Abstract
Fission yeast Cut5/Rad4 and its budding yeast homolog Dpb11 are required for both DNA replication and the S-phase checkpoint. Here, we have investigated the role of the Xenopus homolog of Cut5 in the initiation of DNA replication using Xenopus egg extracts. Xenopus Cut5, which shows sequence similarity to DmMus101 and HsTopBP1, is essential for DNA replication in the egg extracts. It is required for the chromatin binding of Cdc45 and DNA polymerases, but not for the formation of pre-replicative complexes or the elongation stage of DNA replication. The chromatin binding of Cut5 consists of two distinct modes. S-phase cyclin-dependent kinase (S-CDK)-independent binding is sufficient for DNA replication while S-CDK-dependent binding is dispensable. Further, S-CDK acts after the chromatin binding of Cut5 and before the binding of Cdc45. These results demonstrate that the chromatin binding of Cut5 is required for the action of S-CDK, which in turn triggers the formation of pre-initiation complexes of DNA replication.
Keywords: Cdc45/Cut5/DNA replication/Dpb11/S-CDK
Introduction
In eukaryotic cells, the regulated assembly of replication factors onto origins is crucial for the control of the initiation of DNA replication. At the end of M-phase and during G1-phase of the cell cycle, origin recognition complex (ORC), Cdc6, Cdt1 and minichromosome maintenance (MCM) proteins are sequentially assembled onto each replication origin, leading to the formation of pre-replicative complexes (pre-RCs) (reviewed in Nishitani and Lygerou, 2002). At the onset of S-phase, pre-RCs are activated through the action of two kinds of protein kinase, S-phase cyclin-dependent kinase (S-CDK) and Dbf4-dependent Cdc7 kinase (DDK). This converts the pre-RCs into pre-initiation complexes (pre-ICs) (Zou and Stillman, 2000). The exact roles of S-CDK and DDK are not fully understood but accumulating evidence suggests that S-CDK and DDK target specific replication proteins such as Sld2/Drc1 (Masumoto et al., 2002; Noguchi et al., 2002) and MCM proteins (Jiang et al., 1999). Upon the phosphorylation of target proteins, Cdc45 becomes associated with MCM proteins in pre-RCs, leading to the formation of pre-ICs and the unwinding of origins (Mimura and Takisawa, 1998; Zou and Stillman, 1998; Walter and Newport, 2000). Replication protein A (RPA) and DNA polymerase α are sequentially recruited (Tanaka and Nasmyth, 1998; Mimura et al., 2000; Walter and Newport, 2000), and DNA polymerase ε is independently recruited (Mimura et al., 2000) onto the unwound DNA, forming initiation complexes on the origins. After initiation, S-CDK is no longer required for the elongation of DNA replication (Strausfeld et al., 1994; Walter and Newport, 1997) but it serves to inhibit re-initiation during S phase through multiple mechanisms (Nguyen et al., 2001), ensuring that DNA replication occurs only once in each cell cycle.
In addition to the known components of pre-RCs, recent studies have revealed additional factors necessary for the initiation of DNA replication (Takisawa et al., 2000; Bell and Dutta, 2002). In budding yeast, Dpb11 has been shown to play a pivotal role in the loading of DNA polymerases onto replication origins (Masumoto et al., 2000). Dpb11 was originally identified as a multicopy suppressor of the pol2-12 and dpb2-1 mutations, which affect the catalytic and the second largest subunits of DNA polymerase ε, respectively (Araki et al., 1995). The identification of five essential genes called SLD2–5 (synthetically lethal with dpb11-1) further supports the importance of Dpb11 in DNA replication (Kamimura et al., 1998). The fission yeast protein Cut5/Rad4 is a functional homolog of Dpb11 and both mutants display similar phenotypes of abnormal cell division with incomplete DNA replication, suggesting that Cut5/Rad4 and Dpb11 are required for DNA replication itself as well as for the replication checkpoint (Saka and Yanagida, 1993; Saka et al., 1994; Araki et al., 1995). Further, the association between Dpb11/Cut5 and Sld2/Drc1 is required for DNA replication (Kamimura et al., 1998; Wang and Elledge, 1999) and is under the control of CDK (Masumoto et al., 2002; Noguchi et al., 2002). Here, we will use the single name Cut5 to refer to Cut5/Rad4/Dpb11 because Cut5 was the first factor identified as an essential replication protein (Saka and Yanagida, 1993). Human TopBP1 and Drosophila Mus101 share sequence similarities with Cut5 (Yamane et al., 1997; Yamamoto et al., 2000) and TopBP1 is also involved in DNA replication and the DNA damage response (Mäkiniemi et al., 2001; Yamane et al., 2002). Some mutants of Mus101 display mutagen sensitivities, defects in DNA synthesis and chromosome instability (Yamamoto et al., 2000). These results strongly suggest that TopBP1 and Mus101 are the functional homologs of yeast Cut5 but the role of Cut5 in the initiation of DNA replication is only poorly understood in higher eukaryotes.
