Abstract
The outcome of coevolutionary interactions is predicted to vary across landscapes depending on local conditions and levels of gene flow, with some populations evolving more extreme specializations than others. Using a globally distributed parasite of colonial seabirds, the tick Ixodes uriae, we examined how host availability and geographic isolation influences this process. In particular, we sampled ticks from 30 populations of six different seabird host species, three in the Southern Hemisphere and three in the Northern Hemisphere. We show that parasite races have evolved independently on hosts of both hemispheres. Moreover, the degree of differentiation between tick races varied spatially within each region and suggests that the divergence of tick races is an ongoing process that has occurred multiple times across isolated areas. As I. uriae is vector to the bacterium responsible for Lyme disease Borrelia burgdorferi sensu lato, these results may have important consequence for the epidemiology of this disease. With the increased occurrence of novel interspecific interactions due to global change, these results also stress the importance of the combined effects of gene flow and selection for parasite diversification.
Keywords: colonial seabirds, ectoparasite, host-dependent dispersal, Ixodes uriae, population genetic structure, vector-borne disease
1. Introduction
Interspecific interactions are considered to play a primary role in the diversification and organization of life. Due to the effects of climate change and habitat fragmentation on species' distributions, the importance of such interactions has become increasingly recognized (Altizer et al. 2003). In particular, landscape level changes may increase contact between previously isolated populations. In such situations, it is difficult to predict what type of evolutionary outcome will result or how quickly it will evolve. In addition, little information currently exists on coevolutionary changes within the context of communities, where a species may be exposed to several alternative interacting species (Thompson 1999a). Depending on local conditions, the intensity and direction of reciprocal selection can vary, resulting in some populations evolving more extreme specializations than others (Benkman 1999; Buckling & Rainey 2002; Thompson & Cunningham 2002). There are, for example, several cases of incipient speciation occurring via the evolution of host specialization. Many of these examples come from observations of phytophagous insects, where diversification has occurred over relatively small spatial scales and despite a high potential for dispersal (e.g. Bush 1969; Jaenike 1990; Craig et al. 2001). However, what happens in situations where the same interaction occurs in different isolated regions? Will the coevolutionary outcome always be the same? To evaluate these questions, it is necessary to examine the same antagonistic system across multiple communities (Thompson 1999b).
The interaction between the tick Ixodes uriae and its seabird hosts has attributes governed by evolutionary dynamics across broad geographic ranges and represents an ideal system for examining how coevolution operates over large spatial scales. This ectoparasite is specific to colonial seabirds, infesting species in the circumpolar areas of both hemispheres (Guiguen 1988). The seabird hosts are often found in isolated areas and typically form large colonies that can be either monospecific or heterospecific (containing several sympatric species) in nature. High levels of infestation by I. uriae are associated with reduced host reproductive success, colony abandonment, and pathogen transmission (e.g. Chastel 1988; Boulinier & Danchin 1996; Mangin et al. 2003). As this tick requires a prolonged period on the host to complete its blood meal (3–12 days), an intense interaction between parasite and host takes place. Indeed, adaptation to local hosts and a role for the host immune response in this process has been suggested by experimental work (McCoy et al. 2002). Immediately after its blood meal, I. uriae returns to the substrate of the host colony (under rocks, in cliff fissures or dirt burrows) where it moults, over-winters and awaits the return of the seabird hosts the following season. In heterospecific seabird colonies, different host species often breed side-by-side, therefore facilitating the infestation of alternative host types. Although this exchange could swamp selection for specialized adaptations in some communities (Lajeunesse & Forbes 2002), genetic data support the specialization of this tick for two sympatric seabird species of the Northern hemisphere, the black-legged kittiwake Rissa tridactyla and the Atlantic puffin Fratercula arctica (McCoy et al. 2001). It is unknown whether the differentiation of these races represents an isolated case, or whether specialization is a general characteristic of the interaction between I. uriae and its different seabird hosts.
To address how geographic structure and host availability have shaped the evolution of this globally distributed parasite, we tested whether there has been independent evolution of host specific races across different host communities and examined how race formation may have been affected by population isolation. In particular, we analysed the distribution of neutral genetic variation in I. uriae across a range of host species and spatial scales. As the seabird hosts considered are confined to their respective circumpolar areas and few seabirds, in general, utilize land areas of both hemispheres (a requirement for tick dispersal), there is little possibility for parasite gene flow between polar regions. We, therefore, expected that host race formation would be independent in the two hemispheres. If the ticks of each hemisphere have formed host specific races, we expected populations to cluster by host type, regardless of the geographic distance between the sampled colonies. Moreover, if each tick race arose only once, we predicted that the degree of differentiation between races would be relatively similar in all heterospecific colonies because the time since divergence would be the same (assuming complete isolation between races).
