Abstract
Serum mannose-binding proteins (MBPs) are C-type lectins that recognize cell surface carbohydrate structures on pathogens, and trigger killing of these targets by activating the complement pathway. MBPs circulate as a complex with MBP-associated serine proteases (MASPs), which become activated upon engagement of a target cell surface. The minimal functional unit for complement activation is a MASP homodimer bound to two MBP trimeric subunits. MASPs have a modular structure consisting of an N-terminal CUB domain, a Ca2+-binding EGF-like domain, a second CUB domain, two complement control protein modules and a C-terminal serine protease domain. The CUB1-EGF-CUB2 region mediates homodimerization and binding to MBP. The crystal structure of the MASP-2 CUB1-EGF-CUB2 dimer reveals an elongated structure with a prominent concave surface that is proposed to be the MBP-binding site. A model of the full six-domain structure and its interaction with MBPs suggests mechanisms by which binding to a target cell transmits conformational changes from MBP to MASP that allow activation of its protease activity.
Keywords: complement/C1 complex/mannose-binding protein/MASP
Introduction
Complement-mediated neutralization of pathogens by lysis or opsonization can be initiated through multiple pathways. The classical and lectin pathways both start by specific recognition of the target cell. In the classical pathway, antibodies provide the recognition function, which leads to activation of the C1r and C1s proteases, which in turn activate complement components C4 and C2 (Arlaud et al., 2002). Complement component C1q links antibody binding to C1r and C1s activation, by binding to the Fc region of IgM or IgG and to C1r and C1s. In the lectin pathway, mannose-binding protein (MBP, also referred to as mannose-binding lectin) recognizes cell-surface carbohydrate structures and directly activates MBP-associated serine proteases (MASPs), which then activate C4 and C2 (Weis et al., 1998).
The serum form of MBP is composed of two, three or four subunits, each of which is a trimer of identical polypeptides (Wallis and Drickamer, 1999). The trimeric subunits are elongated molecules, consisting of N-terminal collagen-like domains and C-terminal carbohydrate-recognition domains (CRDs). Between these two regions, a short coiled-coil of α-helices holds the CRDs in a fixed orientation so that they can bind carbohydrate arrays on target cell surfaces (Weis and Drickamer, 1994). At the extreme N-terminus of the polypeptide, disulfide bonds link polypeptides within and between trimeric subunits (Wallis and Drickamer, 1999).
There are three known MASPs, denoted MASP-1, -2 and -3 (Takayama et al., 1994; Thiel et al., 1997; Dahl et al., 2001). Each MASP consists of six modular protein domains. The N-terminal three domains, comprising two CUB domains (Bork and Beckmann, 1993) flanking a single epidermal growth factor (EGF)-like domain, are responsible for interaction with MBP (Wallis and Dodd, 2000; Chen and Wallis, 2001). Two complement control repeats (CCPs, also known as sushi domains or short consensus repeats) link this portion of the molecule to the C-terminal serine protease (SP) domain. MASP-1 and MASP-2 are distinct gene products. MASP-3 is an alternatively spliced product of the MASP-1 gene, which is identical to MASP-1 except in the SP domain (Dahl et al., 2001). Although all three MASPs can form complexes with MBP, there is conflicting evidence about their roles in complement activation. There is general agreement that MASP-2 can activate C4 and C2 (Vorup-Jensen et al., 2000; Rossi et al., 2001). The role of MASP-1 is less clear: in one report, MASP-1 displays little to no activity on C2, C4 or C3 (Rossi et al., 2001), whereas MASP-1 was reported to activate C2 and C3 directly in other studies (Matsushita et al., 1998, 2000). The physiological role of MASP-3 is not known, but it may downregulate the activity of MASP-2 by competing for binding to MBP (Dahl et al., 2001).
Dimers of trimeric MBP subunits are the smallest oligomers that fix complement (Chen and Wallis, 2001). The dimers form a 1:1 complex with homodimers of MASP-1 or MASP-2. Thus, a complex consisting of two MBP subunits and two MASP polypeptides represents the minimal complement-fixing unit. Calcium ions play two distinct roles in this complex. Weak Ca2+-binding sites, with dissociation constants of ∼0.1–1 mM (Kawasaki et al., 1983; Taylor and Summerfield, 1987; Weis et al., 1991; Ng and Weis, 1998), form an essential part of the carbohydrate-binding site in the CRDs of MBP. Ca2+ is also required for the interaction between MASPs and MBP (Wallis and Dodd, 2000; Chen and Wallis, 2001; Thielens et al., 2001); in this case, the affinity for Ca2+ is higher. MASP constructs spanning the CUB1-EGF-CUB2 region are proteolytically sensitive in the absence of Ca2+, suggesting that Ca2+ stabilizes the structure and relative orientations of these domains (Wallis and Dodd, 2000).
There are strong parallels between the MBP–MASP complex and the C1qrs complex. Like MBP, C1q consists of multimers of a trimeric subunit that is held together by lateral association of N-terminal collagen-like domains (Arlaud et al., 2002). Interruptions in the collagen-like domains of both MBP and C1q create bends in the collagen triple helices, giving the oligomeric structures the overall appearance of bouquets. The C1r and C1s components associate with the collagen-like domain of C1q and have the same domain organization as the MASPs, and each can form homodimers. C1r and C1s interact to form a C1r2C1s2 tetramer that binds to C1q. Binding of C1q to antibodies activates C1r, which in turn activates C1s for action on the downstream complement components (Arlaud et al., 2002). In contrast, binding of the different MASPs to MBP appears to be mutually exclusive. MASP-2 is activated upon binding of MBP to cell surfaces, and subsequently activates C2 and C4, even in the absence of MASP-1/3 (Vorup-Jensen et al., 2000; Chen and Wallis, 2001).
A recently published structure of the C-terminal portion of C1r has provided insights into the mechanism of activation of the SP domain (Budayova-Spano et al., 2002b). However, no information about the structure of the N-terminal domains of any of this family of molecules has been reported. In the work reported here, the structure of the three-domain fragment of MASP-2 that forms the MBP-binding region has been determined, revealing its probable mechanism for interaction with MBP and providing an explanation for the Ca2+ dependence of this interaction. Combining the various structures that are now known, it is possible to develop a plausible model for the activation complex between MBP and MASP.
Results and discussion
Structure determination
The rat MASP-2 CUB1-EGF-CUB2 region (residues 1–280) was expressed in Chinese hamster ovary cells with a C-terminal His6 tag as described previously (Wallis and Dodd, 2000). Crystals of this material were grown in the presence of Ca2+ near neutral pH, and the structure was determined at 2.7 Å resolution (Table I). The asymmetric unit of the crystals contains one dimer. The CUB1 domain comprises residues 1–118, the EGF-like domain spans residues 119–163 and CUB2 contains residues 164–280. Residues 1–4 and 103–106 in CUB1, 125–131 in the EGF-like domain, and 218–221, 223–224 and 279–280 in CUB2, and the C-terminal His6 tag, are disordered, as are the side chains of seven other residues. Of the two N-linked glycosylation sites, the first GlcNAc residue is observed at Asn84 of CUB1, whereas no carbohydrate is visible at Asn266 of CUB2. The structure confirms the presence of the β-hydroxylation of Asn139 that is characteristic of many Ca2+-binding EGF domains.
Table I. Crystallographic statistics.
| (A) Data collection |
Native |
K2PtCl4 |
| Wavelength (Å) | 0.979 | 1.06238 |
| Resolution (Å) (last shell) | 40–2.7 (2.77–2.7) | 62–3.0 (3.08–3.0) |
| Rsyma | 0.086 (0.346) | 0.074 (0.232) |
| % completeness | 98.7 (99.6) | 97.2 (99.0) |
| % anomalous completeness | 96.0 (97.3) | |
| Average redundancy |
2.8 |
4.8 |
| (B) SIRAS phasing (to 4 Å) |
|
|
| Isomorphous phasing powerc | 0.86 | |
| Anomalous phasing powerc | 0.93 | |
| Figure of merit | 0.358 | |
| Figure of merit (4.17–4.00 Å) |
0.287 |
|
| (C) Refinement |
|
|
| R-factorb | 0.248 | |
| Rfreeb | 0.282 | |
| Average B-factor CUB1/EGF/CUB2 (Å2) | 32.7/34.5/58.6 | |
| Bond length r.m.s.d. (Å) | 0.0069 | |
| Angle r.m.s.d. (°) | 1.4 | |
| Ramachandran plot (% in most favored/allowed/generous/disallowed regions) | 81.3/16.8/1.4/0.5 |
aRsym = ΣhΣi(|Ii(h)| – |<I(h)>|)/ΣhΣi Ii(h), where Ii(h) is the observed intensity and <I(h)> is the mean intensity obtained from multiple measurements.
bR-factor and Rfree = Σ||Fo| – |Fc||/Σ|Fo|, where |Fo| is the observed structure factor amplitude and |Fc| is the calculated structure factor amplitude for the working and test sets, respectively.
cPhasing power = <|FH|>/E, where <|FH|> is the r.m.s. calculated heavy atom scattering amplitude and E is the estimated lack of closure error.
