Abstract
The parasitic Nematomorph hairworm, Spinochordodes tellinii (Camerano) develops inside the terrestrial grasshopper, Meconema thalassinum (De Geer) (Orthoptera: Tettigoniidae), changing the insect's responses to water. The resulting aberrant behaviour makes infected insects more likely to jump into an aquatic environment where the adult parasite reproduces. We used proteomics tools (i.e. two-dimensional gel electrophoresis (2-DE), computer assisted comparative analysis of host and parasite protein spots and MALDI-TOF mass spectrometry) to identify these proteins and to explore the mechanisms underlying this subtle behavioural modification. We characterized simultaneously the host (brain) and the parasite proteomes at three stages of the manipulative process, i.e. before, during and after manipulation. For the host, there was a differential proteomic expression in relation to different effects such as the circadian cycle, the parasitic status, the manipulative period itself, and worm emergence. For the parasite, a differential proteomics expression allowed characterization of the parasitic and the free-living stages, the manipulative period and the emergence of the worm from the host. The findings suggest that the adult worm alters the normal functions of the grasshopper's central nervous system (CNS) by producing certain ‘effective’ molecules. In addition, in the brain of manipulated insects, there was found to be a differential expression of proteins specifically linked to neurotransmitter activities. The evidence obtained also suggested that the parasite produces molecules from the family Wnt acting directly on the development of the CNS. These proteins show important similarities with those known in other insects, suggesting a case of molecular mimicry. Finally, we found many proteins in the host's CNS as well as in the parasite for which the function(s) are still unknown in the published literature (www) protein databases. These results support the hypothesis that host behavioural changes are mediated by a mix of direct and indirect chemical manipulation.
Keywords: extended phenotype, parasite–host systems, parasite manipulation, proteomics
1. Introduction
It is now well established that parasites of all kinds modify the behaviour of their hosts in ways that seem to improve the parasite's chances of completing its life cycle (Poulin 1998; Poulin & Thomas 1999; Combes 2001; Moore 2002; Thomas et al. 2005). These changes enhance host-to-host transmission, ensure the parasite or its propagules are released in an appropriate location, or increase parasite survival. From an evolutionary point of view, these behavioural alterations are classically seen as compelling illustrations of the ‘extended phenotype’ concept as proposed by Dawkins (1982), in which genes in one organism (i.e. the parasite) have phenotypic effects on another organism (i.e. the host). Although there are many impressive examples of host manipulation by parasites (see Moore 2002 and Thomas et al. 2005 for recent reviews), little is known concerning the host–parasite molecular ‘cross-talk’ during the manipulative process. Thus, the proximate mechanisms underlying this intriguing parasitic strategy remain generally poorly understood (Helluy & Holmes 1990; Adamo 1997; Beckage 1997; Adamo & Shoemaker 2000; Overly et al. 2001, Webster 2001; Thomas et al. 2002a; Helluy & Thomas 2003; Klein 2003; Williams et al. 2004).
A recently documented case of the influence of a parasite on host behaviour is that of hairworms (Nematomorpha) infecting insects of the Order Orthoptera (grasshoppers and crickets). As juveniles, nematomorphs are mostly parasites of terrestrial arthropods but become free-living adult worms in aquatic environments such as rivers, streams and lakes (Schmidt-Rhaesa 1997, 2001) where they mate and produce eggs. Hairworms must thus make two critical transitions during their life cycle. The first is that from aquatic larva to the terrestrial definitive host, the second from the definitive host to water. The first transition occurs when hosts ingest parasitic larvae directly or indirectly through a paratenic host (Hanelt & Janovy 1999; Ponton et al., unpublished data). During development, the initially microscopic larvae grow to become very large worms whose size exceeds that of the host by a considerable amount (3–4 times). Because the normal host species are terrestrial arthropods, the second transition is a challenging task for most hairworm species. It has been recently shown that insects harbouring mature hairworms display a behaviour originally absent from the host's repertoire—that is to say, they seek water and jump into it (Thomas et al. 2002a). The adult worm then emerges from the host and actively leaves it by swimming away, to begin the search for a sexual partner.
The aim of the present study was to improve our understanding of the proximate cause(s) of this behavioural manipulation by simultaneously examining the proteome of the host and that of the parasite at three strategic moments of the manipulative process, i.e. before, during and after the host jumps into water. The study of the proteome with two key technologies of proteomics—two-dimensional gel electrophoresis (2-DE) and mass spectrometry—can provide a rapid and comprehensive view of the expression of entire genomes. The study of all proteins encoded by the genome of parasites and hosts using proteomics, or ‘parasitoproteomics’ is being used to investigate global protein synthesis and gene expression (Biron et al. 2005c). Thus, proteomics offers an excellent way to examine the host genome in action, through the evaluation of the host proteome during the host–parasite interaction process. By permitting the study of the host and the parasite in action during the manipulative process, proteomics a priori offers an excellent tool with which to explore the proximate mechanisms responsible for host manipulation (Biron et al. 2005a). Here, we performed such an approach and involving one of the most common insect–hairworm systems of Southern France, the long-horned grasshopper (=oak bush-cricket or drumming katydid), Meconema thalassinum (De Geer 1773) (Orthoptera: Tettigoniidae) parasitized by the hairworm, Spinochordodes tellinii (Camerano 1888) (Nematomorpha: Spinochordodidae).
2. Material and methods
(a) Experimental protocol for the analysis of the host and parasite proteomes
Previous field experiments had shown that water-seeking behaviour exists in M. thalassinum harbouring mature individuals of S. tellinii (Thomas et al. 2002a). Because in this system, the parasite is very big, it is easy to separate the host and the parasite, thereby allowing the simultaneous study of both their proteomes without the risk of contamination. We analysed the proteomics data gained using a holistic and integrative approach. First, the electrophoretic gels of grasshopper and hairworm proteins were classified using heuristic cluster analysis in order to identify grasshopper and hairworm categories displaying similar proteome expression. Second, in order to identify proteins linked to the manipulative process, qualitative (presence/absence (break-even point of detection)) analysis was performed on gels of both host and parasite proteins.
