(2):209–224. doi: 10.1016/S0009-9236(97)90070-9. [DOI] [PubMed] [Google Scholar]
- 16.Lascio LD, Gisolfi A, Albunia A, Galardi G, Meschi F. A fuzzybased methodology for the analysis of diabetic neuropathy. Fuzzy Sets and Systems. 2002;129(2):203–228. [Google Scholar]
- 17.Zahlmann G, Kochner B, Ugi I, et al. Hybrid fuzzy image processing for situation assessment. IEEE Engineering in Medicine and Biology Magazine. 2000;19(1):76–83. doi: 10.1109/51.816246. [DOI] [PubMed] [Google Scholar]
- 18.Sproule BA, Bazoon M, Shulman KI, Turksen IB, Naranjo CA. Fuzzy logic pharmacokinetic modeling: application to lithium concentration prediction. Clinical Pharmacology and Therapeutics. 1997;62(1):29–40. doi: 10.1016/S0009-9236(97)90149-1. [DOI] [PubMed] [Google Scholar]
- 19.Stip E, Dufresne J, Boulerice B, Elie R. Accuracy of the Pepin method to determine appropriate lithium dosages in healthy volunteers. Journal of Psychiatry & Neuroscience. 2001;26(4):330–335. [PMC free article] [PubMed] [Google Scholar]
- 20.Brandt ME, Bohan TP, Kramer LA, Fletcher JM. Estimation of CSF, white and gray matter volumes in hydrocephalic children using fuzzy clustering of MR images. Computerized Medical Imaging and Graphics. 1994;18(1):25–34. doi: 10.1016/0895-6111(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 21.Lu Y, Jiang T, Zang Y. Region growing method for the analysis of functional MRI data. NeuroImage. 2003;20(1):455–465. doi: 10.1016/s1053-8119(03)00352-5. [DOI] [PubMed] [Google Scholar]
- 22.Dickerson JA, Helgason CM. The characterization of stroke subtype and science of evidence-based medicine using fuzzy logic. Journal of Neurovascular Disease. 1997;2(4):138–144. [Google Scholar]
- 23.Helgason CM, Jobe TH. Causal interactions, fuzzy sets and cerebrovascular “accident”: the limits of evidence-based medicine and the advent of complexity-based medicine. Neuroepidemiology. 1999;18(2):64–74. doi: 10.1159/000069409. [DOI] [PubMed] [Google Scholar]
- 24.Papageorgiou EI, Stylios CD, Groumpos PP. An integrated two-level hierarchical system for decision making in radiation therapy based on fuzzy cognitive maps. IEEE Transactions on Biomedical Engineering. 2003;50(12):1326–1339. doi: 10.1109/TBME.2003.819845. [DOI] [PubMed] [Google Scholar]
- 25.Oshita S, Nakakimura K, Sakabe T. Hypertension control during anesthesia. Fuzzy logic regulation of nicardipine infusion. IEEE Engineering in Medicine and Biology Magazine. 1994;13(5):667–670. [Google Scholar]
- 26.Johnson M, Firoozbakhsh K, Moniem M, Jamshidi M. Determining flexor-tendon repair techniques via soft computing. IEEE Engineering in Medicine and Biology Magazine. 2001;20(6):176–183. doi: 10.1109/51.982290. [DOI] [PubMed] [Google Scholar]
- 27.Hassanien AE. Intelligent data analysis of breast cancer based on rough set theory. International Journal on Artificial Intelligence Tools. 2003;12(4):465–479. [Google Scholar]
- 28.Seker H, Odetayo MO, Petrovic D, Naguib RN. A fuzzy logic based-method for prognostic decision making in breast and prostate cancers. IEEE Transactions on Information Technology in Biomedicine. 2003;7(2):114–122. doi: 10.1109/titb.2003.811876. [DOI] [PubMed] [Google Scholar]
- 29.Schneider J, Peltri G, Bitterlich N, et al. Fuzzy logic-based tumor marker profiles including a new marker tumor M2-PK improved sensitivity to the detection of progression in lung cancer patients. Anticancer Research. 2003;23(2A):899–906. [PubMed] [Google Scholar]
- 30.Belacel N, Boulassel MR. Multicriteria fuzzy classification procedure PROCFTN: methodology and medical application. Fuzzy Sets and Systems. 2004;141(2):203–217. [Google Scholar]