Protomer structure and Ca2+ binding
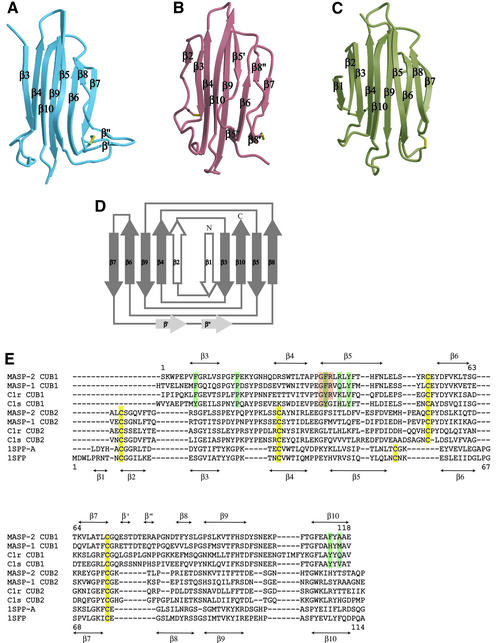
The MASP-2 CUB1-EGF-CUB2 region has the shape of an elongated letter ‘C’ (Figure 1), with approximate dimensions 100 × 35 × 35 Å. Both CUB and EGF-like domains have elongated shapes, and the small interfaces between each pair of domains in the monomer occur at the ends of the domains. The first two domains are very well defined in the electron density, whereas the CUB2 domain is less well defined, as reflected in its much higher average temperature factor (Table IC). The CUB1 and EGF domains have more crystal- or non-crystallographic symmetry-related contacts than does the CUB2 domain, which may explain these differences. The relatively poor order of the CUB2 domain may also reflect limited flexibility between it and the EGF-like domain. In contrast, the interface between the CUB1 and EGF-like domains appears to be made rigid by Ca2+ binding to the latter (see below).
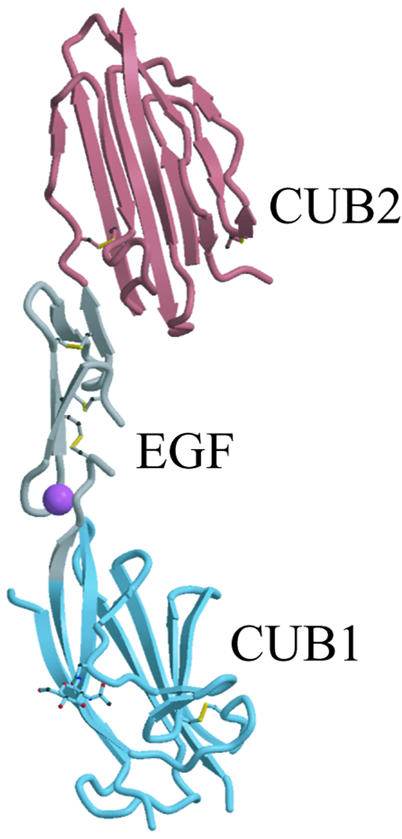
Fig. 1. Overall view of the rat MASP-2 CUB1-EGF-CUB2 protomer. CUB1 is shown in blue, the EGF-like domain in gray, CUB2 in red, Ca2+ in magenta and disulfide bonds in yellow.
The only other known CUB domain structures are those of seminal plasma spermadhesins (Romao et al., 1997; Romero et al., 1997; Varela et al., 1997). Both the spermadhesin and MASP-2 CUB domains are β sandwich structures that contain two four-stranded antiparallel β-sheets (Figure 2A–D). The N-terminal portion of the spermadhesin polypeptides has two additional β strands, each of which forms a fifth parallel strand at the edge of a sheet (Figure 2C and D). Using the spermadhesin strand nomenclature, the MASP-2 CUB1 domain lacks both β1 and β2 strands, whereas the CUB2 domain contains the β2 strand. A small β hairpin is present in the loop between β7 and β8 of CUB1. All of the CUB domains contain a disulfide bond between the β5–β6 and the β7–β8 loops. The spermadhesin and the MASP-2 CUB2 domains have an additional disulfide bond linking the polypeptide preceding β2–β4 (Figure 2B and C). Superposition of the common eight-stranded core of acidic seminal fluid protein (aSFP) (Romao et al., 1997) and the MASP-2 CUB domains gives a root mean square deviation (r.m.s.d.) of 1.16 Å for CUB1 and 1.63 Å for CUB2 (64 Cα positions). Superposition of the two MASP-2 domains gives an r.m.s.d. of 1.45 Å. A structure-based sequence alignment based on comparison of aSFP and the MASP domains is presented in Figure 2E.
Fig. 2. The CUB domain. (A and B) Ribbon diagrams of MASP-2 CUB1 and CUB2. The color scheme is the same as Figure 1. (C) Ribbon diagram of bovine aSFP (Romao et al., 1997) (Protein Data Bank accession code ID 1SFP). (D) CUB domain topology. The MASP-2 CUB1 domain is shown in gray. β strands 1 and 2 appear in aSFP and other spermadhesin CUB domains, and β strand 2 is present in the MASP-2 CUB2 domain. β′ and β” are two short strands formed from an extended loop found in CUB1 between β strands 7 and 8. (E) Structure-based sequence alignment of CUB domains. The secondary structure of MASP-2 CUB1 is shown above the sequences, and the secondary structure of aSFP is shown below. Highlighted residues are: green, compact dimer interface; pink, CUB1-EGF interface; yellow, cysteines in disulfide bonds. Phe36 is in both the dimer and EGF interfaces. The sequences and their SwissProt identifiers are: rat MASP-2, Q9JJS8; rat MASP-1, Q9JJS9; human C1r, P00736; human C1s, P09871; pig seminal plasma glycoprotein chain a (SPP-a), P35495; cow acidic seminal fluid protein, P29392.
The MASP-2 EGF-like domain is very similar to other Ca2+-binding EGF-like domains (Figure 3). These domains feature an extended polypeptide followed by a long β hairpin and often, as in the MASP-2 domain, a second, smaller β hairpin near the C-terminus of this domain. The structure is stabilized by three disulfide bonds found in all EGF-like domains. Superposition of 31 structurally equivalent Cα positions between the MASP-2 and the Ca2+-binding EGF-like domain from factor IX (Rao et al., 1995) gives an r.m.s.d. of 1.65 Å.
Fig. 3. The EGF-like domain. (A and B) The EGF-like domains of MASP-2 and human coagulation factor IX (Protein Data Bank accession code ID 1EDM). Ca2+ is in magenta, disulfide bonds in yellow. Residues that contribute coordination bonds are shown in ball-and-stick representation, with red oxygen atoms and blue nitrogen atoms. In (B), Asn58 from a symmetry-related EGF domain is shown with green bonds. (C) Structure-based sequence alignment. Symbols above and below the alignments are: *, amino acid side chain Ca2+ ligand; ∧, main-chain carbonyl oxygen Ca2+ ligand; #, β-hydroxylated side chain Ca2+ ligand; !, side chain Ca2+ ligand from symmetry-related molecule (factor IX). Highlighted residues are: green, compact dimer interface; pink, CUB1-EGF interface; yellow, cysteines in disulfide bonds. SwissProt identifiers for rat MASP and human C1r and C1s sequences are given in the Figure 2 legend; the human factor IX sequence is SwissProt P00740. (D) The CUB1-EGF interface. The CUB1 domain is shown in blue, the EGF-like domain in gray. Residues involved in Ca2+ binding and packing interactions between the domains are shown in ball-and-stick representation. The Ca2+ is shown as a magenta sphere, with coordination bonds shown in pink.