In this study, we have identified and characterized a Xenopus homolog of Cut5 using the cell-free system of Xenopus egg extracts. We found that Cut5 is required for the association of Cdc45 with chromatin. Furthermore, Cut5 is required for S-CDK to function in the initiation reaction. These results suggest that Cut5 plays an essential role in the formation of pre-ICs upon the action of S-CDK.
Results
Identification of Xenopus Cut5
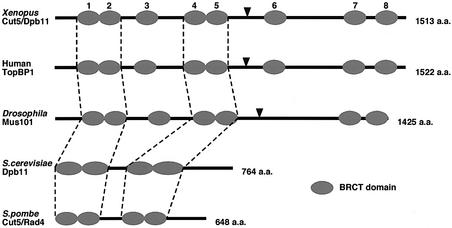
To isolate Xenopus Cut5, we searched the EST database for a Xenopus homolog of human TopBP1, a putative human Cut5, and obtained a Xenopus EST sequence with high homology (accession No. AW635109). Using the EST sequence as a probe, we screened a Xenopus oocyte cDNA library and isolated a Xenopus homolog of TopBP1 (accession No. AB091779). Figure 1 compares the primary structure of the predicted Xenopus protein with those of human TopBP1, Drosophila Mus101 and yeast Cut5/Dpb11. The Xenopus protein consists of 1513 amino acids and contains eight copies of BRCA1 C-terminus (BRCT) domains, which are thought to be involved in protein–protein interactions. Four of the BRCT domains, indicated by dotted lines, are highly conserved from yeast to human while the others are unique to higher eukaryotes. The overall identities between Xenopus and Schizo saccharomyces pombe, Saccharomyces cerevisiae, Drosophila and human are 15%, 16%, 24% and 63%, respectively. These structural features show that the isolated clone is a Xenopus homolog of TopBP1 and Mus101.
Fig. 1. Schematic comparison of Cut5/Dpb11 homologs. Elliptical symbols represent BRCT domains. Dotted lines indicate highly conserved BRCT domains from yeast to human. BRCT domains 1–2 of the Xenopus protein share sequence similarity with the homologs in human (98%), Drosophila (54%), S.cerevisiae (30%) and S.pombe (47%), and BRCT domains 4–5 of the Xenopus protein share similarity with BRCT domains 4–5 of human (91%) and Drosophila (31%), and with BRCT domains 3–4 of S.cerevisiae (34%) and S.pombe (37%). Arrowheads indicate putative target sites of CDK conserved among Xenopus, human and Drosophila. Total numbers of amino acid residues are indicated at the right side.
A previous study showed that TopBP1 is unable to suppress temperature-sensitive mutants of either budding yeast Dpb11 or fission yeast Cut5 (Mäkiniemi et al., 2001). We have examined the suppression of these yeast mutants with the full-length and N-terminal half (amino acids 1–761, a conserved part of Cut5 homologs) of Xenopus Cut5. The overexpression of either protein failed to suppress these yeast mutants (data not shown; see also Materials and methods). This failure in functional complementation is not surprising if we consider the sequence differences between these gene products. Nevertheless, functional similarity among Cut5 homologs supports the view that TopBP1-related proteins in higher eukaryotes are the counterparts of yeast Cut5/Dpb11 (see also Discussion) and we therefore designate the Xenopus homolog of TopBP1, Xenopus Cut5.
Role of Xenopus Cut5 in DNA replication
To investigate the role of Xenopus Cut5 in DNA replication, we expressed His-tagged Xenopus Cut5 in insect cells and produced antibodies against the recombinant protein. The affinity-purified antibody recognized a single polypeptide with an apparent molecular mass of 180 kDa in interphase egg extracts, and also recognized recombinant Cut5 with a similar mobility on SDS–PAGE (Figure 2A).