(b) Sampling
M. thalassinum infected by S. tellinii were captured nocturnally (between 22.00 and 01.00 h) in July and August 2002 around a swimming pool (15 m×10 m) in Avènes les Bains, southern France, about 70 km north of Montpellier, as described by Thomas et al. (2002a). The swimming pool was located near a forest where adult hairworms were commonly found during the summer. Between swimming pool and the forest, a concrete area 5 m wide allowed the direct observation and capture of arriving infected grasshoppers (Thomas et al. 2002a). To avoid the possible effects of multiple infection, or host- and/or parasite sex specific factors on the proteomic expressions (Thomas et al. 2002b), only male grasshoppers infected with one adult male worm were used for the proteomics analysis. Uninfected individuals were also captured in the forest around the swimming pool. Our sampling procedure distinguished five categories of grasshoppers, all nymphs. The first corresponded to manipulated grasshoppers (DM, ‘during manipulation’), i.e. infected individuals captured between 22.00 and 01.00 h near the edge of the swimming pool just before they jumped into water (Thomas et al. 2002a). As a control for this category, we also collected uninfected grasshoppers at night in the nearby forest, which were termed ‘NC (night control) grasshoppers’. Third, in order to obtain grasshoppers harbouring a mature worm without manipulation, DM category grasshoppers were captured and kept until the following day in a terrarium containing wood and leaves from their natural habitat. These insects were dissected between 01.00 and 03.00 h—that is, during a period at which no behavioural change was observed under natural conditions (at least for M. thalassinum; F. Thomas, unpublished data). Since the behavioural change recurred every night, we termed this third category of host as ‘BM (before manipulation) grasshoppers’. As a control for this category, we also collected uninfected grasshoppers and dissected them the following day (13.00–15.00 h); this fourth category we termed ‘CD (day control) grasshoppers’. Last, we considered hosts that had released their worm. Thus arriving infected insects were visually tracked until they entered the swimming pool. After worm emergence, they were then placed in a dry opaque plastic tumbler for 1 h, whereafter most grasshoppers, which were still vigorous, were then dissected. This fifth category of host were termed ‘AM (after manipulation) grasshoppers’. Individuals of each category were placed separately in a micro-centrifuge tube of 1.5 ml capacity. By collecting hairworms from BM, DM and AM grasshoppers, worms were also collected before (BM*), during (DM*) and after (AM*) manipulation. Worms from grasshopper categories BM and DM were recuperated by dissecting the host's abdomen on a sterile ice bath. All worms were stored at −80 °C prior to electrophoretic testing. AM* hairworms were placed after their emergence in a glass of water for 1 h prior to frozen storage.
(c) Two-dimensional electrophoresis (2-DE)
Water-soluble proteins were extracted as detailed by Biron et al. (2005b). The concentration of each protein sample was estimated by measuring the shift of extinction of Coomasie Blue G-250 at 595 nm (Bradford 1976). The concentration of each sample was standardized at 2 μg μl−1 by the addition of the required volume of homogenizing solution. Protein samples were stored at −70 °C prior to electrophoresis. Two dimensional gels (two-dimensional electrophoresis, 2-DE) were run as detailed by Biron et al. (2005b). At least five IPG strips (Immobiline DryStrip gels; BioRad, USA) of pH 5–8 were run per treatment. Gels were stained using the tetrathionate–silver nitrate technique of Oakley et al. (1980; see also Rabilloud et al. 1994).
(d) Computer analyses
At least three well replicated 2D gels were preserved and used for computer analysis of the various grasshopper and hairworm categories described above. Replicated gels for the same treatment were compared using ImageMaster 2D Platinum Software Version 5.0 (Amersham Biosciences, UK; GENEBIO, Switzerland). The best gels obtained for each category were then used to build a 2D master gel for both grasshopper and hairworm, respectively. The point isoelectric and molecular weight scales of 2D gels were determined using a protein standard kit from BioRad (USA). ImageMaster 2D Platinum was used to compare the proteomic results obtained for both grasshopper and hairworm.
The ImageMaster 2D Platinum software was also used for the comparison between the protein patterns expressed in the host and its manipulative parasite during their daily biochemical interactions. The software takes into account the gel variation in making a ratio for each protein spot based on the sum of ‘volume’ for all proteins observed in each gel. To visualize the global effects of each treatment on the expression of the M. thalassinum and S. tellinii proteomes, we used a heuristic clustering analysis that allowed classification of gels (categories of treatments) into two or more groups, along with determin;ation of the characteristic protein spots of each group, i.e. proteins which were differentially expressed (Appel et al. 1988; Navas et al. 2004; Biron et al. 2005c).
Since it is difficult to determine whether loci are homologous among populations and/or species using 2-DE, the generally employed genetic distance methods could therefore not be employed. Instead, we used the Nei and Li coefficient (Nei & Li 1979) for the heuristic classification: F=2nxy/(nx+ny) where nx and ny are the number of protein spots scored in species x and y, respectively, and where nxy is the total number of protein spots shared by both species x and y. The proteome divergence was computed as a genetic distance according the value of 1−F (Thomas & Singh 1992; Tastet et al. 1999, 2000; Biron et al. 2005c). The proteome distance and the overall protein spots similarity (%vol.) were used to perform heuristic analysis to classify gels of M. thallasinum and S. tellinii (treatments). Consequently, for both grasshopper and hairworm, two distinct dendograms were constructed using the Statistica 5.0 software (Statsoft Inc., Tulsa, OK, USA).