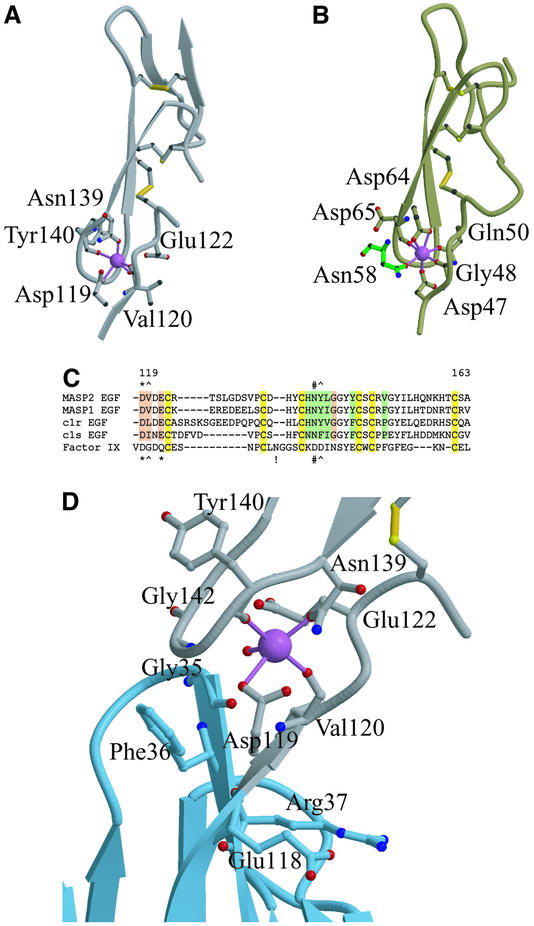
The single Ca2+ is bound near the N-terminal end of the EGF-like domain (Figure 3A). Only five coordination ligands are observed. Relative to the pentagonal bipyramidal Ca2+ coordination geometry observed in the factor IX EGF-like domain (Rao et al., 1995; Figure 3B), the two apical positions are occupied by main chain carbonyl oxygen atoms of Val120 and Tyr140. The side chains of Asp119 and Asn139, residues highly conserved amongst Ca2+-binding EGF-like domains (Stenflo et al., 2000; Figure 3C), each provides a coordination ligand. The second oxygen atom of the Asp119 side chain is too far (2.9 Å) from the Ca2+ to be considered a coordination ligand, but at the 2.7 Å resolution limit of these crystals this possibility cannot be ruled out. In factor IX, both carboxylate oxygen atoms of Asp64, the equivalent of MASP-2 Asn139, coordinate the Ca2+. Surprisingly, Glu122, which is also conserved amongst these domains and the equivalent Gln50 of which provides a coordination ligand in the factor IX domain (Rao et al., 1995), does not interact with the Ca2+; its coordination position is approximately replaced by a water molecule. In the crystal structure of factor IX, the final coordination position is occupied by a symmetry-related asparagine residue, whereas this position is vacant in the present structure.
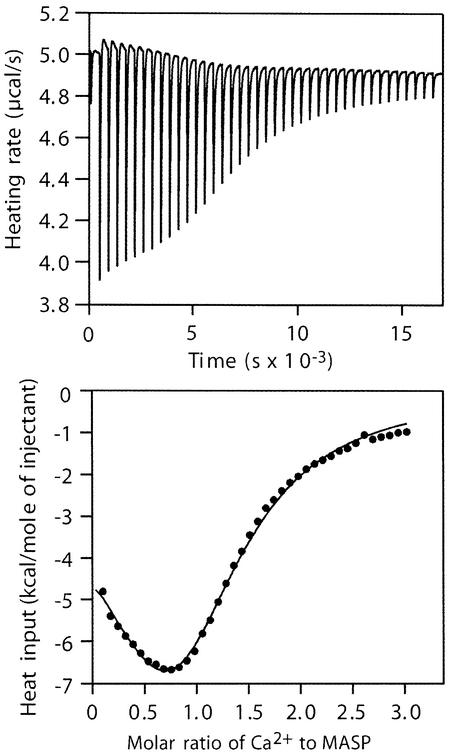
Ca2+-binding EGF-like domains have a low affinity for Ca2+, but the intrinsic mM affinity increases by up to three orders of magnitude in the presence of an adjacent protein domain (Stenflo et al., 2000). For example, the CUB1-EGF construct of C1r displays roughly 300× higher affinity for Ca2+ than the isolated EGF-like module (Thielens et al., 1999). The affinity of the MASP-2 CUB1-EGF-CUB2 fragment for Ca2+ was measured by isothermal titration calorimetry (Figure 4). Stoichiometric binding (occupancy 1.1 ± 0.2) with a Kd of 6.3 ± 2.6 µM, presumably corresponding to the conserved site in the EGF-like domain, is followed by a much weaker partial binding that is manifest as precipitation. Within one protomer, residues Glu118, Asp119, Val120 and Gly142, which lie in or near the Ca2+ site, also participate in interactions with Gly35, Phe36 and Arg37 in the CUB1 domain (Figure 3C and D). The water molecule that serves as a direct Ca2+ coordination ligand also forms hydrogen bonds with residues Gly35 of the CUB1 domain and Glu122 of the EGF-like domain.
Fig. 4. Ca2+-binding to the CUB-1-EGF-CUB-2 fragment of MASP-2 analyzed by isothermal titration calorimetry. Top: representative experiment showing heat release as a solution of CaCl2 was added to the MASP-2 fragment in 40 aliquots over 300 min. Bottom: data were fitted to a model in which there are two Ca2+-binding sites on each MASP fragment: a high-affinity site with an estimated occupancy (1.3) consistent with one Ca2+ on each MASP protomer and a lower affinity site. The occupancy of the low-affinity binding site (Kd > 40 µM) was <0.1 in all experiments, and probably reflects Ca2+-induced protein precipitation.
There are also extensive interactions between the CUB1 and the EGF-like domains in the dimer interface (see below), including a hydrogen bond between Arg39 and the β-OH group of the Ca2+ ligand β-hydroxyAsn139 (Figure 5). Virtually all of the residues observed in the MASP-2 CUB1-EGF interface are conserved in character in MASP-1, C1r and C1s (Figures 2E and 3C). When Ca2+ is removed from Ca2+-binding EGF-like domains, the structure around the Ca2+ site undergoes significant conformational rearrangements (Rao et al., 1995; Stenflo et al., 2000). The intimate involvement of Asp119, Val120 and β-hydroxyAsn139 in both the CUB1 interface and Ca2+ ligation suggests that Ca2+ binding is a critical feature in determining the relative positions of the CUB1 and EGF-like domains in MASPs, C1r and C1s.
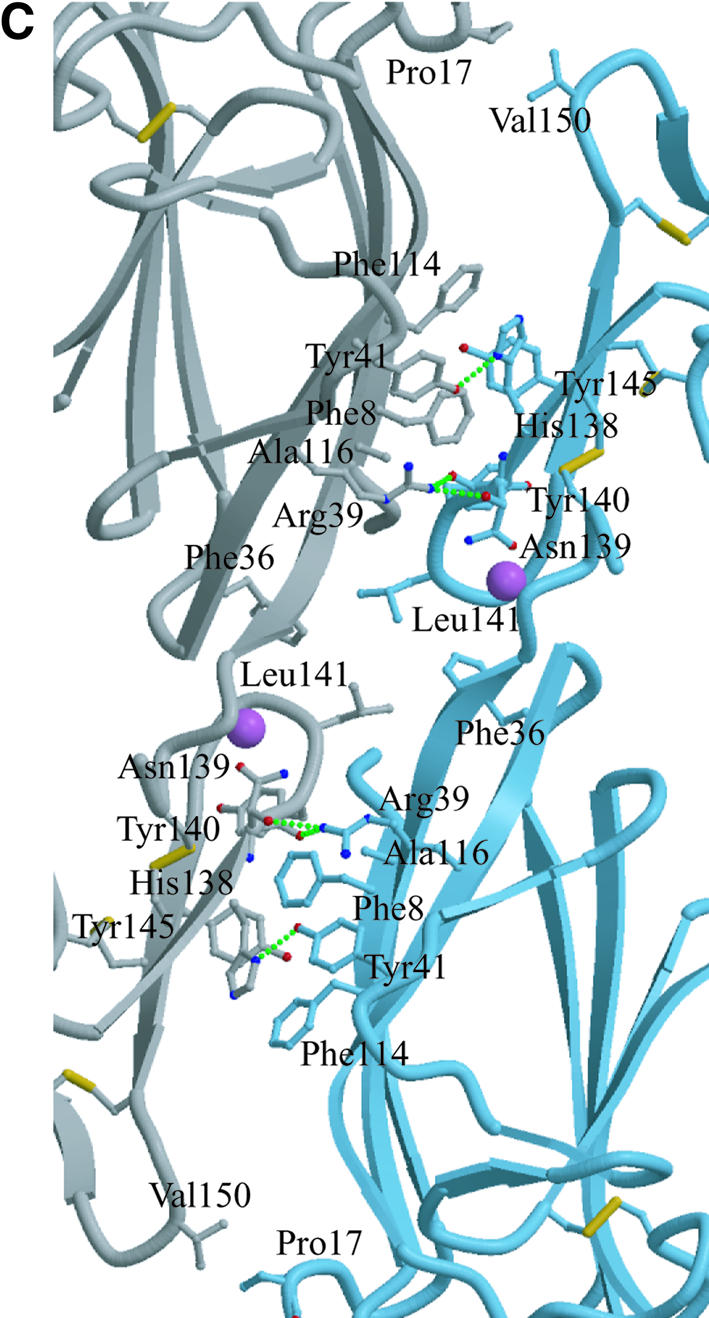
Fig. 5. The CUB1-EGF-CUB2 dimer. (A) The compact dimer. Left, view down the 2-fold symmetry axis [indicated by the ( ) sign]. Right, view perpendicular to the 2-fold axis. The proposed MBP binding sites are indicated by solid or dashed circles, the diameters of which roughly correspond to that of a collagen triple helix. The distance between the sites in the dimer is indicated. (B) The extended dimer. Left, view perpendicular to the 2-fold axis (indicated by the arrow). Right, view down the 2-fold axis. The location of the proposed binding site [(A), solid circles] is shown; the filled circle indicates that one collagen helix would be antiparallel to the other. (C) The compact dimer interface. The two protomers are shown in blue and gray. Side chains participating in direct packing or hydrogen-bonding interactions are shown in ball-and-stick representation. Hydrogen bonds are shown in dashed green lines. The Ca2+ are shown as pink spheres. Water-mediated interactions have been omitted for clarity.