Fig. 2. Time-courses of the chromatin binding of Cut5 and other replication proteins. (A) Specificity of anti-Cut5 antibody. S-phase egg extracts (1.5 µl) and 125 ng of recombinant Xenopus Cut5 protein (rec.Cut5) were resolved with SDS–PAGE and immunoblotted with anti-Xenopus Cut5 antibody. (B) Sperm nuclei were incubated in 50 µl of S-phase egg extract for the times indicated at 23°C. Chromatin fractions were isolated by centrifugation through a 10% sucrose layer and the isolated chromatin fractions and 1.5 µl of the extracts were resolved by SDS–PAGE, then immunoblotted with antibodies against the various replication proteins indicated.
Using this antibody, we first examined the time-course of Cut5 association with chromatin during DNA replication in egg extracts. Upon addition of sperm chromatin to the extracts, components of pre-RCs such as Orc1 and Mcm3 were assembled onto chromatin within 15 min (Figure 2B) and nuclear structures were formed around the chromatin within 30 min, allowing replication initiation. Various replication proteins involved in the formation of initiation complexes, such as Cdc45 and DNA polymerases α and ε, were bound to chromatin upon initiation (30 min) and were gradually dissociated from the chromatin during the progression of DNA replication, while Orc1 binding remained constant throughout the incubation. Cut5 showed essentially the same behavior as the replication proteins involved in the initiation complex (Figure 2B) but we also detected a low level of chromatin binding before initiation (15 min). These results show that Xenopus Cut5 is a chromatin binding protein, possibly involved in DNA replication.
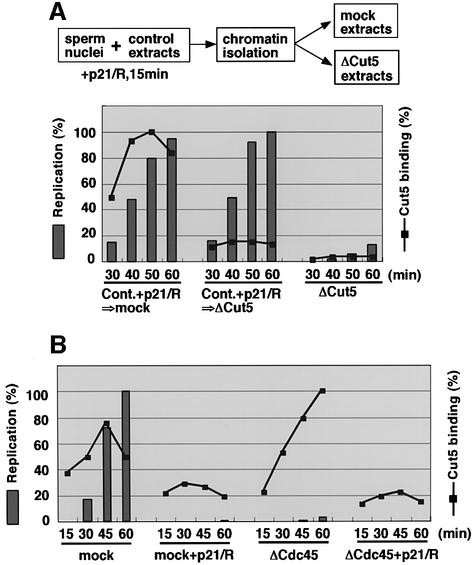
To determine whether or not Cut5 is required for DNA replication, we first depleted Cut5 from the extracts using anti-Xenopus Cut5 antibody (Figure 3A). Endogenous Cut5 could be almost completely eliminated from the egg extracts with the antibody treatment but other replication proteins such as Mcm3 and Cdc45 remained in the depleted extracts (Figure 3A). Addition of 5 ng of recombinant Cut5 to 1 µl of the depleted extracts gave an immunostaining signal similar to that of mock-depleted extracts, indicating that the concentration of endogenous Cut5 is ∼5 µg/ml. We next examined the replication activity of Cut5-depleted extracts (Figure 3B and C). DNA replication activity decreased markedly upon the depletion of Cut5, to ∼14% of the activity of mock-depleted extract (60 min). Recombinant Cut5 could rescue the decreased activity and restore it to >90% of control levels. These results therefore show that the replication defect in the Cut5-depleted extract is due to the selective removal of Cut5, and that no additional factor is required for its restoration.