2. Material and methods
(a) Sampling and genotyping
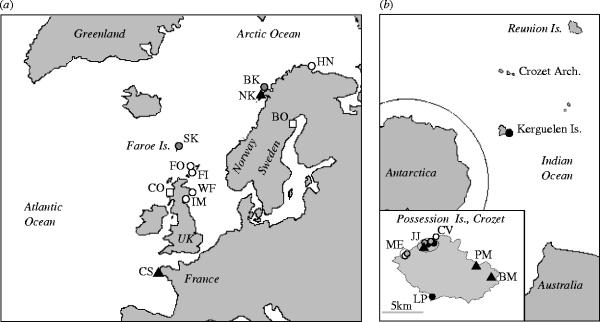
We sampled ticks from six monospecific and five heterospecific colonies of three seabird species in the Northern Hemisphere, the black-legged kittiwake (R. tridactyla), the Atlantic puffin (F. arctica), and the common guillemot (Uria aalge) (figure 1a). All the three of these species breed during the same period (April to August), with some variation in phenology linked to climatic conditions and yearly food availability (e.g. Furness & Barrett 1985). In the Southern Hemisphere, parasites were collected from three additional species, from colonies of king (Aptenodytes patagonicus), macaroni (Eudyptes chrysocome) and rockhopper (Eudyptes chrysolophus) penguins. These samples included ticks from 10 monospecific colonies, and two heterospecific Eudyptes colonies (figure 1b). There were two areas where monospecific colonies were parapatric (areas JJ and ME). Although the three penguin species can be present in the colonies at the same time, there are major differences in the breeding phenologies of the king penguin and the Eudyptes species; these difference can have important implications in terms of the availability of these hosts for tick exploitation (see Frenot et al. 2001).
Figure 1.
(a) In the Northern Hemisphere ticks were sampled in two monospecific colonies for each host: the black-legged kittiwake (black triangles), the Atlantic puffin (grey circles) and the common guillemot (white squares). Parasites were also sampled in heterospecific colonies where at least two of the three species were sympatric (white circles). (b) In the Southern Hemisphere most tick sampling sites consisted of monospecific colonies of the three hosts on Possession Island in the Crozet archipelago; the king penguin (black triangles), the macaroni penguin (black circles), and the rockhopper penguin (gray circles). White circles represent sympatric colonies of Eudyptes. In areas JJ and ME host colonies were parapatric.
In each seabird colony, an effort was made to collect ticks from at least 30 individual birds/nesting areas. In the Northern Hemisphere, ticks were sampled directly from the birds during ringing (except in colony BK where burrows were sampled). In penguin colonies, ticks can be easily found under the rocks surrounding nests. To minimize the disturbance to these birds, ticks were sampled from the off-host environment in the Southern Hemisphere. Due to this sampling method, ticks could not be unambiguously attributed to a given host species in the heterospecific Eudyptes colonies. Collected ticks of both hemispheres were stored in 70–90% ethanol. Over the two hemispheres, a total of 808 ticks from 30 populations were genotyped (477 in the North and 331 in the South) at eight microsatellite loci designed specifically for this parasite (McCoy & Tirard 2000).
DNA extractions, PCR amplifications and genotype visualizations were performed as described in McCoy & Tirard (2000) for most ticks of the Northern Hemisphere. For ticks of the Southern Hemisphere and some populations in the North, genotypes were visualized using an automated sequencer (ABI Prism 310 Genetic Analyser, Applied Biosystem, Perkin-Elmer, USA). Size standards from ticks genotyped using manual methods were used to match allele sizes.
(b) Population genetic analyses
All populations were tested for departure from Hardy–Weinberg equilibrium using exact probability tests (2×106 iterations). To ensure the independence of the markers employed, loci were similarly tested for linkage disequilibrium (Raymond & Rousset 1995). Gene diversity (Nei 1987), observed heterozygosity and allelic richness were calculated for each population. Allelic richness refers to the expected number of alleles given an equal number of individuals sampled in all populations. Differences in gene diversity, heterozygosity and allelic richness between hemispheres were determined by permuting populations between hemispheres (1000 randomizations) using the software FSTAT v. 2.9.3 (Goudet 1995).