(e) Protein identification by MALDI-TOF mass spectrometry
New gels with the candidate protein spots were run and silver stained as detailed by Shevchenko et al. (1996). Candidate protein spots were excised manually and digested ‘in gel’ using trypsin (sequencing grade, Promega, Madison, WI, USA), as previously described (Shevchenko et al. 1996; Lee et al. 2002). Digest products were completely dehydrated in a vacuum centrifuge and re-suspended in 10 μl formic acid (2% v/v), desalted using Zip Tips C18 (Millipore, Bedford, MA, USA), eluted with 10 μl acetonitrile : trifluoroacetic acid, (60 : 0.1%) and concentrated to a volume of 2 μl. Aliquots (0.3 μl) of this solute were mixed with the same volume of α-cyano-4-hydroxy-trans-cinnamic acid (saturated solution is prepared in acetronile : trifluoroacetic acid, 50 : 0.1%, vortexed, sonicated for 30 s), then micro-centrifuged for 30 s, whereafter a (1/3) dilution of the supernatant was used as the matrix. The mixture was deposited on a 384-well MALDI target using the dry-droplet procedure (Karas & Hillenkamp 1988) and then air dried at room temperature. Analysis was performed using an UltraFlex MALDI-TOF mass spectrometer (Bruker-Franzen Analytik, Bremen, Germany) in a ‘reflectron’ mode with an accelerating voltage of 20 kV and a delayed extraction of 70 ns. Mass spectra were acquired in an automatic mode using the AutoXecute module of Flexcontrol (Bruker-Franzen Analytik, Bremen, Germany).
Spectra were analyzed using the FlexAnalysis software (Bruker-Franzen Analytik) and calibrated internally with the auto-proteolysis peptides of trypsin (m/z 842.51, 1045.56, 2211.10). Peptides were selected in the mass range of 800–4000 Da. Identification of proteins was performed using PeptIdent and Protein Prospector software, respectively (available online). A mass deviation of 50 ppm was allowed for database interrogation. Coverage of the full-length protein exceeding 25% was considered to be sufficient—unless there was some obvious conflict(s) between the experimental molecular weight or isoelectric point and those of the identified protein (Wilkins & Williams 1997; Habermann et al. 2004). Matching peptides with missed cleavages were considered as pertinent only when there were two consecutives basic residues or when arginine and lysine residues were followed by acidic residues along the peptide amino acid sequence. Taking into consideration the possibility of molecular cross-talk between grasshopper and hairworm via the synthesis of mimetic proteins, we performed protein searches with all categories of the host–parasite system (Salzet et al. 2000).
3. Results
(a) Heuristic classification
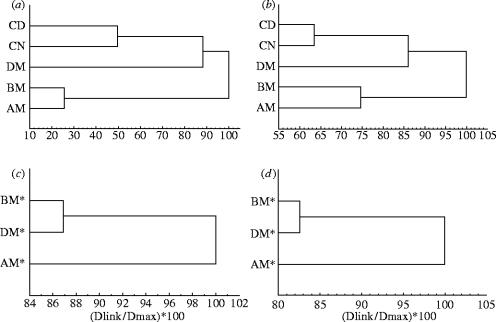
Table 1 shows the number of common protein spots resolved for both M. thalassinum and S. tellinnii, as well as the proteome distances between host and parasite categories. The heuristic cluster based on proteome distances suggests for the host a dendrogram with two groups: the first group contains the two uninfected categories (day control CD and night control CN), the second group two parasitized categories (before, BM, and after, AM, manipulation), whilst the DM category (during manipulation) is separate from both two groups (figure 1a). Similar findings were obtained when considering the relative abundance of total protein spots (figure 1b).
Table 1.
Number of common protein spots (above diagonal) and proteome distances (below diagonal) between M. thalassinum categories also for S. tellinii categories.
| host proteome reaction | |||||
|---|---|---|---|---|---|
| CD | CN | BM | DM | AM | |
| CD | – | 413 | 388 | 368 | 375 |
| CN | 0.062 | – | 402 | 377 | 395 |
| BM | 0.107 | 0.109 | – | 370 | 436 |
| DM | 0.104 | 0.117 | 0.121 | – | 370 |
| AM | 0.148 | 0.135 | 0.032 | 0.132 | – |
| parasite proteome reaction | ||||
|---|---|---|---|---|
| BM* | DM* | AM* | ||
| BM* | – | 614 | 616 | |
| DM* | 0.063 | – | 604 | |
| AM* | 0.074 | 0.071 | – |
Note: Grasshopper categories: day control (CD), night control (CN), before (BM), during (DM) and after manipulation (AM); Hairworm categories: before (BM*), during (DM*) and after (AM*) manipulation.
Figure 1.
Classification of two-dimensional gels resulting from an heuristic analysis of the five grasshopper categories: (a) genetic distance (D), (b) for overall protein spot (presence/absence) and also on the three nematomorph categories: (c) genetic distance (D), (d) for overall protein spots (presence/absence).
For the three hairworm categories, the heuristic clusters based on the proteome distances (figure 1c) and on the relative abundance of overall protein spots observed (figure 1d) reveals a dendrogram with two groups, one containing ‘hairworm in host’ (i.e. before, BM*, and during, DM*, manipulation), the other containing ‘hairworm outside host’ (after manipulation, AM*).
(b) Analysis of 2D-gels
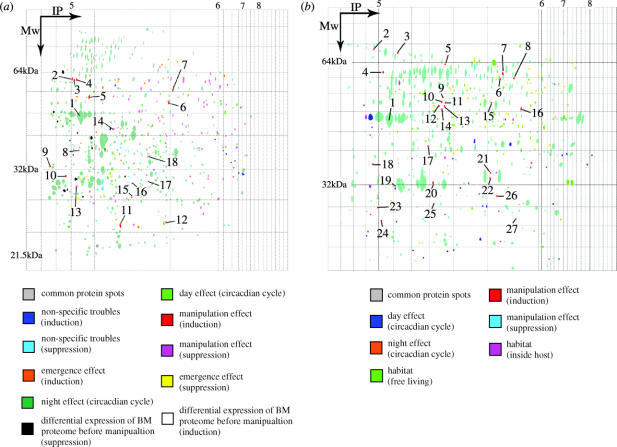
Figure 2 shows the differential grasshopper and hairworm proteome expression at different periods of their interaction and highlights spots that are specific to a subset of categories. Three hundred and forty six protein spots were common to the five grasshopper categories, whilst 220 spots were specific to one or another. Table 2a gives the number of M. thalassinum protein spots present or absent (not detectable (break-even point of detection)) in relation to the different factors characterizing the five categories of host. For instance, we considered that a protein spot was likely to be linked to the manipulative process when its presence or its absence (i.e. not detectable) was specifically observed in DM gels, or in both DM and AM gels. Among the specific protein spots observed, 17.3% were related to a circadian effect, 13.6% to non-specific effects of infection (i.e. common to all parasitized categories), 18.2% to the parasite emergence, 7.7% to the differential expression of BM category before the manipulative process, and 43.2% (i.e. 95) to the manipulation. 592 protein spots were common to the three hairworm categories, whilst 137 were specific (18.8% of the total number of protein spots) (table 2b). Among these specific proteins, 38 (27.7%) were expressed when the host exhibited the water-seeking behaviour.