Both MASP-1 and MASP-2 display increased resistance to trypsinolysis in the presence of Ca2+ (Wallis and Dodd, 2000). The proteolytically sensitive site at Arg173 is in the N-terminal region of the CUB2 domain, and forms part of the interface between the EGF-like and CUB2 domains. These data suggest that the binding of Ca2+ to the EGF-like domain may also influence the position of the CUB2 domain and thereby contribute to the formation of the MBP-binding site.
Dimer structure and MBP binding
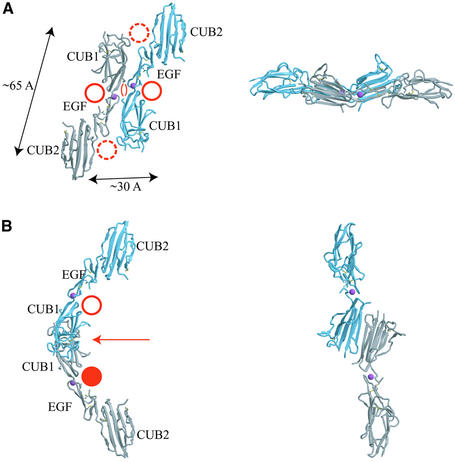
The MASP-2 CUB1-EGF-CUB2 fragment studied here mediates dimerization of the protein and binding to MBP (Wallis and Dodd, 2000; Chen and Wallis, 2001). The crystallographic asymmetric unit contains a dimer and, combined with the packing of molecules in the crystal lattice, there are two distinct dimers present in the crystal. In the first, lateral association of the CUB1 and EGF-like domains about a 2-fold symmetry axis creates a relatively compact dimer (Figure 5A). The other dimer interface involves only the CUB1 domain, where antiparallel pairing of the β4 strands from two protomers creates a highly extended structure (Figure 5B). This pairing is distinct from the spermadhesin PSI/II dimer, which forms by antiparallel strand pairing on the opposite edge of the sandwich, using both sheets (Romero et al., 1997). Sedimentation equilibrium studies demonstrate that dimerization requires at most the first two domains and is independent of Ca2+ (Wallis and Dodd, 2000), but it has not been possible to produce the CUB1 domain in isolation to assess whether it alone can dimerize.
Analytical ultracentrifugation was used to determine which of the two crystallographically observed dimers corresponds to the solution configuration of the CUB1-EGF-CUB2 fragment. The s20,w of the MASP-2 fragment determined by sedimentation velocity is 4.55 ± 0.10 S. Sedimentation coefficients calculated from the atomic coordinates of the two dimers gives values of 4.53 S for the more compact dimer (Figure 5A) and 4.18 S for the extended dimer (Figure 5B), indicating that the compact dimer is the solution species. Inspection of the interfaces also favors the compact dimer. Obligate oligomer interfaces tend to have significant numbers of non-polar interactions (Janin et al., 1988; Jones and Thornton, 1995). The compact dimer interface is larger (1844 Å2 total buried surface area) and features the packing of a number of hydrophobic side chains (Figure 5C), in contrast to the almost exclusively polar interactions that characterize the smaller (1540 Å2 total buried surface area) extended dimer interface (data not shown). The residues in the compact dimer interface are highly conserved among MASPs, C1r and C1s (Figures 2E and 3C). Finally, as discussed below, the symmetry of this dimer also favors its assignment as the solution species. Unless otherwise noted, the remainder of the discussion refers to the compact dimer.
The CUB1-EGF-CUB2 dimer features two prominent concave surfaces that are large enough to accommodate a collagen-like triple helix, which including side chains has a diameter of ∼12 Å (Emsley et al., 2000). The first is formed by the ‘C’ shape of the protomer. A collagen triple helix can be placed in the middle of each protomer surface, where it would contact all three domains (Figure 5A, solid circles). These two sites are separated by ∼30 Å. A second concave surface is formed by CUB1 of one protomer and the EGF-like and CUB2 domains of the other protomer (Figure 5A, dashed circles); these two sites are separated by ∼65 Å. No mutational data are available that would localize the MBP binding site on the CUB1-EGF-CUB2 surface, and either of the proposed arrangements would be consistent with the observation that all three domains are required for MBP binding. However, the fact that the only subfragment of CUB1-EGF-CUB2 that displays even weak binding to MBP is the CUB1-EGF fragment suggests that the primary interactions involve these two domains (Chen and Wallis, 2001; Thielens et al., 2001). Of the two proposed binding sites shown in Figure 5A, the first (solid circles) has more extensive contact with the CUB1 and EGF-like domains, whereas much of the interaction surface in the second site (dashed circles) comes from CUB2, making it less likely.
Electron microscopy of C1 complexes indicates that the C1r2C1s2 complex binds to the collagen-like portion of C1q just C-terminal to a kink created by an interruption in the Gly-X-Y sequence pattern (Strang et al., 1982; Weiss et al., 1986; Arlaud et al., 2002). Given the similarity of the two systems, it is likely that MASPs interact with MBPs in the comparable region. In MBPs, the N-terminal cysteine-rich domains and the first portion of the collagen-like region associate laterally in a parallel orientation, so that past the kink the individual trimeric subunits splay apart from one another but still run in a roughly parallel direction. Therefore, the collagen-like region of MBP must bind to the MASP dimer in an approximately parallel orientation. The 2-fold axis that generates the compact dimer is parallel to either proposed binding site, which would imply a parallel orientation for the bound MBP trimeric stalks. It is worth noting that for the first binding site (Figure 5A, solid circles), the 2-fold rotation axis of the alternative extended dimer in the crystals would put the two collagen-like stalks in a non-physiological, antiparallel arrangement (Figure 5B).
In isolated C1q, the average angle between individual trimeric subunits past the kink is ∼40–50° (Schumaker et al., 1981). MBP bouquets look very similar to C1q, and the interruption in the Gly-X-Y pattern is the point at which the individual trimeric stalks kink and splay away from each other (Lu et al., 1990). Each Gly-X-Y triplet is 8.65 Å long in a standard collagen structure. Ignoring irregularities in the structure near the kink, the 12 Gly-X-Y repeats in the individual collagenous region of the stalks would be 103 Å long. Inspection of MBP sequences indicates that the most highly conserved region lies in the first five repeats following the kink. If MASPs bind in this region and two trimeric stalks are separated by 45°, the sites would be ∼30 Å apart at repeat 5, consistent with the proposed binding site in the middle of the protomer (Figure 5A, solid circles). It is likely that at least one Gly-X-Y repeat is irregular following the kink, so this spacing might be accommodated even closer to the kink. On the other hand, if the site were located near the C-terminal portion of the stalk, the other proposed binding sites (Figure 5A, dashed circles) could be accommodated. Given the sequence conservation and the electron microscopic data for the location of C1r/C1s on C1q (Strang et al., 1982), we favor the first alternative, with its smaller separation of sites as well as the smaller contribution of CUB2 to the binding surface (see above).