Using the genetic data, we first confirmed that ticks from the two hemispheres could indeed be considered as independent replicates for examining the evolution of host specific races. We calculated the average differentiation between populations of the two hemispheres using Wright's F-statistics estimated according to Weir & Cockerham (1984). We also calculated the maximum theoretical value of FST (FST MAX) as a reference for interpreting population differentiation. The maximum value of FST does not equal unity when dealing with highly polymorphic genetic markers, but instead corresponds to the average expected within-population homozygosity (Hedrick 1999).
We next tested whether tick populations clustered genetically according to the host species exploited. This was done using principal component analyses (PCA) to summarize the variance in population allele frequencies so that the genetic relationships between populations could be easily visualized. Using the program PCA-GEN v. 1.2 (J. Goudet, Institute of Ecology, University of Lausanne, Lausanne, Switzerland, 1999), analyses were performed separately for the populations of each hemisphere. Permutation tests included in the program enabled the significance of each component to be tested (5000 genotype randomizations). The robustness of population groupings was further evaluated using assignment tests. In each hemisphere, a Bayesian approach was employed to directly assign each individual tick to the host species/population where the likelihood of its multilocus genotype was the highest (Program GeneClass v. 1.0.02, Cornuet et al. 1999). We then calculated the percentage of correct assignments to each host race and to the population of origin.
Finally, we examined the degree of population differentiation between different sympatric tick races by calculating pairwise estimates of FST across all loci and individually for each locus. To determine how host species may influence patterns of parasite population structure, we also examined patterns of genetic structure among populations of the same host race. The significance of FST estimates was evaluated by permuting genotypes among populations (5000 randomizations) using FSTAT v. 2.9.3 (Goudet 1995).
Overall, the number of randomizations used for statistical tests was limited to that necessary to ensure the stability of the results.
3. Results
After correction for multiple tests (Rice 1989), all tick populations of the Northern Hemisphere were in Hardy–Weinberg equilibrium, except the kittiwake tick and the guillemot tick populations of colony FI (see figure 1a). The disequilibrium in the kittiwake tick population was due to locus T44 (see below), whereas there were slight heterozygote deficits at several loci for the guillemot race potentially indicating substructure within the colony area (i.e. Wahlund effect). In the south, initial tests indicated that almost all populations were in disequilibrium. This was largely due to two loci (T22 and T39) for which we suspected the presence of null alleles. Global equilibrium was obtained for all populations with the elimination of these two loci. All the tests were carried out including and excluding theses two loci and results were unchanged. Because of the mild deviations from equilibrium, alleles were not considered as independent for testing the significance of population differentiation, hence the use of the genotype as the randomization unit in all permutation tests (Goudet 1995). After Bonferroni correction (Rice 1989), no pairwise comparison of loci showed linkage disequilibrium.
The partitioning of genetic variation between northern and southern ticks suggested that these groups are completely isolated; the average estimate of genetic differentiation between the populations of different hemispheres was close to the maximum value possible given marker diversity (FST=0.38; FMAX=0.42). Despite a smaller sample size (north=18 populations, south=12 populations), all measures of genetic variation were significantly greater in the south (gene diversity: north=0.54, south=0.66; observed heterozygosity: north=0.50, south=0.60; allelic richness: north=4.97, south=6.47; all p<0.001).
Tick populations clustered primarily by host type, regardless of the geographic distance between colonies or the nature of the colony (monospecific or heterospecific) (figure 2). Three groups of ticks were present in the Northern Hemisphere, including a newly described race of guillemot ticks. The Southern Hemisphere ticks formed two differentiated groups: king penguin and Eudyptes host races. Assignment tests confirmed the PCA groupings; there was a high probability of assigning individuals to their host of origin (table 1). Host mis-assignments varied with tick race (table 1). In the north, kittiwake ticks showed the lowest specificity with a 18.2% probability of being assigned to another host species, whereas puffin ticks showed the highest specificity with only a 5% probability of being mis-assigned. In the south, assignment tests indicated that 22.2% of ticks collected from king penguins were assigned to the Eudyptes race, whereas only 4.5% of ticks collected from the Eudyptes species were assigned to the king penguin race. The ability to assign ticks to their population of origin was relatively low in both hemispheres (table 1) reflecting low within host species population differentiation.
Figure 2.