Figure 2.
(a) Two-dimensional master gel (pH range 5–8) showing the differential daily expression of the head M. thalassinum proteome during its interaction with its manipulative parasite, S. tellinii. (b) Two-dimensional master gel (pH range 5–8) showing the differential daily expression of the proteome of S. tellinii following its manipulative action on its host, M. thalassinum.
Table 2.
Number of M. thalassinum and S. tellinii protein spots present or absent during host–parasite interaction. Numbers in parentheses give the percentage relative to the total number of protein spots.
| (a) host proteome reaction | ||||||||
|---|---|---|---|---|---|---|---|---|
| gels where protein spots occurred | number of protein spots | |||||||
| biological interpretation | CD | CN | BM | DM | AM | |||
| always expressed (common protein spots) | X | X | X | X | X | 346 (61.1) | ||
| circadian cycle | day | present | X | 10 (1.8) | ||||
| X | X | 1 (0.2) | ||||||
| absent | X | X | X | X | 1 (0.2) | |||
| night | present | X | 21 (3.7) | |||||
| X | X | X | 5 (0.9) | |||||
| non-specific effects of infection | present | X | X | X | 8 (1.4) | |||
| absent | X | X | 1 (0.2) | |||||
| X | X | X | 3 (0.5) | |||||
| X | X | 18 (3.1) | ||||||
| manipulation | present | X | 6 (1.1) | |||||
| X | X | 5 (0.9) | ||||||
| absent | X | X | 43 (7.6) | |||||
| X | X | X | X | 24 (4.2) | ||||
| X | X | X | 14 (2.5) | |||||
| X | X | X | 3 (0.5) | |||||
| differential expression of BM proteome before the manipulative process | present | X | 3 (0.5) | |||||
| absent | X | X | X | X | 14 (2.5) | |||
| emergence effect | present | X | 30 (5.3) | |||||
| absent | X | X | 5 (0.9) | |||||
| X | X | X | X | 5 (0.9) | ||||
| total number of specific protein spots | 220 | |||||||
| total number of protein spots | 566 | |||||||
| (b) parasite proteome reaction | |||||
|---|---|---|---|---|---|
| biological interpretation | gels where protein spots occurred | number of protein spots | |||
| BM* | DM* | AM* | |||
| always expressed (common protein spots) | X | X | X | 592 (81.2) | |
| circadian cycle | day | X | 32 (4.4) | ||
| night | X | X | 12 (1.6) | ||
| habitat | inside host (parasite action) | X | X | 22 (3.0) | |
| outside host (free living) | X | 33 (4.5) | |||
| manipulation | induction | X | 14 (1.9) | ||
| suppression | X | X | 24 (3.3) | ||
| total number of specific protein spots | 137 (18.8) | ||||
| total number of protein spots | 729 | ||||
Note: grasshopper categories: day control (CD), night control (CN), before (BM), during (DM) and after manipulation (AM). Hairworm categories: before (BM*), during (DM*) and after (AM*) manipulation.
(c) Identification of candidate proteins
Table 3 summarizes for both host and the parasite the protein spots for which PMF (peptide mass fingerprints) were obtained and for which a differential expression (presence/absence) was observed during the expression of the water-seeking behaviour by the host. Many of the identified proteins expressed in the grasshopper brain (see table 3) belonged to a family of proteins (i.e. Actin; 1, Tubulin; 1, Band_41; 1, Band_7; 1, Folotillin; 1; Ig; 2, Wnt; 1, SNAP-25; 1, SNARE; 1) directly and/or indirectly involved in the development of the central nervous system (CNS) of insects (see table in the Electronic Appendix for more details concerning the known function of each identified protein). In addition, other identified proteins in the grasshopper brain are linked to: the geotactic behaviour (the orientated locomotion of an organism in response to gravity) such as CG31732-PD (isoform D); the control of development of head structures such as Hunchback proteins belonging to zf-C2H2; the ‘6’ family of proteins; are involved in the repair of the damage cause by stress such as CG8863-PA (isoform A) belonging to the DnaJ family of proteins; the biosynthesis of proteins belonging to Ribosomal_L10e; the ‘1’ family of proteins (see table in the Electronic Appendix for more details concerning the known function of each protein). Six proteins—hM, hN, hO, hP, hQ and hR—were only expressed in the brains of manipulated grasshoppers. However, it is impossible to identify and to link these proteins to a known family and they presently remain unknown in all online protein databases (SwissProt, TREMBL, NCBI).
Table 3.
Proteins secreted in the grasshopper CNS and hairworm showing a differential expression during the observation of the abnormal behaviour of the host and for which PMF (peptide mass fingerprints) were obtained (see table S1 in the Electronic Appendix for further details concerning the identification of each candidate protein).
