Abstract
Mutualisms are balanced antagonistic interactions where both species gain a net benefit. Because mutualisms generate resources, they can be exploited by individuals that reap the benefits of the interaction without paying any cost. The presence of such ‘cheaters’ may have important consequences, yet we are only beginning to understand how cheaters evolve from mutualists and how their evolution may be curtailed within mutualistic lineages. The yucca–yucca moth pollination mutualism is an excellent model in this context as there have been two origins of cheating from within the yucca moth lineage. We used nuclear and mitochondrial DNA markers to examine genetic structure in a moth population where a cheater species is parapatric with a resident pollinator. The results revealed extensive hybridization between pollinators and cheaters. Hybrids were genetically intermediate to parental populations, even though all individuals in this population had a pollinator phenotype. The results suggest that mutualisms can be stable in the face of introgression of cheater genes and that the ability of cheaters to invade a given mutualism may be more limited than previously appreciated.
Keywords: obligate pollination mutualism, hybridization, cheater, exploitation
1. Introduction
Mutualists trade a variety of commodities such as protection, food, and dispersal services in return for a service or resource that is difficult or impossible for mutualists to obtain otherwise (Noë & Hammerstein 1994; Schwartz & Hoeksema 1998). The presence of these commodities also creates ample opportunity for individuals to exploit mutualisms by taking resources without providing services in return. These ‘cheaters’ may be phylogenetically unrelated opportunists from the community or they may evolve from within the mutualist lineage itself. In the latter case, cheating has been suggested to cause breakdown of the mutualism unless there are regulatory mechanisms that prevent the shift to pure parasitism (Trivers 1971; Axelrod & Hamilton 1981; Murray 1985; Bull & Rice 1991; Doebeli & Knowlton 1998; Herre et al. 1999; Weiblen et al. 2001; West et al. 2002; Holland et al. 2004). Even in the absence of such mechanisms, however, theoretical models demonstrate that under some ecological circumstances, cheaters can evolve from mutualists and coexist on the auspices of their mutualist relatives (Hochberg et al. 2000; Law et al. 2001; Ferrière et al. 2002; Morris et al. 2003). These models are revealing the circumstances leading to the stable evolution of exploitation from within mutualistic lineages and have helped to explain the presence of cheater lineages in extant mutualisms such as ants and acacias (Janzen 1975), figs and fig wasps (West & Herre 1994; West et al. 1996), and yuccas and yucca moths (Addicott 1996; Pellmyr et al. 1996). One of the keys to understanding the dynamics of interspecific interactions, then, is to determine the factors causing shifts in the outcome of an interaction along the continuum between antagonism and mutualism.
Two cheater species have evolved from mutualists within the well known obligate pollination mutualism between yuccas and yucca moths (Pellmyr et al. 1996). Yuccas are pollinated exclusively by yucca moths, and the moth larvae only consume yucca seeds (Riley 1892; Powell 1992). The female pollinator lays her eggs into yucca flowers and then actively pollinates using unique tentacular mouthparts. The mutualism derives from the larvae only consuming a small fraction of the developing seeds. In contrast, the cheater moths have lost the tentacles used for pollination, emerge later than the pollinators, lay their eggs directly into fruit, and can inflict heavy costs in host populations in terms of seed consumption (Pellmyr et al. 1996). Although both pollinator and cheater larvae feed on a subset of developing yucca seeds, the cheaters depend completely on the actions of their pollinating relatives for reproduction. The cheater species have invaded many yucca–yucca moth associations (Pellmyr 1999).
For one of these cheater species, Tegeticula intermedia, ecological and phylogeographic studies are revealing the origin and circumstances leading to mutualism reversal (Pellmyr & Leebens-Mack 2000; Segraves & Pellmyr 2004). While conducting a phylogeographic study of T. intermedia and two of its potential pollinator ancestors, we discovered a peculiar pattern in the phylogenetic relationships of mitochondrial DNA (mtDNA) haplotypes from pollinator moths that use Yucca elata in the Big Bend region of western Texas (United States). This pollinator, Tegeticula elatella, has a parapatric distribution with the cheaters in western Texas (figure 1). Five of six pollinators collected from Big Bend National Park, Texas had mtDNA haplotypes phylogenetically nested within the cheater T. intermedia. In contrast, the sixth moth nested with T. elatella collected elsewhere in the host range and differed from T. intermedia by 2.6% sequence divergence, a level consistent with the differences found among yucca moth species (Pellmyr & Leebens-Mack 1999). Interestingly, all individuals collected from Big Bend, irrespective of mtDNA haplotype, had a pollinator phenotype. Females possessed the tentacles necessary for pollination and carried pollen, indicative of the behavioural capacity for pollination.
Figure 1.