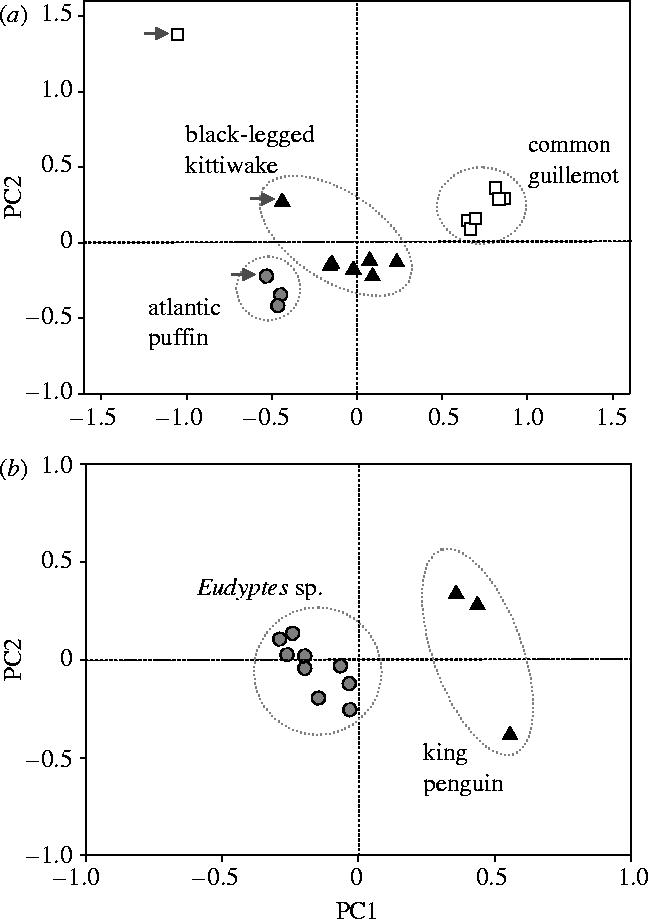
Isolation of I. uriae populations by host species. (a) Principal components analysis of populations in the Northern Hemisphere revealed two significant axes explaining 43.71 and 24.38% inertia respectively for PC1 and PC2 (p<0.001). Only the guillemot tick population of colony HN did not cluster by host. The HN population for all three tick races is indicated by grey arrows. (b) The same analysis of Southern Hemisphere populations also showed two significant axes that explained 38.97 and 19.35% inertia respectively for PC1 and PC2 (p<0.01).
Table 1.
Average percentage of tick assignments (±standard error) to host race and population of origin. Separate analyses were performed for each hemisphere. Host species are labelled as black-legged kittiwake (KT), common guillemot (CG), Atlantic puffin (PF), king penguin (KP) and the combined Eudyptes species of rockhopper and macaroni penguins (RMP).
| assigned to | ||||||
|---|---|---|---|---|---|---|
| tick race | KT | CG | PF | KP | RMP | popa |
| north | ||||||
| KT | 81.8 (3.15) | 9.3 (3.01) | 8.6 (1.76) | — | — | 38.8 (8.15) |
| CG | 9.1 (3.46) | 88.7 (3.83) | 2.3 (0.80) | — | — | 44.4 (10.74) |
| PF | 3.3 (1.35) | 1.6 (1.61) | 95.1 (2.79) | — | — | 28.0 (11.20) |
| south | ||||||
| KP | — | — | — | 77.8 (2.94) | 22.2 (2.94) | 50.0 (3.87) |
| RMP | — | — | — | 4.5 (1.09) | 95.5 (1.09) | 14.9 (3.76) |
population of origin.
In all cases, local sympatric (or parapatric) ticks of different races were significantly differentiated (table 2). However, the degree of differentiation was colony and locus dependent. Indeed, fixed differences were found between guillemot ticks and the other two races at locus T44 in many of the heterospecific colonies, whereas differentiation at the other seven loci was more variable among sites. When pairwise differentiation was calculated without locus T44, population structuring between races sympatric with guillemot ticks was strongly reduced in all colonies, except colony HN where significant structure was evident at all eight loci (table 2). These results suggest that locus T44 is linked to genes under selection for host specialization and that the divergence of the guillemot race in colony HN evolved earlier than in other colonies.
Table 2.