| in grasshopper's CNS | in hairworm | ||
|---|---|---|---|
| assignation number in figure 2a | protein name (protein spot identity; family of protein) | assignation number in figure 2b | protein name (protein spot identity; family of protein) |
| 1 | act2, (hA; Actin; 1) | 1 | actin (pA; 1; actin; 1) |
| 2 | alpha-tubulin (hB; Tubulin; 1) | 2 | hypothetical protein Y49E10.23a (pB; 2; CARD; 1) |
| 3 | CG31732-PD, isoform D (hC; unknown) | 3 | hypothetical protein C47D12.6b (pC; 3; HGTP_anticodon; 1) |
| 4 | Hunchback protein (hD; zf-C2H2; 6) | 4 | heat shock protein 60 (pD; 4; Cpn60_TCP1; 1) |
| 5 | moesin/ezrin/radixin homologue 1 (hE; Band_41; 1) | 5 | tyrosine 3-monooxygenase (pE; 5; biopterin_H; 1) |
| 6 | flotillin-2 (hF; (Band_7; 1, Flotillin; 1)) | 6 | unknown (pF; 6; unknown) |
| 7 | CG8863-PA, isoform A (hG; DnaJ; 1) | 7 | putative acetylcholine regulator unc-18 (pG; 7; Sec1; 1) |
| 8 | neural/ectodermal development factor IMP-L2 [precursor] (hH; Ig; 2) | 8 | intermediate filament protein [fragment] (pH; 8; filament; 1) |
| 9 | wingless [fragment] (hI; Wnt; 1) | 9 | beta-tubulin (pI; 9; (tubulin; 1; tubulin_C; 1)) |
| 10 | synaptosome-associated protein SNAP-25-1 (hJ; (SNAP-25; 1. SNARE; 1)) | 10 | unknown (pJ; 10; unknown) |
| 11 | wingless [fragment] (hK; Wnt; 1) | 11 | guanine nucleotide-binding protein alpha-16 subunit (pK; 11; G-alpha; 1) |
| 12 | similar to Drosophila melanogaster qm [fragment] (hL; Ribosomal_L10e; 1) | 12 | bestrophin 1 (pL; 12; Bestrophin; 1) |
| 13 | unknown (hM; unknown) | 13 | arginine kinase (pM; 13; ATP-gua_Ptrans; 1) |
| 14 | unknown (pN; unknown) | 14 | unknown (pN; 14; unknown) |
| 15 | unknown (pO; unknown) | 15 | hypothetical protein CBG14575 (pO; 15;unknown) |
| 16 | unknown (pP; unknown) | 16 | heat shock protein 60 (pP; 16; Cpn60_TCP1; 1) |
| 17 | unknown (pQ; unknown) | 17 | Wnt5A protein [fragment] (pQ; 17; Wnt; 1) |
| 18 | unknown (pR; unknown) | 18 | hypothetical protein CBG08254 [fragment] (pR; 18; unknown) |
| 19 | DNA binding protein [fragment] (pS; 19; zf-C2H2; 8) | ||
| 20 | Troponin t protein 4, isoform b (pT; 20; Troponin; 1) | ||
| 21 | probable deoxyhypusine synthase (pU; 21; DS; 1) | ||
| 22 | NOA36-like protein (pV; 22; NOA36; 1) | ||
| 23 | unknown (pX; 23; unknown) | ||
| 24 | Wnt-4 protein [fragment] (pY; 24; wnt; 1) | ||
| 25 | hypothetical protein C54D10.10 (pZ; 25; Kunitz_BPTI; 2) | ||
| 26 | binding protein 2 like protein [fragment] (pA1; 26; FKBP_C; 1) | ||
| 27 | hypothetical protein CBG15114 [fragment] (pB1; unknown) | ||
With regard to the hairworm proteome reaction during the expression of the water-seeking behaviour by the host, many of the identified proteins are linked to protein biosynthesis (pC, pD, pP, pA1), to the release and secretion of neurotransmitter (pA, pG), to functions on the CNS (pB, pG, pI, pM, pQ, pY), and to endopeptidase inhibition (pZ), whilst some of these identified proteins have as yet no known function (pL, pO, pR, pB1). Four proteins—pF, pJ, PN and X—were only expressed in the hairworm, but it was impossible to identify these proteins with PMF and to link them to a known family. Again, these parasite proteins are unknown in all available protein databases examined by us to date (see table 1 and table in the Electronic Appendix). We observed that the parasite produced ‘host-like’ proteins, illustrating a case of a molecular mimicry (Salzet et al. 2000; Taylor et al. 2004). More specifically, an overproduction of two proteins (pQ and pY) from the Wnt family acting directly in the development of the CNS was observed (Kalderon 2002; Packard et al. 2003). MALDI-TOF signals suggest that these two proteins are synthesized by hairworms but are mimetic to proteins observed in the class Insecta.
4. Discussion
Examples of behavioural manipulation by parasites are numerous, although the mechanisms underlying these ethological changes are by no means well characterized (Webster 2001; Adamo 2002; Klein 2003; Thomas et al. 2005). The change in host behaviour is usually an indirect effect of the parasite, or a mix of direct and indirect effects of the parasite on its host's CNS (Thompson & Kavaliers 1994; Adamo 2002; Helluy & Thomas 2003; Beckage & Gelman 2004). While most research to elucidate the biochemical mechanisms of parasitic manipulation limits the approach to the quantification of only a few molecules considered in advance as candidates, proteomics makes no assumption on the identity of molecules involved, and thus offers an undoubtedly more powerful alternative to investigate the molecular cross-talk established during the manipulative processes (Thomas et al. 2005; Biron et al. 2005c).
This is the first study to document the differential expression of the proteomes of a parasitic hairworm and its insect host during manipulation. Adult hairworms modulate the behaviour of their host with precise timing and in very subtle ways. Indeed, this behavioural change involves the sudden appearance at night of behaviour originally absent from the host's repertoire (i.e. leaving the natural terrestrial habitat to find and then jump into water) to ensure the continuation of the life cycle of the infecting parasite. The proteomics results clearly show a differential expression of the grasshopper's CNS and of the hairworm's proteomes during the expression of the water seeking behaviour of the host (table 1; figures 1 and 2). Furthermore, the study also shows that proteomics tools are sensitive enough to disentangle proteome alterations linked to factors as various as the circadian cycle, the parasitic status and parasitic emergence (table 1; figures 1 and 2).