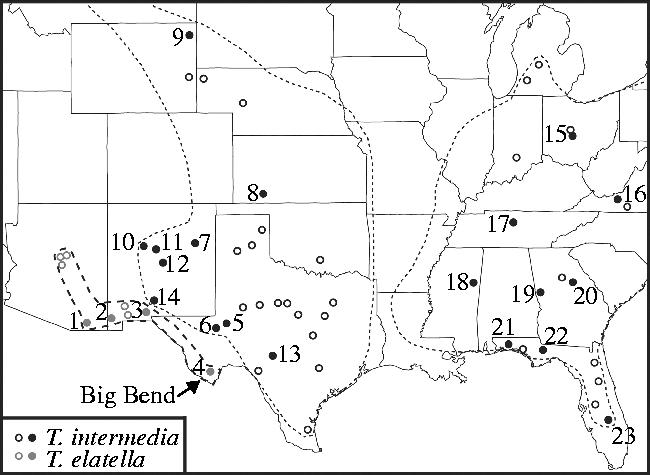
Distribution of the pollinator Tegeticula elatella and the cheater T. intermedia in the United States. Pollinators are shown in grey circles and cheaters are shown in black. Site localities are solid circles, and open circles represent known localities that were not included in the present study. The numbers correspond to the site information presented in table 1. The fine dotted line represents a rough approximation of the cheater's host range (all host species combined), and the coarse dotted line is the host range for T. elatella (Y. elata). There are no known cheater populations or T. intermedia hosts in the area between sites 4 and 6.
This pattern of phenotypic and genetic relationships suggested that gene flow between cheater and pollinator lineages may explain the mtDNA patterns. We tested this hypothesis by using a combination of mtDNA sequences and amplified fragment length polymorphism (AFLP) markers to compare the mitochondrial and nuclear genomes of ‘pure’ T. elatella pollinators, pure T. intermedia cheaters, and individuals collected from the Big Bend population.
2. Material and methods
(a) Natural history
A series of phylogenetic analyses have established that the cheater yucca moths have arisen from mutualistic ancestors and that there are at least two independent origins of cheater moths (Pellmyr et al. 1996; Pellmyr & Leebens-Mack 1999, 2000). These phylogenetic studies have also revealed the closest relative for one of the cheaters, T. intermedia. This cheater species has a broad geographic range and feeds on four species of yuccas (Yucca filamentosa, Yucca glauca, Yucca constricta and Yucca baileyi) that are pollinated by three species of pollinator moths. In the southeastern United States, T. intermedia co-occurs with its pollinating sister species, Tegeticula cassandra, and these two species are similar in most regards with the exception of the behavioural and morphological modifications required for pollinator and cheater life habits. For example, both T. intermedia and T. cassandra have similar genitalia morphology and oviposit superficially in the plant tissue. In other parts of its range, T. intermedia co-occurs with the pollinators Tegeticula yuccasella and Tegeticula altiplanella. These two pollinator species differ in genitalia morphology from T. intermedia, and rather than depositing their eggs superficially, they pierce through the floral tissue to lay eggs next to the ovules.
Although T. cassandra is currently geographically restricted to the southeastern United States, a recent phylogeographic study showed that T. intermedia originated a long time ago in the western United States and has only more recently spread eastward (Segraves & Pellmyr 2004). There are no known populations of T. cassandra in the western United States, suggesting that the pollinators once had a much more extensive geographic range. As T. intermedia migrated eastward, secondary contact with T. cassandra in the southeastern United States may have resulted in hybridization between these two species (Segraves & Pellmyr 2004). All purported instances of hybridization indicated gene flow from T. intermedia to T. cassandra with no evidence for bidirectional introgression. In this case, hybridization occurred between two close relatives (∼1.3% uncorrected sequence divergence). In contrast, there is no evidence for hybridization between T. intermedia and its two other co-occurring pollinator species even though these moths are members of a recent and rapid radiation ranging on the order of approximately 2–2.5% uncorrected sequence divergence among most pairs of species.
In an effort to find an extant pollinator relative in the western United States, we identified T. elatella as a potential candidate because of its geographic range and similarity in oviposition habit to T. intermedia. Tegeticula elatella also lays eggs superficially within yucca flowers, and has similar genitalia morphology to both T. intermedia and T. cassandra. These factors suggested the possibility that the currently circumscribed species of T. elatella may harbour a cryptic species that is the closest ancestor to the cheater T. intermedia (table 1).
Table 1.
Site localities, host information and sample sizes for the molecular analysis of Tegeticula elatella and T. intermedia.