Pairwise population differentiation between sympatric host races of Ixodes uriae. Host race abbreviations are as follows: KT – kittiwake ticks, PF – puffin ticks, CG – guillemot ticks, KP – king penguin ticks, and RMP – Eudyptes ticks. FST estimates and p-values in brackets indicate values excluding locus T44.
| colony | host races | FST | p-value |
|---|---|---|---|
| north | |||
| Hornøya (HN) | KT–PF | 0.061 (0.062) | <0.0002 (<0.0002) |
| KT–CG | 0.161 (0.163) | <0.0002 (<0.0002) | |
| PF–CG | 0.227 (0.231) | <0.0002 (<0.0002) | |
| Foula (FO) | KT–CG | 0.153 (0.036) | <0.0002 (0.0016) |
| Fair Isle (FI) | KT–PF | 0.075 (0.078) | <0.0002 (<0.0002) |
| KT–CG | 0.054 (0.013) | 0.001 (0.109) | |
| PF–CG | 0.163 (0.124) | <0.0002 (<0.0002) | |
| Whinnyfold (WF) | KT–CG | 0.089 (0.005) | <0.0002 (0.058) |
| Isle of May (IM) | KT–CG | 0.127 (0.013) | <0.0002 (0.034) |
| south | |||
| Jardin Japonais (JJ) | KP–RMP | 0.037 (0.043) | <0.0002 (<0.0002) |
KP and RMP populations were parapatric at site JJ.
Host specific patterns of tick gene flow were evident from levels of population structuring within each host race. In the north, guillemot tick populations were the most strongly structured of the three host races (kittiwake ticks: FST=0.039, p<0.0002, n=7; guillemot ticks: FST=0.161, p<0.0002, n=7; puffin ticks: FST=0.011, p<0.0002, n=4). However, this pattern was largely due to the inclusion of guillemot ticks from colony HN. This population was genetically isolated from all others, including other guillemot tick populations and the kittiwake and puffin tick populations found sympatrically (figure 2a). With the removal of this population, guillemot tick populations showed a level of structuring intermediate to that of the other two races (FST=0.022). In the south, ticks exploiting king penguins showed greater population structure than those exploiting the two Eudyptes hosts (king penguin ticks: FST=0.025, p<0.0002, n=3; Eudyptes ticks: FST=0.004, p=0.002, n=5) despite the inclusion of the Eudyptes tick population on Kerguelen Island (>1000 km from Crozet).
4. Discussion
We analysed the distribution of neutral genetic variation in I. uriae populations across a range of host species and spatial scales to address how host availability and geographic structure has shaped the evolution of this globally distributed parasite. In particular, we tested whether there has been independent and repeated evolution of host specific races across different host communities and how this process may have been affected by population isolation. Using the genetic data, we first confirm that ticks from the two hemispheres can indeed be considered as independent replicates for examining the evolution of host specific races. The partitioning of genetic variation between northern and southern ticks suggests that these groups are completely isolated and should likely be considered as different species. This idea is further supported by the presence of null alleles in Southern Hemisphere ticks; null alleles are often associated with the cross-amplification of distantly related species (Paetkau & Strobeck 1995). Interestingly, levels of genetic variation were significantly greater in southern ticks. This is consistent with a phylogenetic study that suggests an ancestral origin of I. uriae in this hemisphere (Norris et al. 1999).
In both hemispheres, we found that tick populations clustered primarily by host type, regardless of the geographic distance between colonies or the nature of the colony (monospecific or heterospecific) (figure 2). Three host groups of ticks were detected in the Northern Hemisphere, including a newly described race of guillemot ticks. Southern Hemisphere ticks formed two differentiated groups: king penguin and Eudyptes host races. These results suggest that selection for host specialization in this system overrides the potential benefits of remaining a host generalist. Selection of this type could originate from coevolutionary interactions with the host immune response; the fitness of ticks will be greatly affected by their ability to modulate host immunity during the blood meal (Wikel 1996). The lack of specialized races for each Eudyptes species was not surprising given that these hosts are sister species with similar breeding characteristics and phenologies (Warham 1975). This contrasts with the king penguins, which have a very different breeding cycle (14 to 16 months, rather than annual). As ticks are unlikely to survive on the host during long periods spent at sea, these differences in phenology alter the periods of host availability for ticks exploiting each species (Frenot et al. 2001) and may thus impact the evolution of tick specialization. Interestingly, assignment tests indicated that king penguin ticks were more likely to be assigned to the Eudyptes race (22.2%) than the reverse (4.5%). This could suggest that (1) Eudyptes ticks are less specialized than king penguin ticks (i.e. asymmetrical exploitation), (2) the king penguin tick race is the result of a recent host switch from Eudyptes hosts and still harbours some ancestral alleles or (3) king penguins are a ‘habitat’ sink for Eudyptes ticks. Regardless of the explanation, this result was surprising given the potential for a longer history of association between the parasite and its hosts in this hemisphere (see above).