What is the purpose of such differential proteome expressions? Although this study aims to directly correlate protein expression with parasite manipulation and host insect behaviour, interestingly, we have found certain protein families which show differential expression at key periods of the manipulative process. Thus for example, six protein families (Band_41; 1, Band_7; 1, Flottilin; 1, Ig; 2, DnaJ; 1, zf-C2H2; 6) were specifically expressed in the CNS of the manipulated grasshopper. These protein families are directly and/or indirectly involved in the proper development of the CNS in this group of insects. This result suggests a reaction of the grasshopper brain in order to inhibit parasite attack on the host CNS during the manipulative processes. During this process, one protein involved in control of geotactic behaviour (CG31732-PD (isoform D)) is expressed only in the host insect's CNS. In addition, S. tellinii secretes two families of proteins linked to the release of neurotransmitters (Sec; 1, Actin; 1) and one family of proteins linked to the regulation of apoptosis (CARD; 1) during the manipulative process. Many parasites demonstrate an ability to modulate host apoptosis pathways to their own advantage (James & Green 2004). Previous results suggest that S. tellinii controls apoptosis within the host's CNS. Thus in the wood cricket–hairworm system Nemobius sylvestris (Bosc d'Antic 1792) (Orthoptera: Gryllidae)–Paragordius tricuspidatus (Dufour 1828) (Nematomorpha: Chordodidae), Thomas et al. (2003) found using a histological approach, a two-fold increase in neurogenesis in the brain of infected crickets but not in the host's CNS in the M. thallassinum–S. tellinii system (unpubl. data). In the vertebrate–manipulative parasite (micro- and macro-parasite) systems, parasites can modify host behaviour by inducing apoptosis within the host's CNS, causing inflammatory immune responses and thereby altering the chemical signals in the brain and CNS (Klein 2003). The present proteomics results are in general agreement with the findings from previous histological studies and with the strategy of parasites employed in vertebrate host–parasite systems. We propose that S. tellinii disturbs the normal functions of the host grasshopper's CNS during the manipulative process leading to aberrant behavioural responses.
One of the most fascinating results of the present study is the higher synthesis in the grasshopper's CNS of two proteins (see table 2, hI and hK) from the Wnt family. Moreover, these two proteins show a higher synthesis during the induction of the water-seeking behaviour of the host and this higher production is correlated with a higher synthesis in the parasite proteome of two proteins (see table 2; pQ and pY) from the Wnt family. The searches in protein databases revealed that Wnt proteins synthesized in the parasite are homologous to, and probably mimetic with, proteins known in the class Insecta. The Wnt proteins identified here (see table S1, Electronic Appendix) play an important role in the development of the CNS. In a proteomics study of a cricket–hairworm system, N. sylvestris–P. tricuspidatus, Biron et al. (submitted) found that P. tricuspidatus can produce potent concentrations of mimetic molecules (e.g. Wnt family) acting directly on the CNS of N. sylvestris in order to alter the host's behaviour. The proteomics results of this study agree with those of that related study. The complete demonstration that the worm is secreting these mimetic proteins into the grasshopper will, however, require us to also study the haemolymph proteome of manipulated hosts.
Zooparasites have elaborated many biochemical strategies for invading hosts, for escaping immune responses and for taking advantage of host growth factors (Vincendeau et al. 2003; James & Green 2004). Hoek et al. (1997) suggested that the avian schistosome, Trichobilharzia ocellata (La Valette 1855) Brumpt, 1931 can regulate the expression of neuropeptide genes from its gastropod host, Lymnaea stagnalis (L.) and thereby elicit physiological and behavioural changes. Nevertheless, there is little proof that manipulative zooparasites can change host behaviour by secreting molecules as proteins or peptides that act directly on the host's CNS. Because of this, it is usually argued that parasites should mainly exploit indirect and less energetically expensive methods to alter host behaviour (Adamo 2002; Thomas et al. 2005). Given the very large size of the adult hairworms S. tellinii and P. tricuspidatus, it is possibly not too expensive for such parasites to produce potent concentrations of mimetic and effective molecules acting directly on the CNS of the arthropod host to alter its behaviour.
In conclusion, the main results of this proteomics study suggest that the adult hairworm, S. tellinii, can alter the behaviour of the grasshopper, M. thallasinum, such that biochemical interactions occur which result in the production of effective molecules which in turn act directly on the functioning of the host's CNS. In this way, the novel water-seeking behaviour of the host should be seen as an ‘extended phenotypic’ effect of S. tellinii genes. The next step will be to study many other arthropod–hairworm systems with proteomics tools. These studies will accelerate knowledge of the manipulative strategies of such parasites and open the way to create, for the first time, protein databases directly relating to the manipulative tactics of parasites. As such then, proteomics tools offer a new prospect for the study of parasitic manipulation for many parasite taxa. Furthermore, such approaches will promote the reconstruction of the molecular phylogeny of proteins—for instance, those involved in parasitic manipulation. The level of conservation of these proteins in a particular parasite genus can be determined over evolutionary time and the convergence of molecular mechanisms studied. It is possible that proteomics studies will contribute to enhancing general knowledge of molecular cross-talk in host–parasite relationships and hence ultimately assist researchers in the quest for new drugs and vaccines.
Acknowledgements
We thank Shelley Adamo from the Department of Psychology, Neuroscience Institute, Dalhousie University for her very helpful comments on this manuscript; the staff at the Proteomics and Robotics Platform of Montpellier Genopole® for their help concerning the mass spectrometry analysis; Mr J. L. Lafaurie and staff at the thermal station of Avènes-Les-Bains for their cooperation during the field study; and Y. Elie and J. L. Fauquier of VB films for making the movie associated with this work (available on request from D. G. Biron and F. Thomas). This work was supported by an ACI ‘jeunes chercheurs’ grant to F. Thomas.
Supplementary Material
Identification of proteins showing a differential expression in the grasshopper CNS and in the hairworm during the observation of the abnormal behaviour of the host.