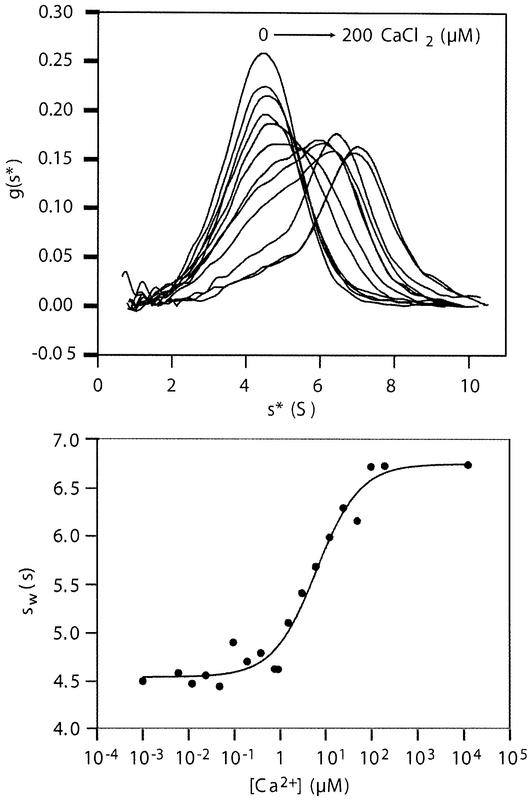
High affinity binding to MBPs requires the first three MASP domains and is strictly dependent on Ca2+ (Wallis and Dodd, 2000; Chen and Wallis, 2001; Thielens et al., 2001). Analytical ultracentrifugation was used to examine quantitatively the Ca2+ dependence of MBP binding by the MASP-2 CUB1-EGF-CUB2 fragment (Figure 6). Addition of Ca2+ results in formation of the more rapidly sedimenting MASP-2–MBP complex. The KCa for association is 6.3 µM, which is the same as the intrinsic Ca2+ affinity of the isolated MASP obtained by calorimetry. Thus, the interaction with MBP appears to require the single Ca2+ site present in the EGF-like domain.
Fig. 6. Ca2+ dependence of MASP-2 binding to MBP-A dimers analyzed by sedimentation velocity. Top: apparent sedimentation distributions [g(s*)] of mixtures of the CUB-1-EGF-CUB-2 fragment of MASP-2 (0.38 mg/ml) and MBP-A dimers (0.075 mg/ml) in the presence of increasing concentrations of CaCl2. The apparent sedimentation coefficients (s*) of the uncomplexed components were 4.5 S and 5.0 S for the MASP-2 fragment and MBP-A dimer, respectively. Bottom: the average sedimentation coefficient of each mixture plotted as a function of Ca2+ concentration.
It is very likely that an essential role of the Ca2+ is to orient the three domains correctly for optimal interaction with MBP. The proposed binding site for MBP (Figure 5A, solid circles) places the collagen-like region in contact with the MASP EGF-like domain in the vicinity of the Ca2+ site. It was noted above that in the present structure, one coordination position is vacant. It is possible that MBP provides the last coordination ligand for the Ca2+, and there are a number of highly conserved glutamine residues in the collagen-like repeats just C-terminal to the kink that might fulfill this role. Precedent for such a binding mode comes from the interaction of a collagen peptide with the I domain of integrin α2β1, where a glutamate from the collagen completes the coordination shell of a bound metal ion (Emsley et al., 2000).
Model of MASP-2–MBP complexes and SP activation
Engagement of a pathogenic cell surface by MBP leads to cleavage of the MASP SP domain, converting the inactive zymogen to a catalytically active protease. Like other SP zymogens, uncleaved MASP-2 shows significant activity towards small substrates (C.-B.Chen and R.Wallis, unpublished data). Presumably, the catalytic regions of the two MASP SP domains are brought into proximity upon binding of MBP to a target cell surface, whereupon cleavage of one zymogen by the other produces an active protease, which subsequently activates the partner.
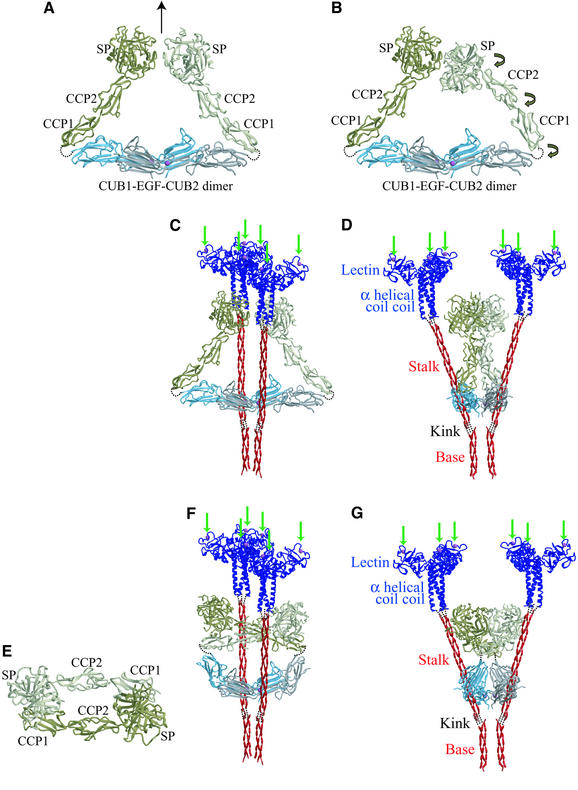
The structure of the CCP1-CCP2-SP region from the homologous C1r containing the inactive zymogen form of the SP domain (Budayova-Spano et al., 2002b) was used to construct a full six-domain model for MASP-2 (Figure 7). The CCP1-CCP2-SP structure features end-to-end interactions of the elongated CCP domains, producing an overall elongated structure. The last residue visible in the CUB1-EGF-CUB2 structure is 278, and the first residue seen in the C1r CCP1-CCP2-SP structure is equivalent to MASP-2 residue 279 (Budayova-Spano et al., 2002b). In C1r and C1s, the junction between CUB2 and CCP1 appears to be flexible, as the γB fragments generated by autocatalysis or plasmin digestion comprise the CCP1-CCP2-SP region (Villiers et al., 1985). The structural data suggest a limited amount of flexibility between the EGF-like and CUB2 domains (see above). NMR studies have indicated that CCP modules can adopt different relative orientations (Wiles et al., 1997), and comparison of the CCP2-SP orientations in several crystal forms indicates a certain amount of flexiblity in this interface (Budayova-Spano et al., 2002a). Thus, there are a number of potential interdomain hinge points that could facilitate the conformational rearrangements required to bring the two SP domains into close apposition. The C1r CCP1-CCP2-SP structure was positioned near the CUB1-EGF-CUB2 dimer (Figure 2A), so that (i) its N-terminus is within bonding distance of the C-terminus of CUB2, (ii) the SP domains contact one another, and (iii) the last three domains are related by the 2-fold symmetry axis of the CUB1-EGF-CUB2 dimer (Figure 7A). Small rotations were then applied in the CUB2-CCP1, CCP1-CCP2 and CCP2-SP interfaces in order to bring the bond to be cleaved within one SP domain close to the catalytic center of the partner SP domain (Figure 7B). The resulting structure is roughly triangular, with the CUB1-EGF-CUB2 dimer forming the base, and the CCP1-CCP2-SP domains of the protomers comprising the other two sides.