- 31.Stanley RJ, Moss RH, Van Stoecker W, Aggarwal C. A fuzzybased histogram analysis technique for skin lesion discrimination in dermatology clinical images. Computerized Medical Imaging and Graphics. 2003;27(5):387–396. doi: 10.1016/s0895-6111(03)00030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Axer H, Jantzen J, Keyserlingk DG, Berks G. The application of fuzzy-based methods to central nerve fiber imaging. Artificial Intelligence in Medicine. 2003;29(3):225–239. doi: 10.1016/s0933-3657(02)00071-4. [DOI] [PubMed] [Google Scholar]
- 33.Matt GE, Turingan MR, Dinh QT, Felsch JA, Hovell MF, Gehrman C. Improving self-reports of drug-use: numeric estimates as fuzzy sets. Addiction. 2003;98(9):1239–1247. doi: 10.1046/j.1360-0443.2003.00444.x. [DOI] [PubMed] [Google Scholar]
- 34.Zouridakis G, Boutros NN, Jansen BH. A fuzzy clustering approach to study the auditory P50 component in schizophrenia. Psychiatry Research. 1997;69(2-3):169–181. doi: 10.1016/s0165-1781(96)02979-4. [DOI] [PubMed] [Google Scholar]
- 35.Massad E, Ortega NR, Struchiner CJ, Burattini MN. Fuzzy epidemics. Artificial Intelligence inMedicine. 2003;29(3):241–259. doi: 10.1016/s0933-3657(02)00070-2. [DOI] [PubMed] [Google Scholar]
- 36.Im EO, Chee W. Fuzzy logic and nursing. Nursing Philosophy. 2003;4(1):53–60. doi: 10.1046/j.1466-769x.2003.00116.x. [DOI] [PubMed] [Google Scholar]
- 37.Zhu QM, Sun XW, Pipe AG. A fuzzy controller to overcome EA accommodation. In: Proceedings of IFAC conference on new technologies for computer control; Hong Kong, China: 2001. pp. 493–498. [Google Scholar]
- 38.Torres A, Nieto JJ. Fuzzy logic and technology in medicine and psychiatry. preprint, 2004.
- 39.Bourbakis NG. Bio-imaging and bio-informatics. IEEE Transactions on Systems, Man and Cybernetics, Part B: Cybernetics. 2003;33(5):726–727. doi: 10.1109/TSMCB.2003.816908. [DOI] [PubMed] [Google Scholar]
- 40.Fuchs R. From sequence to biology: the impact on bioinformatics. Bioinformatics. 2002;18(4):505–506. doi: 10.1093/bioinformatics/18.4.505. [DOI] [PubMed] [Google Scholar]
- 41.Valencia A. Bioinformatics: biology by other means. Bioinformatics. 2002;18(12):1551–1552. doi: 10.1093/bioinformatics/18.12.1551. [DOI] [PubMed] [Google Scholar]
- 42.Kulikova T, Aldebert P, Althorpe N, et al. The EMBL nucleotide sequence database. Nucleic Acids Research. 2004;32(database issue):D27–D30. doi: 10.1093/nar/gkh120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang BC, Halgamuge SK. Protein motif extraction with neuro-fuzzy optimization. Bioinformatics. 2002;18(8):1084–1090. doi: 10.1093/bioinformatics/18.8.1084. [DOI] [PubMed] [Google Scholar]
- 44.Torres A, Nieto JJ. The fuzzy polynucleotide space: basic properties. Bioinformatics. 2003;19(5):587–592. doi: 10.1093/bioinformatics/btg032. [DOI] [PubMed] [Google Scholar]
- 45.Tomida S, Hanai T, Honda H, Kobayashi T. Analysis of expression profile using fuzzy adaptive resonance theory. Bioinformatics. 2002;18(8):1073–1083. doi: 10.1093/bioinformatics/18.8.1073. [DOI] [PubMed] [Google Scholar]
- 46.Schlosshauer M, Ohlsson M. A novel approach to local reliability of sequence alignments. Bioinformatics. 2002;18(6):847–854. doi: 10.1093/bioinformatics/18.6.847. [DOI] [PubMed] [Google Scholar]
- 47.Cordón O, Gomide F, Herrera F, Hoffmann F, Magdalena L. Ten years of genetic fuzzy systems: current framework and new trends. Fuzzy Sets and Systems. 2004;141(1):5–31. [Google Scholar]
- 48.Belacel N, Čuperlović-Culf M, Laflamme M, Ouellette R. Fuzzy J-Means and VNS methods for clustering genes from microarray data. Bioinformatics. 2004;20(11):1690–1701. doi: 10.1093/bioinformatics/bth142. [DOI] [PubMed] [Google Scholar]
- 49.Huang Y, Li Y. Prediction of protein subcellular locations using fuzzy k-NN method. Bioinformatics. 2004;20(1):21–28. doi: 10.1093/bioinformatics/btg366. [DOI] [PubMed] [Google Scholar]
- 50.Carleos C, Rodriguez F, Lamelas H, Baro JA. Simulating complex traits influenced by genes with fuzzy-valued effects in pedigreed populations. Bioinformatics. 2003;19(1):144–148. doi: 10.1093/bioinformatics/19.1.144. [DOI] [PubMed] [Google Scholar]