| species | host | location | longitude/latitude | N (mtDNA) | N (AFLP) |
|---|---|---|---|---|---|
| T. elatella | Y. elata | Sierra Vista, AZ (1) | 31°33′16″N, 110°18′11″W | 4 | 7 |
| Rodeo, NM (2) | 31°50′07″N, 109°01′50″W | 0 | 11 | ||
| Jornada LTER, NM (3) | 33°56′00″N, 106°34′00″W | 15 | 14 | ||
| Big Bend Natl Park, TX (4) | 29°15′00″N, 103°15′00″W | 50 | 31 | ||
| T. intermedia (Western US) | Y. glauca | 11 km SW Odessa, TX (5) | 31°50′44″N, 102°17′51″W | 4 | 2 |
| Royalty, TX (6) | 31°22′20″N, 102°52′00″W | 5 | 5 | ||
| Cuervo, NM (7) | 35°01′52″N, 104°24′29″W | 5 | 6 | ||
| Fowler, KS (8) | 37°23′08″N, 100°11′43″W | 6 | 4 | ||
| Sundance, WY (9) | 44°24′23″N, 104°22′31″W | 1 | 1 | ||
| Y. baileyi | Correo, NM (10) | 34°57′18″N, 107°11′03″W | 14 | 8 | |
| Los Lunas, NM (11) | 34°48′22″N, 106°43′58″W | 5 | 0 | ||
| Punta de Agua, NM (12) | 34°36′00″N, 106°17′00″W | 5 | 5 | ||
| Y. constricta | I-10 and SR 290 jctn., TX(13) | 30°17′42″N, 99°32′16″W | 1 | 0 | |
| Y. elata | White Sands Natl Monument, NM (14) | 32°46′00″N, 106°20′00″W | 1 | 1 | |
| T. intermedia (Eastern US) | Y. filamentosa | Georgesville, OH (15) | 39°53′27″N, 83°13′19″W | 2 | 2 |
| Goldbond, VA (16) | 37°22′48″N, 80°30′40″W | 1 | 1 | ||
| Vine, TN (17) | 36°01′52″N, 86°21′28″W | 3 | 3 | ||
| Columbus, MS (18) | 33°29′44″N, 88°25′38″W | 2 | 2 | ||
| Camp Meeting Rock, GA (19) | 33°18′19″N, 85°07′30″W | 3 | 3 | ||
| Union Point, GA (20) | 33°36′56″N, 83°04′29″W | 1 | 1 | ||
| Eglin Air Force Base, FL (21) | 30°43′15″N, 86°44′18″W | 6 | 6 | ||
| Apalachicola Bluffs Preserve, FL (22) | 30°34′08″N, 84°56′52″W | 5 | 5 | ||
| Archbold Biological Station, Lake Placid, FL (23) | 27°14′00″N, 81°24′00″W | 7 | 7 |
Sample sizes are given separately for the mitochondrial DNA sequence analysis and the AFLP analysis. The numbers following the site location correspond to the site numbers on figure 1. The row indicating T. intermedia on the host plant Y. elata is a new record and an extension of its host range.
(b) Genetic analyses
Samples of T. elatella were collected from four populations of Y. elata. Tegeticula intermedia were collected from Y. glauca, Y. constricta and Y. baileyi in the western United States (table 1). We also included cheaters from Y. filamentosa in the eastern United States for making comparisons between eastern and western populations. DNA was extracted from 131 individuals using Isoquick Extraction Kits (Orca Research, Inc., Bothell, WA). Before extraction, the head, wings, and genitalia were removed and kept as vouchers.
779 bp of the mitochondrial cytochrome oxidase I (COI) gene were sequenced for 69 T. elatella individuals. All of the T. intermedia samples were sequenced previously (GenBank accession no. AY563474–AY563506; Segraves & Pellmyr 2004). Sequences were determined using standard methods for automated DNA sequencing on an ABI 377 (Segraves & Pellmyr 2001). The resulting sequences were aligned by eye and examined using phylogenetic analyses implemented in Paup* (Swofford 2003). To simplify analyses, individuals within species groups bearing identical sequence haplotypes were condensed into a single haplotype designation. The model of sequence evolution was selected using hierarchical likelihood-ratio tests (LRTs) involving an initial neighbour-joining search with LogDet distances to identify a tree (Huelsenbeck & Crandall 1997; Sullivan et al. 1997). This model of evolution was then used in a subsequent tree search under maximum likelihood to find the best tree. One hundred bootstrap replicates were generated to estimate support for the resulting tree topology.
AFLPs were determined for a subset of the DNA samples used for sequencing plus some additional samples. A single T. intermedia from Cuervo, NM was included in the AFLP analysis that would not amplify for sequencing. Also T. elatella samples from the Rodeo, NM and the Sierra Vista, AZ populations were included to broaden the geographic range of sampling and to increase sample sizes for T. elatella outside of the Big Bend region. In total, 63 T. elatella and 61 T. intermedia were included in the AFLP analysis. AFLP reactions were conducted following Segraves & Pellmyr (2004). Presence/absence of fragments was assessed visually using the GeneScan software version 3.1.2 (Applied Biosystems). In total, 308 fragments were scored for the 124 individuals included in the AFLP analysis.
Principal components analysis (PCA) on the AFLP data matrix was used to identify clusters of individuals with similar banding profiles (Young et al. 2001). Parental species were predicted to have distinct clusters within the ‘AFLP-space’ whereas hybrids would appear intermediate to the parental species. PCA was implemented in Jmp 3.2.1 (1998) on the correlations among bands within individuals. An assignment test was used to determine the probability of assignment of individuals to a species group. The assignment test was conducted using Doh (Brzustowski 2002) with ploidy set to 1 (Bensch et al. 2002). Parental species were predicted to have divergent likelihood scores whereas hybrid individuals would have likelihood scores similar to both parents.