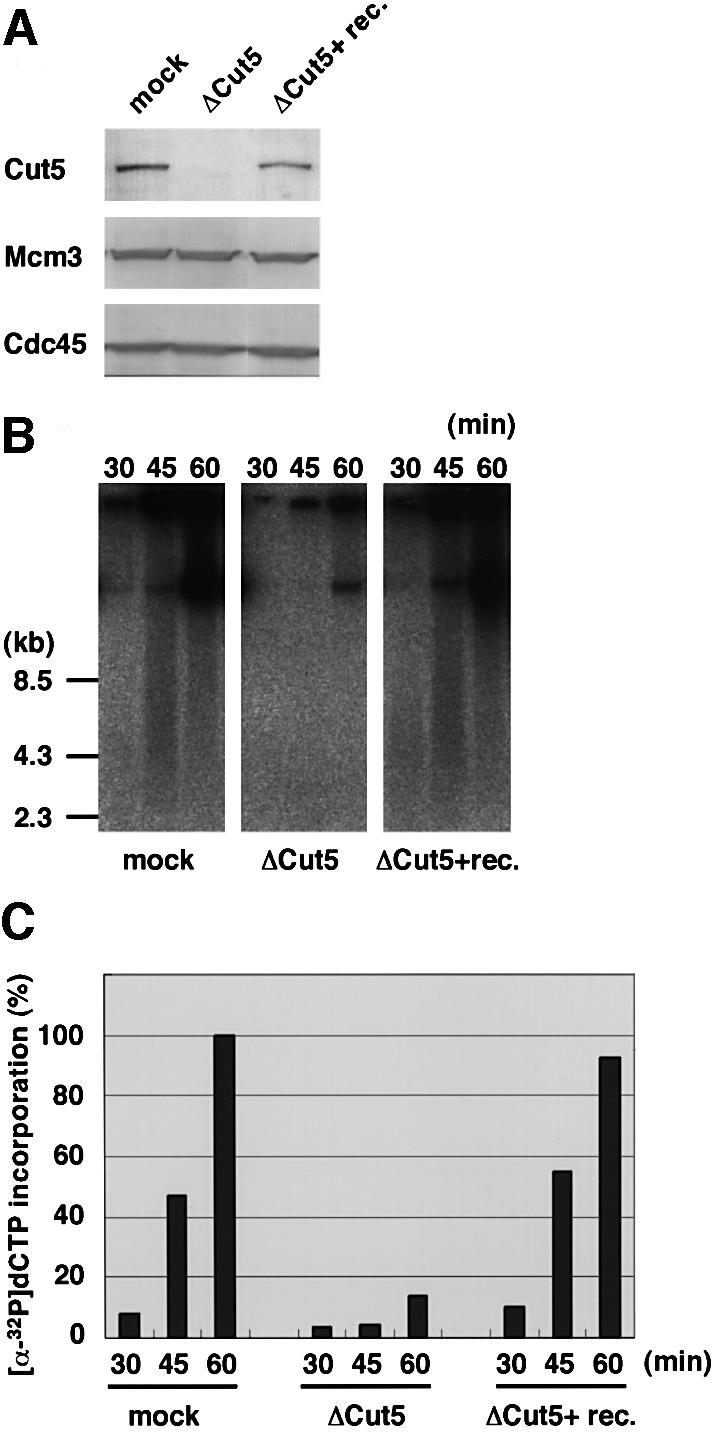
Fig. 3. Requirement of Cut5 for DNA replication. (A) Immunodepletion of Cut5 from S-phase egg extracts. The egg extracts were treated with control and anti-Cut5 antibodies bound to a protein A matrix. Aliquots of mock-depleted (mock) and Cut5-depleted extracts with (ΔCut5+ rec.Cut5) or without (ΔCut5) 5 ng/µl of recombinant Cut5 were resolved by SDS–PAGE, then immunoblotted with antibodies against the proteins indicated in the figure. (B) Replication activity of Cut5- depleted extracts. Sperm nuclei were incubated in the treated egg extracts for the indicated times at 23°C. Replication activity was monitored as the incorporation of [α-32P]dCTP into sperm DNA. Replication products were subjected to agarose gel electrophoresis followed by autoradiography. (C) Relative intensities of autoradiography were analyzed and plotted against time, taking the highest value of the mock-depleted extracts as 100%.
Analysis of replication products by agarose gel electrophoresis further suggests that Cut5 is required for the initiation stage of DNA replication (Figure 3B). In mock-depleted extracts, replication products consisted mainly of high-molecular-weight DNA (Blow and Laskey, 1986). With Cut5-depleted extracts, similar sizes but much smaller amounts of the products were observed and low-molecular-weight replication products of <10 kb did not accumulate, suggesting that the elongation reaction was not affected in the depleted extracts (Waga et al., 2001). To test further whether Cut5 is required for the elongation stage of DNA replication, we examined directly the elongation activity of isolated replicating chromatin fractions in the presence of CDK inhibitors, which inhibit initiation but not elongation reactions (Strausfeld et al., 1994; Walter and Newport, 1997). Replicating chromatin fractions were isolated from the extracts by first incubating sperm chromatin for 40 min to allow maximal loading of DNA polymerases onto chromatin (see Figure 2B). When the chromatin fractions were then washed with a buffer containing 0.1 M NaCl and NP-40, almost all Cut5 bound to the chromatin was removed but MCM proteins and Cdc45, both of which are required for the elongation stage of DNA replication, remained on the chromatin (Figure 4A). NaCl treatment did not affect the replication activity of the chromatin in mock-depleted extracts, indicating that elongation is not affected by NaCl treatment (Figure 4B, mock). Furthermore, we found that the elongation activity of Cut5-depleted chromatin fractions in Cut5-depleted extracts (+NaCl chromatin) was essentially the same as in mock-depleted extracts (Figure 4B, right panel). In contrast, the replication of sperm chromatin in Cut5-depleted extracts in the absence of inhibitors but without 40 min pre-incubation in control extracts was <20% of that of replicating chromatin (data not shown, see also Figure 3C). These results suggest that Cut5 is not required for the elongation activity of replicating chromatin.