The degree of differentiation between different sympatric tick races was colony dependent. For example, in most sympatric colonies, significant pairwise FST estimates between guillemot ticks and the other two races were largely due to fixed differences at a single microsatellite locus that may be linked to genes under selection for host specialization (see §3). However, in colony HN, significant structure was evident at all eight loci and the guillemot ticks of this colony clearly separated from all other populations in the PCA (see figure 2a). As tick dispersal must occur via the host, these results suggest that guillemots in this population do not transport ticks to colonies further south and, as a consequence, that the formation of the guillemot tick race likely occurred independently and evolved earlier in this geographic region. More generally, these patterns suggest that the divergence of tick races is an ongoing process and has occurred multiple times within the Northern Hemisphere.
Within host species, the ability to assign ticks to their correct population of origin was relatively low in both hemispheres, but varied among tick races. For example, in the north where sampling was evenly distributed in space for the three host species, puffin ticks had a lower probability of being assigned to the correct population compared to the other two races (see table 1). These host-specific patterns of structure and gene flow underline the importance of the exploited host for the large-scale dispersal of this parasite (McCoy et al. 2003). Indeed, all the three of these seabirds are considered to show strong breeding site fidelity, but differences in the behaviour of non-breeding birds may modify the probability of tick dispersal at different spatial scales (McCoy et al. 2003, 2005). As the outcomes of interspecific interactions can depend on the spatial isolation of both host and parasite, differences in host life history traits such as dispersal will be important dictators of parasite diversification potential (Weiblen & Bush 2002).
The isolation of ticks among hosts and between hemispheres will have important consequences for the microparasites that they vector. For example, I. uriae is a vector of the bacterium responsible for human Lyme disease, Borrelia burgdorferi sensu lato (Olsen et al. 1993). Studies suggest that this bacterium has a relatively high prevalence in seabirds of both hemispheres (Olsen et al. 1993; Gauthier-Clerc et al. 1999, Gasparini et al. 2001), but little is known about the degree of transhemispheric exchange (Olsen et al. 1995) or the potential interaction between marine and terrestrial disease cycles. Our results suggest that the exchange of this disease agent between hemispheres should be strongly limited and that transmission between different seabird hosts, even those in sympatry, will be low and often uni-directional. Clearly, the consequences of host specialized vectors will be important to consider for the epidemiology and evolution of many vector-borne diseases.
5. Conclusions and perspectives
Theory suggests that interspecific interactions are in continual flux as they evolve independently in different isolated populations (Thompson 1999a). Selection mosaics combined with gene flow can create novel dynamics that can alter the coevolutionary trajectories produced by local interactions alone. Indeed, despite the strong potential for between-host gene flow in the I. uriae-seabird system, we found that specialized races of this ectoparasite have evolved multiple times and differ in their degree of isolation. Given time, the host races of this parasite may become further genetically distinct due to the effects of genetic drift in already isolated populations, or through an increasing degree of reciprocal selection created by specialization (Woolhouse et al. 2002). Inversely, current levels of structure between different races may be maintained over time due to occasional gene flow linked to optimal degrees of host specialization. Determining which traits of the interacting taxa have coevolved to shape the patterns of diversification in this system and testing whether these traits, and the intensity of selection, are the same for each isolated interaction will help resolve these issues. Although specialization appears to be highly favoured in the I. uriae–seabird system, our results clearly show that geographic structure can also play a significant role in the production of large-scale ecological patterns and in the evolution of biodiversity.
Acknowledgments
We thank M. P. Harris, S. Wanless, B. Olsen, Ă. Gylfe and E. Danchin for collecting ticks at some sites and J. Gasparini, D. Jardine, R.W. Furness, D. Shaw, R. Barrett, T. Tveraa, N. G. Yoccoz and the Norwegian Lighthouse Authority for field-related assistance. T. Burg, T. Day, T. deMeeûs and R. Kassen provided helpful comments on a previous version of the manuscript. Funding for this research was provided by the French Polar Institute (IPEV, programmes n°0333 & n°0137), the Centre National de la Recherche Scientifique (Réseau Interactions Durables), and the Institut Français de la Biodiversité. KDM was supported by a Chateaubriand post-doctoral fellowship (French Embassy of Canada) and the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Present address: Génétique et Evolution des Maladies Infectieuses, CNRS IRD UMR 2724, IRD, 911 Avenue Agropolis, B.P. 64501 F-34394 Montpellier, France.