3. Results
The DNA sequencing of mtDNA revealed 45 haplotypes that differed by one to 21 nucleotide substitutions. There were no indels inferred, and 54 of 779 sites were variable. The hierarchical LRTs indicated that the HKY85+I model was the simplest model fitting the data. The Ti/Tv ratio was 4.02, the proportion of invariable sites was 0.85, and the base frequencies were A=0.319, C=0.134, G=0.152 and T=0.395.
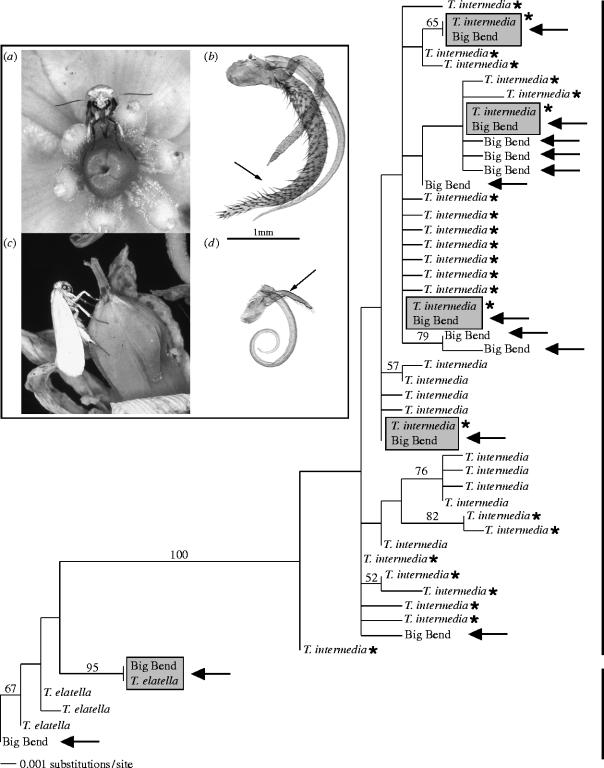
The maximum likelihood search resulted in two similar trees (figure 2; −ln L=1520.84). Both trees indicated strong support for monophyly of T. intermedia, with the exception of pollinators from the Big Bend population (figure 1, site 4) that fell into both pollinator and cheater clades. Big Bend pollinators shared identical haplotypes with T. elatella from the Sierra Vista, Rodeo and Jornada populations (figure 1, sites 1–3) or had mtDNA haplotypes closely related to these populations. Other Big Bend pollinators, however, had mtDNA haplotypes that fell into the cheater clade. These pollinators either shared identical mtDNA haplotypes with T. intermedia from the western United States, or were most closely related to western T. intermedia. Of the 50 phenotypic pollinators sequenced from the Big Bend population, 36 nested in the cheater clade.
Figure 2.
Maximum likelihood tree for Tegeticula intermedia, T. elatella and pollinators from the Big Bend population. Bootstrap support is shown above the branches. The brackets highlight the two major clades corresponding to the pollinator and cheater species groups. Arrows indicate the placement of Big Bend haplotypes. Shaded boxes show instances where Big Bend individuals shared the same haplotype with either T. intermedia or T. elatella from outside of the Big Bend region. Asterisks indicate T. intermedia haplotypes from the western United States. Inset: (a) Oviposition by a pollinator female. (b) Pollinator mouthparts (arrow indicates tentacle). (c) Oviposition by a cheater female. (d) Cheater mouthparts (arrow indicates non-functional tentacle rudiment).
PCA of 308 AFLP markers showed that T. elatella and T. intermedia were distinct clusters and that individuals from the Big Bend population occupied the intermediate AFLP space between the pollinator and cheater clusters (figure 3a). With the exception of the Big Bend population, the assignment test correctly assigned all individuals to the species that they had been previously classified as based on genitalia and mouthpart morphology (figure 2 inset). Furthermore, all but two of the T. intermedia individuals were correctly assigned to their regional grouping of either eastern (figure 1, sites 15–23) or western (figure 1, sites 6–14) United States. The Big Bend moths harbouring a T. intermedia mtDNA were nearly equally split between the T. intermedia and T. elatella species group whereas the Big Bend individuals bearing a T. elatella mtDNA were more frequently assigned to the T. elatella parental group (table 2). Moths from Big Bend tended to have intermediate likelihood scores whereas T. elatella and T. intermedia individuals had divergent scores (figure 3b). Analyses of the AFLP data using the program NewHybrids 1.1 beta (Anderson & Thompson 2002) further supported the idea that the Big Bend population consists of hybrids and indicated that these individuals were a mixture of F1s, F2s, and backcrosses between T. intermedia and T. elatella (results not shown).
Figure 3.