References
- Adamo S.A. How parasites alter the behaviour of their insect hosts. In: Beckage N.E, editor. Parasites and pathogens: effects on host hormones and behaviour. Chapman & Hall; London: 1997. pp. 231–245. [Google Scholar]
- Adamo S. Modulating the modulators: parasites, neuromodulators and host behavioural change. Brain Behav. Evol. 2002;60:370–377. doi: 10.1159/000067790. 10.1159/000067790 [DOI] [PubMed] [Google Scholar]
- Adamo S.A, Shoemaker K.L. Effects of parasitism on the octopamine content of the central nervous system of Manduca sexta: a possible mechanism underlying host behavioural change. Can. J. Zool. 2000;78:1580–1587. 10.1139/cjz-78-9-1580 [Google Scholar]
- Appel R, Hochstrasser D, Roch C, Funk M, Muller A.F, Pellegrini C. Automatic classification of two-dimensional gel electrophoresis pictures by heuristic clustering analysis: a step forward machine learning. Electrophoresis. 1988;9:136–142. doi: 10.1002/elps.1150090307. 10.1002/elps.1150090307 [DOI] [PubMed] [Google Scholar]
- Beckage N.E. New insights: how parasites and pathogens alter the endocrine physiology and development of insect hosts. In: Beckage N.E, editor. Parasites and pathogens: effects on host hormones and behaviour. Chapman & Hall; London: 1997. pp. 3–36. [Google Scholar]
- Beckage N.E, Gelman D.B. Wasp parasitoid disruption of host development: implications for new biologically based strategies for insect control. Ann. Rev. Entomol. 2004;49:299–330. doi: 10.1146/annurev.ento.49.061802.123324. 10.1146/annurev.ento.49.061802.123324 [DOI] [PubMed] [Google Scholar]
- Biron D.G, Joly C, Galeotti N, Ponton F, Marché L. The proteomics: a new prospect for studying parasitic manipulation. Behav. Process. 2005;68:249–253. doi: 10.1016/j.beproc.2004.08.016. 10.1016/j.beproc.2004.08.016 [DOI] [PubMed] [Google Scholar]
- Biron D.G, Joly C, Marché L, Galéotti N, Calcagno V, Schmidt-Rhaesa A, Renault L, Thomas F. First analysis of the proteome in two nematomorph species, Paragoridus tricuspidatus (Chordodidae) and Spinochordodes tellinii (Spinochordodidae) Inf. Genet. Evol. 2005;5:167–175. doi: 10.1016/j.meegid.2004.09.003. 10.1016/j.meegid.2004.09.003 [DOI] [PubMed] [Google Scholar]
- Biron D.G, Moura H, Marché L, Hughes A.L, Thomas F. Towards a new conceptual approach to ‘parasitoproteomics’. Trends Parasitol. 2005c;21:162–168. doi: 10.1016/j.pt.2005.02.009. 10.1016/j.pt.2005.02.009 [DOI] [PubMed] [Google Scholar]
- Biron, D. G. et al Submitted. Differential expression of the proteomes of a host and its manipulative parasite at different stages of their biochemical interaction.
- Bradford M.M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Combes C. The ecology and evolution of intimate interactions. The University of Chicago Press; 2001. Parasitism. [Google Scholar]
- Dawkins R. Oxford University Press; Oxford: 1982. The extended phenotype. [Google Scholar]
- Habermann G, Oegema J, Sunyaev J, Shevchenko A. The power and the limitations of cross-species protein identification by mass spectrometry driven sequence similarity searches. Mol. Cell Proteomics. 2004;3:238–249. doi: 10.1074/mcp.M300073-MCP200. 10.1074/mcp.M300073-MCP200 [DOI] [PubMed] [Google Scholar]
- Hanelt B, Janovy J. The life cycle of a horsehair worm, Gordius robustus (Nematomorpha: Gordioidea) J. Parasitol. 1999;85:139–142. [PubMed] [Google Scholar]
- Helluy S, Holmes J.C. Serotonin, octopamine, and the clinging behaviour induced by the parasite Polymorpha paradoxus (Acanthocephala) in Gammarus lacustris (Crustacea) Can. J. Zool. 1990;8:1214–1220. [Google Scholar]
- Helluy S, Thomas F. Effects of Microphallus papillorobustus (Platyheminthes: Trematode) on serotonergic immunoreactivity and neuronal architecture in the brain of Gammarus insensibilis (Crustacea: Amphipoda) Proc. R. Soc. B. 2003;270:563–568. doi: 10.1098/rspb.2002.2264. 10.1098/rspb.2002.2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek R.M, Van Kesteren R.E, Smit A.B, De Jong-Brink M, Geraerts P.M. Altered gene expression in the host brain caused by a trematode parasite: neuropeptide genes are preferentially affected during parasitosis. Proc. Natl Acad. Sci. USA. 1997;94:14 072–14 076. doi: 10.1073/pnas.94.25.14072. 10.1073/pnas.94.25.14072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James E.R, Green D.R. Manipulation of apoptosis in the host–parasite interaction. Trends Parasitol. 2004;20:280–287. doi: 10.1016/j.pt.2004.04.004. 10.1016/j.pt.2004.04.004 [DOI] [PubMed] [Google Scholar]
- Kalderon D. Similarities between the Hedgehog and Wnt signalling pathways. Trends Cell. Biol. 2002;12:523–531. doi: 10.1016/s0962-8924(02)02388-7. 10.1016/S0962-8924(02)02388-7 [DOI] [PubMed] [Google Scholar]
- Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. 10.1021/ac00171a028 [DOI] [PubMed] [Google Scholar]
- Klein S.L. Parasite manipulation of the proximate mechanisms that mediate social behaviour in vertebrates. Physiol. Behav. 2003;79:441–449. doi: 10.1016/s0031-9384(03)00163-x. 10.1016/S0031-9384(03)00163-X [DOI] [PubMed] [Google Scholar]
- Lee K, Bae D, Lim D. Evaluation of parameters in peptide mass fingerprinting for protein identification by MALDI-TOF mass spectrometry. Mol. Cells. 2002;13:175–184. [PubMed] [Google Scholar]
- Moore J. Oxford series in ecology and evolution. Oxford University Press; London: 2002. Parasites and the behaviour of animals. [Google Scholar]
- Navas A, Albar J.P. Application of proteomics in phylogenetic and evolutionary studies. Proteomics. 2004;4:299–302. doi: 10.1002/pmic.200300603. 10.1002/pmic.200300603 [DOI] [PubMed] [Google Scholar]