References
- Altizer S, Harvell D, Friedle E. Rapid evolutionary dynamics and disease threats to biodiversity. Trends Ecol. Evol. 2003;18:589–596. 10.1016/j.tree.2003.08.013 [Google Scholar]
- Benkman C.W. The selection mosaic and diversifying coevolution between crossbills and lodgepole pine. Am. Nat. 1999;153:S75–S91. doi: 10.1086/303213. 10.1086/303213 [DOI] [PubMed] [Google Scholar]
- Boulinier T, Danchin E. Population trends in kittiwake Rissa tridactyla colonies in relation to tick infestation. Ibis. 1996;138:326–334. [Google Scholar]
- Buckling A, Rainey P.B. The role of parasites in sympatric and allopatric host diversification. Nature. 2002;420:496–499. doi: 10.1038/nature01164. 10.1038/nature01164 [DOI] [PubMed] [Google Scholar]
- Bush G.L. Sympatric host race formation and speciation in frugivorous flies of the genus Rhagoletis (Diptera, Tephritidae) Evolution. 1969;23:237–251. doi: 10.1111/j.1558-5646.1969.tb03508.x. [DOI] [PubMed] [Google Scholar]
- Chastel C. Tick-borne virus infections of marine birds. In: Harris K.F, editor. Advances in disease vector research. Springer; New York: 1988. pp. 25–60. [Google Scholar]
- Cornuet J.-M, Piry D, Luikart G, Estoup A, Solignac M. New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics. 1999;153:1989–2000. doi: 10.1093/genetics/153.4.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig T.P, Horner J.D, Itami J.K. Genetics, experience and host-plant preference in Eurosta solidaginis: implications for host shifts and speciation. Evolution. 2001;55:773–782. doi: 10.1554/0014-3820(2001)055[0773:geahpp]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Frenot Y, de Oliveira E, Gauthier-Clerc M, Deunff J, Bellido A, Vernon P. Life cycle of the tick Ixodes uriae in penguin colonies: relationships with host breeding activity. Int. J. Parasitol. 2001;31:1040–1047. doi: 10.1016/s0020-7519(01)00232-6. 10.1016/S0020-7519(01)00232-6 [DOI] [PubMed] [Google Scholar]
- Furness R.W, Barrett R.T. The food requirements and ecological relationships of a seabird community in North Norway. Ornis Scand. 1985;16:305–313. [Google Scholar]
- Gasparini J, McCoy K.D, Haussy C, Tveraa T, Boulinier T. Induced maternal response to the Lyme disease spirochaete Borrelia burgdorferi sensu lato in a colonial seabird, the kittiwake Rissa tridactyla. Proc. R. Soc. B. 2001;268:647–650. doi: 10.1098/rspb.2000.1411. 10.1098/rspb.2000.1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier-Clerc M, Jaulhac B, Frenot Y, Bachelard C, Monteil H, Le Maho Y, Handrich Y. Prevalence of Borrelia burgdorferi (the Lyme disease agent) antibodies in king penguin Aptenodytes patagonicus in Crozet Archipelago. Polar Biol. 1999;22:141–143. 10.1007/s003000050402 [Google Scholar]
- Goudet J. FSTAT (vers.1.2): a computer program to calculate F-statistics. J. Hered. 1995;86:485–486. [Google Scholar]
- Guiguen, C. 1988 Anthropozoonoses et oiseaux marins: contribution à l'étude des ectoparasites hématophages des espèces nicheuses sur les côtes françaises continentales et insulaires. Ph.D. thesis, Faculté de Médecine de Marseille.