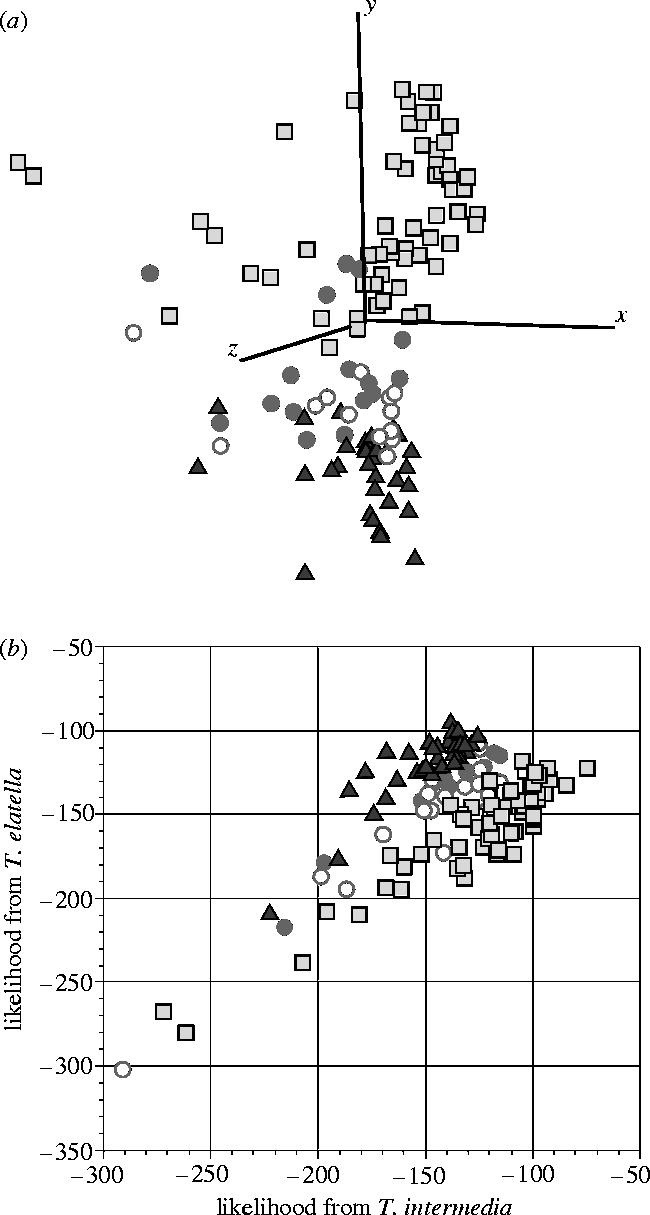
AFLP results. (a) Three-dimensional diagram of PCA results for Tegeticula elatella, T. intermedia and Big Bend individuals. Black triangles are T. elatella, open squares are T. intermedia, solid grey circles are Big Bend moths with T. elatella mtDNA and open grey circles are Big Bend moths with T. intermedia mtDNA haplotypes. (b) Likelihood scores calculated when individuals were placed into each parental group. Parental species were predicted to have divergent likelihood scores whereas hybrids would have scores more similar to the two parents.
Table 2.
Assignment test results for Tegeticula intermedia, T. elatella and individuals from the Big Bend population.
| species | mtDNA haplotype | number individuals assigned to | |
|---|---|---|---|
| T. intermedia | T. elatella | ||
| T. intermedia | T. intermedia | 61 | 0 |
| T. elatella outside Big Bend | T. elatella | 0 | 32 |
| T. elatella in Big Bend | T. intermedia | 9 | 11 |
| T. elatella in Big Bend | T. elatella | 3 | 8 |
4. Discussion
The observed genetic patterns are consistent with the hypothesis of interspecific hybridization between pollinator and cheater moths. Moths in the Big Bend population were pollinators with a mix of mtDNA haplotypes from both the pollinator and cheater clades. The AFLP analyses showed that the Big Bend moths were intermediate to the parental populations as expected under the hypothesis of hybridization. Hybridization was further supported by the AFLP-based assignment test where parental individuals were assigned to the predicted species group based on morphology. The individuals from the Big Bend population, however, were nearly equally assigned between the two species despite that they were all morphologically classified as pollinators. The only exceptions for the assignment test were the Big Bend moths that shared mtDNA haplotypes with T. elatella. The AFLP results indicated that these moths were more similar to the T. elatella group, and thus may represent pure parental individuals or backcrosses with T. elatella.
Although incomplete lineage sorting may also produce a similar genetic pattern, this alternative is highly unlikely considering that T. intermedia and T. elatella are distantly related (∼2.5% sequence divergence) within the yucca moth species complex (Pellmyr & Leebens-Mack 1999). More closely related species pairs, such as T. intermedia and the pollinator T. yuccasella (∼2% sequence divergence), have achieved reciprocal monophyly in their mtDNA (Althoff et al. submitted). Furthermore, Big Bend haplotypes that clustered with the cheaters were always close relatives of western cheaters (rather than both western and eastern cheaters) as expected under the hypothesis of hybridization between western T. intermedia and Big Bend T. elatella. Genetic evidence for hybridization has been reported for two yucca species (Hanson 1993) and hypothesized for many others (Webber 1953), but this is the first definitive evidence for hybridization among yucca moths.