- 51.Dembélé D, Kastner P. Fuzzy C-means method for clustering microarray data. Bioinformatics. 2003;19(8):973–980. doi: 10.1093/bioinformatics/btg119. [DOI] [PubMed] [Google Scholar]
- 52.Heger A, Holm L. Sensitive pattern discovery with ‘fuzzy’ alignments of distantly related proteins. Bioinformatics. 2003;19(suppl 1):i130–i137. doi: 10.1093/bioinformatics/btg1017. [DOI] [PubMed] [Google Scholar]
- 53.Woolf PJ, Wang Y. A fuzzy logic approach to analyzing gene expression data. Physiological Genomics. 2000;3(1):9–15. doi: 10.1152/physiolgenomics.2000.3.1.9. [DOI] [PubMed] [Google Scholar]
- 54.Blankenbecler R, Ohlsson M, Peterson C, Ringnér M. Matching protein structures with fuzzy alignments. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(21):11936–11940. doi: 10.1073/pnas.1635048100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang DH, Lee NK, Dillon TS. Extraction and optimization of fuzzy protein sequence classification rules using GRBF neural networks. Neural Information Processing—Letters and Reviews. 2003;1(1):53–59. [Google Scholar]
- 56.Ressom H, Reynolds R, Varghese RS. Increasing the efficiency of fuzzy logic-based gene expression data analysis. Physiological Genomics. 2003;13(2):107–117. doi: 10.1152/physiolgenomics.00097.2002. [DOI] [PubMed] [Google Scholar]
- 57.Lukac R, Plataniotis KN, Smolka B, Venetsanopoulos AN. cDNA microarray image processing using fuzzy vector filtering framework. Fuzzy Sets and Systems. 2005;152(1):17–35. [Google Scholar]
- 58.Bandyopadhyay S. An efficient technique for superfamily classification of amino acid sequences: feature extraction, fuzzy clustering and prototype selection. Fuzzy Sets and Systems. 2005;152(1):5–16. [Google Scholar]
- 59.Nieto JJ, Torres A. Midpoints for fuzzy sets and their application in medicine. Artificial Intelligence in Medicine. 2003;27(1):81–101. doi: 10.1016/s0933-3657(02)00080-5. [DOI] [PubMed] [Google Scholar]
- 60.Nieto JJ, Torres A, Vázquez-Trasande MM. A metric space to study differences between polynucleotides. AppliedMathematics Letters. 2003;16(8):1289–1294. [Google Scholar]
- 61.Zaus M. Crisp and Soft ComputingWith Hypercubical Calculus. Heidelberg, Germany: Physica; 1999. [Google Scholar]
- 62.Nieto JJ, Torres A, Georgiou DN, Karakasidis T. Fuzzy polynucleotide spaces andmetrics. to appear in Bulletin of Mathematical Biology. [DOI] [PubMed]
- 63.Kasabov NK. Foundations of Neural Networks, Fuzzy Systems and Knowledge Engineering. Cambridge, Mass: MIT Press; 1996. [Google Scholar]
- 64.Lewinsohn PM, Rohde P, Brown RA. Level of current and past adolescent cigarette smoking as predictors of future substance use disorders in young adulthood. Addiction. 1999;94(6):913–921. doi: 10.1046/j.1360-0443.1999.94691313.x. [DOI] [PubMed] [Google Scholar]
- 65.Nelson CB, Wittchen HU. DSM-IV alcohol disorders in a general population sample of adolescents and young adults. Addiction. 1998;93(7):1065–1077. doi: 10.1046/j.1360-0443.1998.937106511.x. [DOI] [PubMed] [Google Scholar]
- 66.Casasnovas J, Rosselló F. Averaging fuzzy biopolymers. Fuzzy Sets and Systems. 2005;152(1):139–158. [Google Scholar]
- 67.Castelli V, Aury J-M, Jaillon O, et al. Whole genome sequence comparisons and “full-length” cDNA Sequences: a combined approach to evaluate and improve Arabidopsis genome annotation. Genome Research. 2004;14(3):406–413. doi: 10.1101/gr.1515604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cole ST, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393(6685):537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 69.Deckert G, Warren PV, Gaasterland T, et al. The complete genome of the hyperthermophilic bacterium . Aquifex aeolicus Nature. 1998;392(6674):353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]