- Nei M, Li W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl Acad. Sci. USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley B.R, Kirsch B.R, Moris N.R. A simplified ultrasensitive silver stain for detecting protein polyacrylamide gels. Anal. Biochem. 1980;105:361–363. doi: 10.1016/0003-2697(80)90470-4. 10.1016/0003-2697(80)90470-4 [DOI] [PubMed] [Google Scholar]
- Overli O, Pall M, Borg B, Jobling M, Winberg S. Effects of Schistocephalus solidus infection on brain monoaminergic activity in female three-spined sticklebacks Gasterosteus aculeatus. Proc. R. Soc. B. 2001;268:1411–1415. doi: 10.1098/rspb.2001.1668. 10.1098/rspb.2001.1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard M, Mathew D, Budnik V. Wnts and TGF beta in synaptogenesis: old friends signalling at new places. Nat. Rev. Neurosci. 2003;4:113–120. doi: 10.1038/nrn1036. 10.1038/nrn1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin R. Chapman & Hall; New York: 1998. Evolutionary ecology of parasites: from individuals to communities. [Google Scholar]
- Poulin R, Thomas F. Phenotypic variability induced by parasites. Parasitol. Today. 1999;15:28–32. doi: 10.1016/s0169-4758(98)01357-x. 10.1016/S0169-4758(98)01357-X [DOI] [PubMed] [Google Scholar]
- Rabilloud T, Vuillard L, Gilly C, Lawrence J.J. Silver-staining of proteins in polyacrylamide gels: a general overview. Cell. Mol. Biol. 1994;40:57–75. [PubMed] [Google Scholar]
- Salzet M, Capron A, Stefano G.B. Molecular crosstalk in host–parasite relationships: schistosome- and leech–host interactions. Parasitol. Today. 2000;16:536–540. doi: 10.1016/s0169-4758(00)01787-7. 10.1016/S0169-4758(00)01787-7 [DOI] [PubMed] [Google Scholar]
- Schmidt-Rhaesa A. Nematomorpha. In: Schwoerbel J, Zwick P, editors. Subwasserfauna Mitteleuropas. Fischer-Verlag; Stuttgart: 1997. pp. 1–124. [Google Scholar]
- Schmidt-Rhaesa A. The life cycle of horsehair worms (Nematomorpha) Acta Parasitol. 2001;46:151–158. [Google Scholar]
- Shevechenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. 10.1021/ac950914h [DOI] [PubMed] [Google Scholar]
- Tastet C, Bossis M, Gauthier J.P, Renault L, Mugniéry D. Meloidogyne chitwoodi and M. fallax protein variation assessed by two-dimensional electrophoregram computed analysis. Nematology. 1999;1:301–314. 10.1163/156854199508171 [Google Scholar]
- Tastet C, Bossis M, Renault L, Mugniéry D. Protein variation in tropical Meloidogyne spp. as shown by two-dimensional electrophoregram computed analysis. Nematology. 2000;2:343–353. 10.1163/156854100509105 [Google Scholar]
- Taylor J.E, Hatcher P.E, Paul N.D. Crosstalk between plant responses to pathogens and herbivores: a view from the outside in. J. Exp. Biol. 2004;55:159–168. doi: 10.1093/jxb/erh053. [DOI] [PubMed] [Google Scholar]
- Thomas S, Singh R.S. A comprehensive study of genic variation in natural populations of Drosophila melanogaster. VII. Varying rates of genic divergence as revealed by two-dimensional electrophoresis. Mol. Biol. Evol. 1992;9:507–525. doi: 10.1093/oxfordjournals.molbev.a040738. [DOI] [PubMed] [Google Scholar]
- Thomas F, Schmidt-Rhaesa A, Martin G, Manu C, Durand P, Renaud F. Do hairworms (Nematomorpha) manipulate the water-seeking behaviour of their terrestrial hosts? J. Evol. Biol. 2002;15:356–361. 10.1046/j.1420-9101.2002.00410.x [Google Scholar]
- Thomas F, Brown S.P, Sukhedo M, Renaud F. Understanding parasite strategies: a state-dependent approach? Trends Parasitol. 2002;18:387–390. doi: 10.1016/s1471-4922(02)02339-5. 10.1016/S1471-4922(02)02339-5 [DOI] [PubMed] [Google Scholar]
- Thomas F, Ulitsky P, Augier R, Dusticier N, Samuel D, Strambi C, Biron D.G, Cayre M. Biochemical and histological changes in the brain of the cricket Nemobius sylvestris infected by the manipulative parasite Paragordius tricuspidatus (Nematomorpha) Int. J. Parasitol. 2003;33:435–443. doi: 10.1016/s0020-7519(03)00014-6. 10.1016/S0020-7519(03)00014-6 [DOI] [PubMed] [Google Scholar]
- Thomas F, Adamo S, Moore J. Parasitic manipulation: where are we and where should we go? Behav. Process. 2005;68:185–199. doi: 10.1016/j.beproc.2004.06.010. 10.1016/j.beproc.2004.06.010 [DOI] [PubMed] [Google Scholar]
- Thompson S.N, Kavaliers M. Physiological bases for parasite-induced alterations of host behaviour. Parasitology. 1994;109:S119–S138. doi: 10.1017/s0031182000085139. [DOI] [PubMed] [Google Scholar]
- Vincendeau P, Gobert A.P, Daulouède S, Moynet D, Mossalayi M.D. Arginases in parasitic diseases. Trends Parasitol. 2003;19:9–12. doi: 10.1016/s1471-4922(02)00010-7. 10.1016/S1471-4922(02)00010-7 [DOI] [PubMed] [Google Scholar]
- Webster J.P. Rats, cats, people and parasites: the impact of latent toxoplasmosis on behaviour. Microbes Infect. 2001;3:1037–1045. doi: 10.1016/s1286-4579(01)01459-9. 10.1016/S1286-4579(01)01459-9 [DOI] [PubMed] [Google Scholar]
- Wilkins M.R, Williams K.L. Cross-species protein identification using amino acid composition, peptide mass fingerprinting, isoelectric point and molecular mass: a theoretical evaluation. J. Theor. Biol. 1997;186:7–15. doi: 10.1006/jtbi.1996.0346. 10.1006/jtbi.1996.0346 [DOI] [PubMed] [Google Scholar]
- Williams C.M, Poulin R, Sinclair B.J. Increased haemolymph osmolality suggests a new route for behavioural manipulation of Talorchestia quoyana (Amphipoda: Talitridae) by its mermithid parasite. Funct. Ecol. 2004;18:685–691. 10.1111/j.0269-8463.2004.00910.x [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of proteins showing a differential expression in the grasshopper CNS and in the hairworm during the observation of the abnormal behaviour of the host.