Introgression resulting in the exclusive formation of a pollinator phenotype suggests that the cheater phenotype may be unstable in the hybrid population. We hypothesize that a phenological shift towards later flowering of Y. elata in comparison to the cheater's hosts in adjacent areas causes overlap in cheater and pollinator emergence on Y. elata. As a result, hybrids between cheater and pollinator would have an emergence phenology resembling that of a pollinator. Hybrid cheater phenotypes would emerge too early during the flowering season and lack suitable oviposition sites. Individuals ovipositing into flowers would be highly unsuccessful because they would be unable to pollinate to increase the probability of flower retention and offspring survival. Thus, the only stable strategy would be one of pollination. We lack good phenological records of flowering and moth emergence, although limited data suggest that if flowering is shifted in Y. elata, it is only slightly later than nearby populations of other yucca species (Kerley et al. 1993; James et al. 1994).
Even in the absence of a phenological shift in flowering, cheaters may have a difficult time invading a population in the face of hybridization. The closest extant population of cheaters is about 130 km away. Although this is not an unreasonable distance for small, flying insects to travel (e.g. Gardner & Early 1996; Nason et al. 1998) it may impede gene flow. The absence of cheater phenotypes in the Big Bend population suggests that the southward migration of T. intermedia onto populations of Y. elata may be rare. If cheaters were rare in the population, there would be a high probability of interspecific mating for an invading cheater moth. This continual swamping by pollinator genes may erode the viability of cheaters attempting to establish in the population. No matter the reason, the absence of cheaters in the hybrid population reinforces the view that under some ecological circumstances, cheating is an unstable strategy.
Although behavioural life habits have been considered important for delineating species boundaries among yucca moths, ovipositor morphology may be the key factor driving reproductive isolation. Yucca moths have two basic modes of oviposition: locule-ovipositing species use a long, narrow ovipositor to pierce through the pistil into the locule, whereas superficially ovipositing species use a short, stout ovipositor to lay eggs in the tissue surface (Pellmyr 1999, 2003). Mechanical mismatching of genitalia may prevent mating between moth species differing in oviposition habit. The cheater T. intermedia oviposits superficially, and western populations only coexist with locule-ovipositing pollinators (Pellmyr 1999). Some eastern populations of T. intermedia, however, co-occur with a superficially ovipositing pollinator, and genetic evidence supports hybridization between these species (Segraves & Pellmyr 2004). The present study documents hybrids between T. intermedia and the superficial-ovipositing pollinator T. elatella, further lending support to the idea that introgression may be limited by reproductive morphology. The importance of oviposition habit to species coexistence is also apparent in the current distribution of pollinating yucca moths. In all instances where two pollinator species coexist on a single yucca species, they differ in oviposition habit (Pellmyr & Leebens-Mack 2000). As a result, hybridization may occur when two species with similar oviposition habits occur in sympatry.
Cheaters have been classically viewed as disruptors of mutualistic interactions that may trigger mutualism extinction. Contrary to this view, cheaters are not uncommon in mutualisms and we have only a limited understanding of the ecological and evolutionary dynamics governing their stable coexistence with mutualists (Bronstein 2001; Bronstein et al. 2003). The present study suggests that mutualisms can be very resilient and may persist even when facing incorporation of a cheater genome into a mutualist lineage. Hybridization, then, may serve as a general mechanism that limits the spread of cheaters.
Acknowledgments
We thank A. Brody, J. Bronstein, J. Burke, D. Funk, A. Herre, J. Leebens-Mack and D. McCauley for helpful comments on the manuscript. B. Crabb and J. Leebens-Mack collected some of the moths used in this study. M. Gitzendanner and J. Koontz provided AFLP advice. Big Bend National Park, Black Gap Wildlife Management Area, Jornada Long Term Ecological Research site and White Sands National Monument provided access to field sites. Funding was provided by the Xerces Society, the American Museum of Natural History, and the National Science Foundation (grants 0206033, 0075944 and 0075803).