- Hedrick P.W. Perspective: highly variable loci and their interpretation in evolution and conservation. Evolution. 1999;53:313–318. doi: 10.1111/j.1558-5646.1999.tb03767.x. [DOI] [PubMed] [Google Scholar]
- Jaenike J. Host specialization in phytophagous insects. Annu. Rev. Ecol. Syst. 1990;21:243–273. 10.1146/annurev.es.21.110190.001331 [Google Scholar]
- Lajeunesse M.J, Forbes M.R. Host range and local parasite adaptation. Proc. R. Soc. B. 2002;269:703–710. doi: 10.1098/rspb.2001.1943. 10.1098/rspb.2001.1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangin S, Gauthier-Clerc M, Frenot Y, Gendner J.-P, Le Maho Y. Ticks Ixodes uriae and the breeding performance of a colonial seabird, king penguin Aptenodytes patagonicus. J. Avian Biol. 2003;34:30–34. 10.1034/j.1600-048X.2003.02916.x [Google Scholar]
- McCoy K.D, Boulinier T, Schjørring S, Michalakis Y. Local adaptation of the ectoparasite Ixodes uriae to its seabird host. Evol. Ecol. Res. 2002;4:441–456. [Google Scholar]
- McCoy K.D, Boulinier T, Tirard C, Michalakis Y. Host specificity of a generalist parasite: genetic evidence of sympatric host races in the seabird tick Ixodes uriae. J. Evol. Biol. 2001;14:395–405. 10.1046/j.1420-9101.2001.00290.x [Google Scholar]
- McCoy K.D, Boulinier T, Tirard C, Michalakis Y. Host-dependent genetic structure of parasite populations: differential dispersal of seabird tick host races. Evolution. 2003;57:288–296. doi: 10.1111/j.0014-3820.2003.tb00263.x. [DOI] [PubMed] [Google Scholar]
- McCoy K.D, Boulinier T, Tirard C. Comparative host-parasite population structures: disentangling prospecting and dispersal in the black-legged kittiwake Rissa tridactyla. Mol. Ecol. 2005;14:2825–2838. doi: 10.1111/j.1365-294X.2005.02631.x. [DOI] [PubMed] [Google Scholar]
- McCoy K.D, Tirard C. Isolation and characterisation of microsatellites in the seabird ectoparasite Ixodes uriae. Mol. Ecol. 2000;9:2213–2214. [PubMed] [Google Scholar]
- Nei M. Columbia University Press; New York: 1987. Molecular evolutionary genetics. [Google Scholar]
- Norris D.E, Klompen J.S.H, Black W.C.I. Comparison of the mitochondrial 12S and 16S ribosomal DNA genes in resolving phylogenetic relationships among hard ticks (Acari: Ixodidae) Annu. Entomol. Soc. Am. 1999;92:117–129. [Google Scholar]
- Olsen B, Duffy D.C, Jaenson T.G.T, Gylfe A, Bonnedahl J, Bergström S. Transhemispheric exchange of Lyme disease spirochetes by seabirds. J. Clin. Microbiol. 1995;33:3270–3274. doi: 10.1128/jcm.33.12.3270-3274.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B, Jaenson T.G.T, Noppa L, Bunikis J, Bergström S. A Lyme borreliosis cycle in seabirds and Ixodes uriae ticks. Nature. 1993;362:340–342. doi: 10.1038/362340a0. [DOI] [PubMed] [Google Scholar]
- Paetkau D, Strobeck C. The molecular basis and evolutionary history of a microsatellite null allele in bears. Mol. Ecol. 1995;4:519–520. doi: 10.1111/j.1365-294x.1995.tb00248.x. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP(version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 1995;86:248–249. [Google Scholar]
- Rice W.R. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Thompson J.N. The evolution of species interactions. Sci. 1999a;284:2116–2118. doi: 10.1126/science.284.5423.2116. 10.1126/science.284.5423.2116 [DOI] [PubMed] [Google Scholar]
- Thompson J.N. Specific hypotheses on the geographic mosaic of coevolution. Am. Nat. 1999b;153(Suppl.):S1–S14. 10.1086/303208 [Google Scholar]
- Thompson J.N, Cunningham B.M. Geographic structure and dynamics of coevolutionary selection. Nature. 2002;417:735–738. doi: 10.1038/nature00810. 10.1038/nature00810 [DOI] [PubMed] [Google Scholar]
- Warham J. The crested penguins. In: Stonehouse B, editor. The biology of penguins. MacMillan Press Ltd; London: 1975. pp. 189–269. [Google Scholar]
- Weiblen G.D, Bush G.L. Speciation in fig pollinators and parasites. Mol. Ecol. 2002;11:1573–1578. doi: 10.1046/j.1365-294x.2002.01529.x. 10.1046/j.1365-294X.2002.01529.x [DOI] [PubMed] [Google Scholar]
- Weir B.S, Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wikel S.K. Host immunity to ticks. Annu. Rev. Entomol. 1996;41:1–22. doi: 10.1146/annurev.en.41.010196.000245. 10.1146/annurev.en.41.010196.000245 [DOI] [PubMed] [Google Scholar]
- Woolhouse M.E.J, Webster J.P, Domingo E, Charlesworth B, Levin B.R. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet. 2002;32:569–577. doi: 10.1038/ng1202-569. 10.1038/ng1202-569 [DOI] [PubMed] [Google Scholar]