Fig. 7. Model of MASP-2 and interaction with the MBP dimer. (A) The N-termini of two molecules of human C1r CCP1-CCP2-SP (Protein Data Bank accession code ID 1GPZ) were positioned near the C-termini of the CUB1-EGF-CUB2 dimer, preserving symmetry about the 2-fold axis of the dimer (arrow). The CUB1-EGF-CUB2 dimer is colored blue and gray, Ca2+ in magenta, and the CCP1-CCP2-SP chains in dark and light green. (B) Changes were applied to the junctions between domains as indicated by the arrows in order to bring the catalytic sites of the SP domains into proximity of one another. (C) Model of a dimer of trimeric MBP subunits bound to the MASP2 model shown in (A). Coordinates of the trimeric coiled-coil neck and lectin region of rat MBP-A (blue, PDB ID 1RTM) were placed at the C-terminus of a model of the collagen-like region (red) generated from a collagen peptide [(Pro-Pro-Gly)10]3 (Berisio et al., 2002) (Protein Data Bank accession code ID 1K6F); the dashed lines represent the flexible joint between these two regions of the MBP primary structure. The green arrows point to the Ca2+ (magenta) at which carbohydrate ligands from a pathogen bind. The gap in the collagen connected by three dashed lines represents the interruption in the collagen consensus sequence, where the trimeric stalks splay away from the kink. Other dashed lines in the model represent protein links between the different domains. (D) Same as (C), but rotated ∼90° about the vertical axis. (E) The dimer observed in the crystal structure of the human C1r CCP1-CCP2-SP zymogen (Protein Data Bank accession code ID 1GPZ), proposed to correspond to an inactive form due to blockage of the catalytic site of one SP by CCP1 of the other protomer (Budayova-Spano et al., 2002b); colored as in (A)–(D). (F) Model of inactive MASP-2 made by moving CUB2 relative to CUB1-EGF (curved arrow) such that the N-terminal ends of the C1r CCP1-CCP2-SP zymogen dimer are near the C-termini of the CUB1-EGF-CUB2, bound to two MBP trimers. The MBP dimer model is that shown in (C) and (D). (G) Same as (F), but rotated ∼90° about the vertical axis.
A model for the dimer of trimeric MBP subunits was constructed from coordinates of a collagen peptide and generating the correct number of Gly-X-Y repeats before and after the kink. The region N-terminal to the kink was arranged in a close-packed, parallel orientation, as expected from the lateral association observed in the electron microscope and the inter-trimer disulfide bonds present in the preceding N-terminal cysteine-rich domain (Wallis and Drickamer, 1999). A small gap was left for the kink, then the following 12 Gly-X-Y repeats were placed at a relative angle of ∼40°. The known structure of the MBP-A trimeric α-helical neck and lectin domains (Weis and Drickamer, 1994) was placed at the C-terminus of the collagenous sequence such that the carbohydrate-binding sites of the two trimers are parallel to one another, as though interacting with a flat target surface (Figure 7C and D).
The MBP dimer model was docked onto the MASP-2 model in the center of the ‘C’ (Figure 5A, solid circles). The MASP model fits ‘inside’ the MBP dimer, with the SP domain near the level of the collagen–α-helical neck junction but not contacting MBP (Figure 7C and D). Without knowing the binding site on MBP, a more precise model for the MBP–MASP complex cannot be made. Nonetheless, this exercise demonstrates that the experimentally observed MASP-2 CUB1-EGF-CUB2 dimer and the homologous C1r CCP1-CCP2-SP structures can be accommodated on the MBP dimer with enough room to permit conformational changes required to activate the protease upon binding of MBP to a cell surface. Moreover, this arrangement would provide access of C1 inhibitor, which has been shown to block activity of MASP-2 (Rossi et al., 2001), through the wider part of the cone-shaped MBP oligomer, as proposed for C1 (Schumaker et al., 1986).
The conformation of MASP-2 in the model presented here is distinct from those proposed for the homologous C1r and C1s based on electron microscopy of the isolated proteins, but are roughly consistent with those based on pictures of the C1 complex. Monomeric C1r and C1s appear as a dumbbell-shaped molecules with a slight curvature, ∼170–180 Å long (Villiers et al., 1985; Weiss et al., 1986). The MASP-2 CUB1-EGF-CUB2 monomer is 100 Å long, and the CCP1-CCP2-SP region of the C1r zymogen is 115 Å long (Budayova-Spano et al., 2002b). An end-to-end arrangement of these two halves would therefore give a total length of 215 Å, longer than the observed monomer but consistent with the idea that inter-domain hinges can change the overall shape of the molecule. Dimeric C1r appears as ‘X’ or ‘croissant’ shaped particles (Villiers et al., 1985; Weiss et al., 1986), some of which look similar to, but more open than, the proposed structure for MASP-2 (Figure 7). Electron micrographs of crosslinked C1 complexes do not show evidence of extended C1r–C1s dimers seen in isolation, suggesting the proteases adopt a more compact conformation when bound to C1q (Strang et al., 1982). It is likely that binding to the MBP or C1q oligomer locks the associated protease into a conformation that allows facile approach of the SP domains upon target binding.
In the C1r CCP1-CCP2-SP zymogen crystals, a dimer is formed by contacts between the CCP1 module of one monomer and the SP of another (Budayova-Spano et al., 2002b) (Figure 7E). This interaction blocks access to the bond that must be cleaved for activation. It was suggested that this might represent the inactive conformation of C1r, and that binding of C1q to IgG or IgM would induce conformational changes that would mechanically alter the relative domain dispositions and thereby remove the block to the activation cleavage site. To assess whether this structure can be accommodated in the homologous MBP–MASP complex, the C1r CCP1-CCP2-SP dimer was arranged such that the N-termini of the CCP1 domains were near the C-termini of the CUB2 domains of the MASP-2 CUB1-EGF-CUB2 (Figure 7F). This required changing the angle between the EGF-like and CUB2 domains. CUB1-EGF-CUB2 was docked on the MBP dimer model as described above. This model can also be accommodated on MBP (Figure 7F and G). However, the SP domains are now much closer to MBP (Figure 7G), so that movements required to change from this inactive dimer to the active form would probably require significant movements of MBP.
MBPs have two flexible joints or hinge points: the interruption in the collagen-like Gly-X-Y pattern at which the individual stalks splay away from each other (Lu et al., 1990), and the junction between the collagen-like and α-helical regions. The kink generated by the interruption in the Gly-X-Y pattern in the stalk of C1q exhibits limited flexibility, as shown by a sharply peaked distribution with significant variation of the angle between the stalk and N-terminal bundle of collagen helices observed by electron microscopy of the isolated molecule (Schumaker et al., 1981). The junction between the collagen and α-helical region must accommodate the mismatch of the collagen screw symmetry with the pure 3-fold rotational symmetry of the α-helices. This region is sensitive to proteases (Weis and Drickamer, 1994), and was not observed in a crystal structure of a trimeric MBP-A fragment containing the last four Gly-X-Y repeats of the collagenous sequence (Ng et al., 1998). Flexibility in the joint between the α-helical coiled-coil and collagen-like region of class A scavenger receptors has been directly observed in electron micrographs (Resnick et al., 1996). Flexibility at this junction would allow CRDs in different subunits of the MBP oligomer to bind to a flat surface even through the collagenous domains are splayed from each other (Figure 7D).
The modeling presented here demonstrates that MASPs can fit on MBPs in the region between the two hinge points. Engagement of a cell surface by two or more trimeric lectin heads probably imparts changes to these joints that can be transmitted to the bound protease and alter the relative dispositions of subunits to allow activation to occur. Details of such mechanisms must await further structural analysis of MBP–MASP complexes.
Materials and methods
Structure determination
Crystals of MASP-2 CUB1-EGF-CUB2 were grown using the hanging drop method (0.4:0.4 or 0.6:0.6 µl protein/µl reservoir in a drop). The protein solution contained 2–5 mg/ml protein, 50 mM Tris pH 8.2, 10 mM CaCl2 and 0.5 M NaCl. The reservoir solution contained 0.8–1.0 M Na2tartrate, 80 mM Li2SO4 and 0.1 M MES pH 6.5. The crystals used for the Pt2+ derivative data set were soaked in reservoir solution containing 5 mM K2PtCl4 for 3 h. Crystals were briefly transferred to reservoir solution containing 20% ethylene glycol before freezing in liquid nitrogen, and were maintained at 100 K during data collection.
Native and derivative data sets were measured on an ADSC Q315 CCD detector at SSRL beam line 11-1 (Table IA). The Pt2+ data were measured with inverse beam geometry to insure complete anomalous data, at a wavelength of 1.062 Å to enhance the value of Δf”. Diffraction data were processed with MOSFLM (Leslie, 1993) and SCALA (CCP4, 1994). The space group is P21, with unit cell parameters for the native crystal a = 70.4 Å, b = 103.9 Å, c = 70.5 Å, β = 119.9°. The native and Pt2+ data scaled to each other with an R-factor of 15.7% for data to 3.5 Å Bragg spacings.