Footnotes
Present Address: Department of Biology, Syracuse University, 130 College Place, Syracuse, NY 13244, USA
References
- Addicott J.F. Cheaters in yucca/moth mutualism. Nature. 1996;380:114–115. 10.1038/380114a0 [Google Scholar]
- Althoff, D. M., Segraves, K. A., Leebens-Mack, J. & Pellmyr, O. Submitted. Patterns of speciation in the yucca moths: parallel species radiations within the Tegeticula yuccasella complex. [DOI] [PubMed]
- Anderson E.C, Thompson E.A. A model-based method for identifying species hybrids using multilocus genetic data. Genetics. 2002;160:1217–1229. doi: 10.1093/genetics/160.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod R, Hamilton W. The evolution of cooperation. Science. 1981;211:1390–1396. doi: 10.1126/science.7466396. [DOI] [PubMed] [Google Scholar]
- Bensch S, Helbig A.J, Salomon M, Seibold I. Amplified fragment length polymorphism analysis identifies hybrids between two subspecies of warblers. Mol. Ecol. 2002;11:473–481. doi: 10.1046/j.0962-1083.2001.01455.x. 10.1046/j.0962-1083.2001.01455.x [DOI] [PubMed] [Google Scholar]
- Bronstein J. The exploitation of mutualisms. Ecol. Lett. 2001;4:277–287. 10.1046/j.1461-0248.2001.00218.x [Google Scholar]
- Bronstein J.L, Wilson W.G, Morris W.E. Ecological dynamics of mutualist/antagonist communities. Am. Nat. 2003;162:S24–S39. doi: 10.1086/378645. 10.1086/378645 [DOI] [PubMed] [Google Scholar]
- Brzustowski, J. 2002 Doh assignment test calculator. http://www.biology.ualberta.ca/jbrzusto/Doh.php
- Bull J.J, Rice W.R. Distinguishing mechanisms for the evolution of co-operation. J. Theor. Biol. 1991;149:63–74. doi: 10.1016/s0022-5193(05)80072-4. [DOI] [PubMed] [Google Scholar]
- Doebeli M, Knowlton N. The evolution of interspecific mutualisms. Proc. Natl Acad. Sci. USA. 1998;95:8676–8680. doi: 10.1073/pnas.95.15.8676. 10.1073/pnas.95.15.8676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrière R, Bronstein J.L, Rinaldi S, Law R, Gauduchon M. Cheating and the evolutionary stability of mutualisms. Proc. R. Soc. B. 2002;269:773–780. doi: 10.1098/rspb.2001.1900. 10.1098/rspb.2001.1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R, Early J. The naturalisation of banyan figs (Ficus spp., Moraceae) and their pollinating wasps (Hymenoptera: Agaonidae) in New Zealand. New Zeal. J. Bot. 1996;34:103–110. [Google Scholar]
- Hanson, M. 1993 Dispersed unidirectional introgression from Yucca schidigera into Yucca baccata (Agavaceae). Ph.D. thesis, The Claremont Graduate School, Pomona, CA.
- Herre E, Knowlton N, Mueller U, Rehner S. The evolution of mutualisms: exploring the paths between conflict and cooperation. Trends Ecol. Evol. 1999;14:49–53. doi: 10.1016/s0169-5347(98)01529-8. 10.1016/S0169-5347(98)01529-8 [DOI] [PubMed] [Google Scholar]
- Hochberg M.E, Gomulkiewicz R, Holt R.D, Thompson J.N. Weak sinks could cradle mutualistic symbiosis—strong sources should harbour parasitic symbioses. J. Evol. Biol. 2000;13:213–222. 10.1046/j.1420-9101.2000.00157.x [Google Scholar]
- Holland J.N, DeAngelis D.L, Schultz S.T. Evolutionary stability of mutualism: interspecific population regulation as an evolutionarily stable strategy. Proc. R. Soc. B. 2004;271:1807–1814. doi: 10.1098/rspb.2004.2789. 10.1098/rspb.2004.2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Crandall K.A. Phylogeny estimation and hypothesis testing using maximum likelihood. Annu. Rev. Ecol. Syst. 1997;28:437–466. 10.1146/annurev.ecolsys.28.1.437 [Google Scholar]
- James C.D, Hoffman M.T, Lightfoot D.C, Forbes G.S, Whitford W.G. Fruit abortion in Yucca elata and its implications for the mutualistic association with yucca moths. Oikos. 1994;69:207–216. [Google Scholar]
- Janzen D. Pseudomyrmex nigropilosa: a parasite of a mutualism. Science. 1975;188:936–937. doi: 10.1126/science.188.4191.936. [DOI] [PubMed] [Google Scholar]
- Kerley G, Tiver F, Whitford W. Oecologia. Vol. 93. 1993. Herbivory of clonal populations: cattle browsing affects reproduction and population structure of Yucca elata. Washington, DC. [DOI] [PubMed] [Google Scholar]
- Law R, Bronstein J, Ferrière R. On mutualists and exploiters: plant–insect coevolution in pollinating seed-parasite systems. J. Theor. Biol. 2001;212:373–389. doi: 10.1006/jtbi.2001.2383. 10.1006/jtbi.2001.2383 [DOI] [PubMed] [Google Scholar]
- Morris W.F, Bronstein J.L, Wilson W.G. Three-way coexistence in obligate mutualist–exploiter interactions: the potential role of competition. Am. Nat. 2003;161:860–875. doi: 10.1086/375175. 10.1086/375175 [DOI] [PubMed] [Google Scholar]