The program SOLVE (Terwilliger and Berendzen, 1999) was used to find two heavy atom sites. Phasing, density modification, refinement and map calculations were performed using CNS (Brünger et al., 1998). The single isomorphous replacement with anomalous scattering (SIRAS) phases (Table IB) were improved using density modification and phase extension to 3.5 Å (figure of merit 0.91). The non-crystallographic symmetry (NCS) 2-fold axis was found using the program GETAX (CCP4, 1994). A partial polyalanine model was built and refined with strict NCS using the MLHL target, guided by the known homologous CUB (Romao et al., 1997; Romero et al., 1997; Varela et al., 1997) and EGF-like (Rao et al., 1995) domain structures. Bulk solvent and anisotropic temperature factor corrections were applied during the refinement. The resolution was gradually increased to 2.7 Å, using the MLF target. The strict NCS was not dropped at any stage of the refinement, as moving to restrained NCS or removing the NCS entirely did not change the statistics or improve the map quality. Grouped and later individual temperature factors improved the map quality and lowered Rfree significantly. Water molecules were added to peaks >3σ in Fo – Fc maps and were within hydrogen bond distance to the protein or other water molecules. The final model has 53 water molecules per protomer.
Buried surface area calculations were performed in CNS, using a 1.4 Å probe radius. For CUB domain superpositions, the Cα positions corresponding to the following CUB1 residues were used: 7–13, 25–32, 35–46, 56–62, 65–71, 86–89, 92–101 and 110–118. The EGF-like domain superpositions used MASP-2 residues 119–124, 132–134, 136–154, 158–159 and 161.
Coordinates and structure factors for the rat MASP-2 CUB1-EGF-CUB2 region have been deposited in the Protein Data Bank (accession code ID 1NT0). Coordinates of the MBP–MASP complex model (Figure 7) are available upon request.
Isothermal titration calorimetry
To remove trace amounts of Ca2+ prior to isothermal titration calorimetry and analytical ultracentrifugation experiments, samples of the MASP fragment were dialyzed against at least three changes of buffer in the presence of Chelex-100 resin (Bio-Rad). Buffer (50 mM Tris–HCl pH 7.5, containing 150 mM NaCl) was pretreated by stirring for 2 weeks in the presence of Chelex-100 resin in a dialysis bag. Ca2+ concentration was monitored with a Ca2+-sensitive dye (Tsien, 1980). Experiments were carried out using an MCS calorimeter (MicroCal, Inc., Northampton, MA), at 25°C. A solution of CaCl2 (862 µM) was added in 5 µl injections to the MASP-2 fragment (34 or 23 µM) in the calorimeter cell (1.3 ml). Data were fitted by non-linear least-squares regression (Origin package; MicroCal, Inc.), with stoichiometry, association constant and change of enthalpy of interaction as free variables, after subtracting the heats resulting from the addition of CaCl2 into buffer alone and protein into buffer alone. Data represent the average value ± SE from two separate experiments.
Analytical ultracentrifugation
Sedimentation velocity experiments were carried out in a Beckman XLA-70 centrifuge at 40 000 r.p.m. and 20°C using Epon aluminum-filled centerpieces. Scans were collected at 2–4 min intervals at 230 nm. Samples consisting of a mixture of MBP-A dimers (0.075 mg/ml; prepared as described by Wallis and Drickamer, 1999) and MASP-2 fragment (0.028 mg/ml) were analyzed in 50 mM Tris–HCl pH 7.5, containing 150 mM NaCl and increasing concentrations of CaCl2 ranging from 0 to 200 mM. Apparent sedimentation distributions [g(s*)] and average sedimentation coefficients (sw) were determined using the program DCDT (Stafford, 1992) at each concentration of CaCl2. The data were fitted using the Origin package.
The sedimentation coefficient of the MASP-2 fragment was determined by the second moment method and was corrected for the effects of buffer density and viscosity as described previously (Chen and Wallis, 2001). Data represent the average value ± SE from two separate experiments. The predicted sedimentation coefficients of the two alternative dimer conformations were calculated from the atomic coordinates, using the program HYDROPRO (Garcia de la Torre et al., 2000). These calculations used the mol. wt of 73 500 Da determined by equilibrium sedimentation and a partial specific volume of 0.714 cm3/g calculated from the amino acid and carbohydrate composition (Wallis and Dodd, 2000). The solvent viscosity was 0.01 poise, the solvent density was 1 g/cm3, and the temperature was 293 K. The effect of hydration is included in an effective atomic radius of 3.2 Å for all non-hydrogen atoms, which is the average value calculated from experimentally determined hydrodynamic properties of 13 different proteins (Garcia de la Torre et al., 2000).
Acknowledgments
Acknowledgements
We thank Dawn Torgersen for assistance in protein production and Tony Day for access to the calorimeter. The analytical ultracentrifugation facility is supported by the Wellcome Trust and the Biotechnology and Biological Sciences Research Council. Some of this work is based upon research at SSRL, which is funded by the US Department of Energy and the National Institutes of Health. J.C.M.U. was supported by postdoctoral fellowship ALTF59-2000 from the European Molecular Biology Organization. This work was supported by grants 041845 and 054508 from the Wellcome Trust to K.D., and grant GM50565 from the National Institutes of Health to W.I.W.
References
- Arlaud G.J., Gaboriaud,C., Thielens,N.M., Budayova-Spano,M., Rossi,V. and Fontecilla-Camps,J.C. (2002) Structural biology of the C1 complex of complement unveils the mechanisms of its activation and proteolytic activity. Mol. Immunol., 39, 383–394. [DOI] [PubMed] [Google Scholar]
- Berisio R., Vitagliano,L., Mazzarella,L. and Zagari,A. (2002) Crystal structure of the collagen triple helix model [(Pro-Pro-Pro)10]3. Protein Sci., 11, 262–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P. and Beckmann,G. (1993) The CUB domain: a widespread module in developmentally regulated proteins. J. Mol. Biol., 231, 549–545. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR System (CNS): A new software system for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Budayova-Spano M., Brabarse,W., Thielens,N.M., Hillen,H., Lacroix,M., Schmidt,M., Fontecilla-Camps,J.C., Arlaud,G.J. and Gaboriaud,C. (2002a) Monomeric structures of the zymogen and active catalytic domain of complement protease C1r: further insights into the C1 activation mechanism. Structure, 10, 1509–1519. [DOI] [PubMed] [Google Scholar]
- Budayova-Spano M., Lacroix,M., Thielens,N.M., Arlaud,G.J., Fontecilla-Camps,J.C. and Gaboriaud,C. (2002b) The crystal structure of the zymogen catalytic domain of complement protease C1r reveals that a disruptive mechanical stress is required to trigger activation of the C1 complex. EMBO J., 21, 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chen C.B. and Wallis,R. (2001) Stoichiometry of complexes between mannose-binding protein and its associated serine proteases: defining functional units for complement activation. J. Biol. Chem., 276, 25894–25902. [DOI] [PubMed] [Google Scholar]
- Dahl M.R., Thiel,S., Matsushita,M., Fujita,T., Willis,A.C., Christensen,T., Vorup-Jensen,T. and Jensenius,J.C. (2001) MASP-3 and its association with distinct complexes of the mannan-binding lectin complement activation pathway. Immunity, 15, 127–135. [DOI] [PubMed] [Google Scholar]
- Emsley J., Knight,C.G., Farndale,R.W., Barnes,M.J. and Liddington,R.C. (2000) Structural basis of collagen recognition by integrin α2β1. Cell, 101, 47–56. [DOI] [PubMed] [Google Scholar]
- Garcia de la Torre J., Huertas,M.L. and Carrasco,B. (2000) Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J., 78, 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janin J., Miller,S. and Chothia,C. (1988) Surface, subunit interfaces and interior of oligomeric proteins. J. Mol. Biol., 204, 155–164. [DOI] [PubMed] [Google Scholar]
- Jones S. and Thornton,J.M. (1995) Protein–protein interactions: a review of protein dimer structures. Prog. Biophys. Mol. Biol., 63, 31–65. [DOI] [PubMed] [Google Scholar]
- Kawasaki N., Kawasaki,T. and Yamashina,I. (1983) Isolation and characterization of a mannan-binding protein from human serum. J. Biochem. (Tokyo), 94, 937–947. [DOI] [PubMed] [Google Scholar]
- Leslie A.G.W. (1993) Autoindexing of rotation diffracton images and parameter refinement. In Sawyer,L., Isaacs,N. and Bailey,S. (eds), Proceedings of the CCP4 Study Weekend: ‘Data Collection and Processing’, 29–30 January 1993. SERC Daresbury Laboratory, Daresbury,UK, pp. 44–51.