- Murray M. Figs (Ficus spp.) and fig wasps (Chalcidoidea, Agaonidae): hypotheses for an ancient symbiosis. Biol. J. Linn. Soc. 1985;26:69–81. [Google Scholar]
- Nason J, Herre E, Hamrick J. The breeding structure of a tropical keystone plant resource. Nature. 1998;391:685–687. 10.1038/35607 [Google Scholar]
- Noë R, Hammerstein P. Biological markets: supply and demand determine the effect of partner choice in cooperation, mutualism, and mating. Behav. Ecol. Sociobiol. 1994;35:1–11. [Google Scholar]
- Pellmyr O. Systematic revision of the yucca moths in the Tegeticula yuccasella complex (Lepidoptera: Prodoxidae) north of Mexico. Syst. Entomol. 1999;24:243–271. 10.1046/j.1365-3113.1999.00079.x [Google Scholar]
- Pellmyr O. Yuccas, yucca moths, and coevolution: a review. Ann. Mo. Bot. Gard. 2003;90:35–55. [Google Scholar]
- Pellmyr O, Leebens-Mack J. Forty million years of mutualism: evidence for Eocene origin of the yucca–yucca moth association. Proc. Natl Acad. Sci. USA. 1999;96:9178–9183. doi: 10.1073/pnas.96.16.9178. 10.1073/pnas.96.16.9178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellmyr O, Leebens-Mack J. Adaptive radiation in yucca moths and the reversal of mutualism. Am. Nat. 2000;156:S62–S76. doi: 10.1086/303416. 10.1086/303416 [DOI] [PubMed] [Google Scholar]
- Pellmyr O, Leebens-Mack J, Huth C.J. Non-mutualistic yucca moths and their evolutionary consequences. Nature. 1996;380:155–156. doi: 10.1038/380155a0. 10.1038/380155a0 [DOI] [PubMed] [Google Scholar]
- Powell J. Interrelationships of yuccas and yucca moths. Trends Ecol. Evol. 1992;7:10–15. doi: 10.1016/0169-5347(92)90191-D. 10.1016/0169-5347(92)90191-D [DOI] [PubMed] [Google Scholar]
- Riley C. The yucca moth and yucca pollination. Ann. Mo. Bot. Gard. 1892;3:99–158. [Google Scholar]
- SAS Institute. SAS Institute; Cary, NC: 1998. JMP Statistical Software, v. 5.0.1.2. [Google Scholar]
- Schwartz M.W, Hoeksema J.D. Specialization and resource trade: biological markets as a model of mutualisms. Ecology. 1998;79:1029–1038. [Google Scholar]
- Segraves K.A, Pellmyr O. Phylogeography of the yucca moth Tegeticula maculata: the role of historical biogeography in reconciling high genetic structure with limited speciation. Mol. Ecol. 2001;10:1247–1253. doi: 10.1046/j.1365-294x.2001.01275.x. 10.1046/j.1365-294X.2001.01275.x [DOI] [PubMed] [Google Scholar]
- Segraves K.A, Pellmyr O. Testing the out-of-Florida hypothesis on the origin of cheating in the yucca–yucca moth mutualism. Evolution. 2004;58:2266–2279. doi: 10.1111/j.0014-3820.2004.tb01602.x. [DOI] [PubMed] [Google Scholar]
- Sullivan J, Markert J.A, Kilpatrick C.W. Phylogeography and molecular systematics of the Peromyscus aztecus species group (Rodentia: Muridae) inferred using parsimony and likelihood. Syst. Biol. 1997;46:426–440. doi: 10.1093/sysbio/46.3.426. [DOI] [PubMed] [Google Scholar]
- Swofford D. Sinauer Associates; Sunderland, MA: 2003. PAUP*. Phylogenetic analysis using parsimony (* and other methods), v. 4.0.610. [Google Scholar]
- Trivers R. The evolution of reciprocal altruism. Q. Rev. Biol. 1971;46:35–57. 10.1086/406755 [Google Scholar]
- Webber, J. 1953 Yuccas of the southwest Agricultural monograph no. 17. Washington, DC: US Department of Agriculture.
- Weiblen G, Yu D, West S. Pollination and parasitism in functionally dioecious figs. Proc. R. Soc. B. 2001;268:651–659. doi: 10.1098/rspb.2000.1389. 10.1098/rspb.2000.1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S.A, Herre E.A. The ecology of the New World fig-parasitizing wasps Idarnes and implications for the evolution of the fig-pollinator mutualism. Proc. R. Soc. B. 1994;258:67–72. [Google Scholar]
- West S.A, Herre E.A, Windsor D.M, Green P.R.S. The ecology and evolution of the New World non-pollinating fig wasp communities. J. Biogeogr. 1996;23:447–458. [Google Scholar]
- West S.A, Kiers E.T, Simms E.L, Denison R.F. Sanctions and mutualism stability: why do rhizobia fix nitrogen? Proc. R. Soc. B. 2002;269:685–694. doi: 10.1098/rspb.2001.1878. 10.1098/rspb.2001.1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young W.P, Ostberg C.O, Keim P, Thorgaard G.H. Genetic characterization of hybridization and introgression between anadromous rainbow trout (Oncorhynchus mykiss irideus) and coastal cutthroat trout (O. clarki clarki) Mol. Ecol. 2001;10:921–930. doi: 10.1046/j.1365-294x.2001.01247.x. 10.1046/j.1365-294X.2001.01247.x [DOI] [PubMed] [Google Scholar]