- Lu J., Thiel,S., Wiedemann,H., Timpl,R. and Reid,K.B.M. (1990) Binding of the pentamer/hexamer forms of mannan-binding protein to zymosan activates the proenzyme C1r2C1s2 complex of the classical pathway of complement, without involvement of C1q. J. Immunol., 144, 2287–2294. [PubMed] [Google Scholar]
- Matsushita M., Endo,Y. and Fujita,T. (1998) MASP1 (MBL-associated serine protease 1). Immunobiol., 199, 340–347. [DOI] [PubMed] [Google Scholar]
- Matsushita M., Thiel,S., Jensenius,J.C., Terai,I. and Fujita,T. (2000) Proteolytic activities of two types of mannose-binding lectin-associated serine protease. J. Immunol., 165, 2637–2642. [DOI] [PubMed] [Google Scholar]
- Ng K.K.-S. and Weis,W.I. (1998) Coupling of prolyl peptide bond isomerization and Ca2+ binding in a C-type mannose-binding protein. Biochemistry, 37, 17977–17989. [DOI] [PubMed] [Google Scholar]
- Ng K.K.-S., Park-Snyder,S. and Weis,W.I. (1998) Ca2+-dependent structural changes in C-type mannose-binding proteins. Biochemistry, 37, 17965–17976. [DOI] [PubMed] [Google Scholar]
- Rao Z., Handford,P., Mayhew,M., Knott,V., Brownlee,G.G. and Stuart,D. (1995) The structure of a Ca2+-binding epidermal growth factor-like domain: its role in protein–protein interactions. Cell, 82, 131–141. [DOI] [PubMed] [Google Scholar]
- Resnick D., Chatterton,J.E., Schwartz,K., Slayter,H. and Krieger,M. (1996) Structures of class A scavenger receptors. J. Biol. Chem., 271, 26924–26930. [DOI] [PubMed] [Google Scholar]
- Romao M.J., Kölln,I., Dias,J.M., Carvalho,A.L., Romero,A., Varela,P.F., Sanz,L., Töpfer-Petersen,E. and Calvete,J.J. (1997) Crystal structure of acidic seminal fluid protein (aSFP) at 1.9 Å resolution: a bovine polypeptide of the spermadhesin family. J. Mol. Biol., 274, 650–660. [DOI] [PubMed] [Google Scholar]
- Romero A., Romao,J.J., Varela,P.F., Kollin,I., Dias,J.M., Carvalho,A.L., Sanz,L., Topfer-Petersen,E. and Calvete,J.J. (1997) The crystal structures of two spermadhesins reveal the CUB domain fold. Nat. Struct. Biol., 4, 783–788. [DOI] [PubMed] [Google Scholar]
- Rossi V., Cseh,S., Bally,I., Thielens,N.M., Jensenius,J.C. and Arlaud,G.J. (2001) Substrate specificities of recombinant mannan-binding lectin-associated serine proteases-1 and -2. J. Biol. Chem., 276, 40880–40887. [DOI] [PubMed] [Google Scholar]
- Schumaker V.N., Poon,P.H., Seegan,G.W. and Smith,C.A. (1981) Semi-flexible joint in the C1q subunit of the first component of human complement. J. Mol. Biol., 148, 191–197. [DOI] [PubMed] [Google Scholar]
- Schumaker V.N., Hanson,D.C., Kilchherr,E., Phillips,M.L. and Poon,P.H. (1986) A molecular mechanism for the activation of the first component of complement by immune complexes. Mol. Immunol., 23, 557–565. [DOI] [PubMed] [Google Scholar]
- Stafford W.D. (1992) Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal. Biochem., 203, 295–301. [DOI] [PubMed] [Google Scholar]
- Stenflo J., Stenberg,Y. and Muryani,A. (2000) Calcium-binding EGF-like modules in coagulation proteinases: function of the calcium ion in module interactions. Biochim. Biophys. Acta, 1477, 51–63. [DOI] [PubMed] [Google Scholar]
- Strang C.J., Siegel,R.C., Phillips,M.L., Poon,P.H. and Schumaker,V.N. (1982) Ultrastructure of the first component of human complement: electron microscopy of the crosslinked complex. Proc. Natl Acad. Sci. USA, 79, 586–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y., Takada,F., Takahashi,A. and Kawakami,M. (1994) A 100-kDa protein in the C4-activating component of Ra-reactive factor is a new serine protease having module organization similar to C1r and C1s. J. Immunol., 152, 2308–2316. [PubMed] [Google Scholar]
- Taylor M.E. and Summerfield,J.A. (1987) Carbohydrate-binding proteins of human serum: isolation of two mannose/fucose-specific lectins. Biochim. Biophys. Acta, 915, 60–67. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated structure solution for MIR and MAD. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel S. et al. (1997) A second serine protease associated with mannan-binding lectin that activates complement. Nature, 386, 506–510. [DOI] [PubMed] [Google Scholar]
- Thielens N.M., Enrie,K., Lacroix,M., Jaquinod,M., Hernandez,J.-F., Esser,A.F. and Arlaud,G.J. (1999) The N-terminal CUB-epidermal growth factor module pair of human complement protease C1r binds Ca2+ with high affinity and mediates Ca2+-dependent interaction with C1s. J. Biol. Chem., 274, 9149–9159. [DOI] [PubMed] [Google Scholar]
- Thielens N.M., Cseh,S., Thiel,S., Vorup-Jensen,T., Rossi,V., Jensenius,J.C. and Arlaud,G.J. (2001) Interaction properties of human mannan-binding lectin (MBL)-associated serine proteases-1 and -2, MBP-associated protein 19 and MBL. J. Immunol., 166, 5068–5077. [DOI] [PubMed] [Google Scholar]
- Tsien R.Y. (1980) New calcium indicators and buffers with high selectivity against magnesium and protons: design, synthesis and properties of prototype structures. Biochemistry, 19, 2396–2404. [DOI] [PubMed] [Google Scholar]
- Varela P.F., Romero,A., Sanz,L., Romao,M.J., Töpfer-Petersen,E. and Calvete,J.J. (1997) The 2.4 Å resolution crystal structure of boar seminal plasma PSP-I/PSP-II: a zona pellucida-binding glycoprotein heterodimer of the spermadhesin family built by a CUB domain architecture. J. Mol. Biol., 274, 635–649. [DOI] [PubMed] [Google Scholar]
- Villiers C.L., Arlaud,G.J. and Colomb,M.G. (1985) Domain structure and associated functions of subcomponents C1r and C1s of the first component of human complement. Proc. Natl Acad. Sci. USA, 82, 4477–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorup-Jensen T. et al. (2000) Distinct pathways of mannan-binding lectin (MBL)- and C1-complex autoactivation revealed by reconstitution of MBL with recombinant MBL-associated serine protease-2. J. Immunol., 165, 2093–2100. [DOI] [PubMed] [Google Scholar]
- Wallis R. and Dodd,R.B. (2000) Interaction of mannose-binding protein with associated serine proteases. J. Biol. Chem., 275, 30962–30969. [DOI] [PubMed] [Google Scholar]
- Wallis R. and Drickamer,K. (1999) Molecular determinants of oligomer formation and complement fixation in mannose-binding proteins. J. Biol. Chem., 274, 3580–3589. [DOI] [PubMed] [Google Scholar]
- Weiss V., Fauser,C. and Engel,J. (1986) Functional model of subcomponent C1 of human complement. J. Mol. Biol., 189, 573–581. [DOI] [PubMed] [Google Scholar]
- Weis W.I. and Drickamer,K. (1994) Trimeric structure of a C-type mannose-binding protein. Structure, 2, 1227–1240. [DOI] [PubMed] [Google Scholar]
- Weis W.I., Crichlow,G.V., Murthy,H.M.K., Hendrickson,W.A. and Drickamer,K. (1991) Physical characterization and crystallization of the carbohydrate-recognition domain of a mannose-binding protein from rat. J. Biol. Chem., 266, 20678–20686. [PubMed] [Google Scholar]
- Weis W.I., Taylor,M.E. and Drickamer,K. (1998) The C-type lectin superfamily in the immune system. Immunol. Rev., 163, 19–34. [DOI] [PubMed] [Google Scholar]
- Wiles A.P., Shaw,G., Bright,J., Perczel,A., Campbell,I.D. and Barlow,P.N. (1997) NMR studies of a viral protein that mimics the regulators of complement activation. J. Mol. Biol., 272, 253–265. [DOI] [PubMed] [Google Scholar]