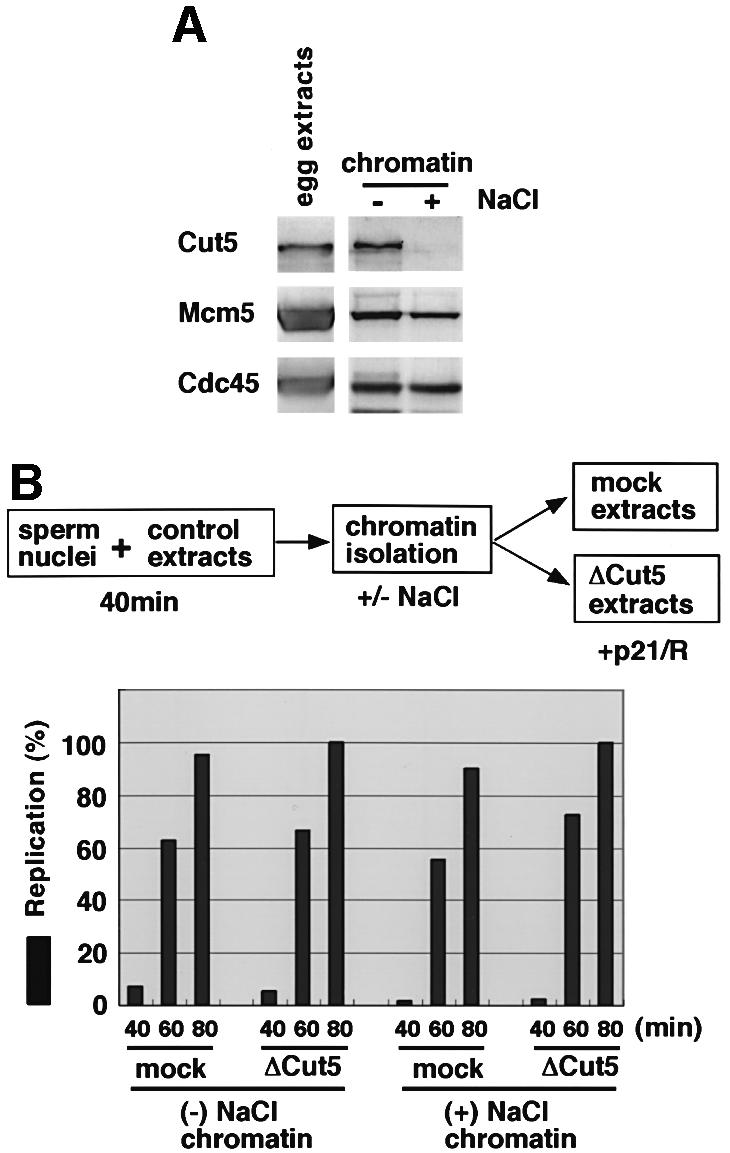
Fig. 4. Requirement of Cut5 for elongation stages of DNA replication. (A) Selective removal of Cut5 from chromatin. Sperm nuclei were incubated in 50 µl of control egg extracts for 40 min at 23°C. The egg extracts (1.5 µl) and the chromatin fractions, isolated and washed with EB containing 0.0025% NP-40 in the absence (–) or presence (+) of 0.1 M NaCl, were subjected to immunoblotting. (B) Elongation activity of the isolated replicating chromatin fractions. The chromatin fractions prepared as in (A) were resuspended and incubated in mock- and Cut5-depleted egg extracts in the presence of 50 µg/ml GST–p21 and 100 µM roscovitine (+p21/R). Elongation activity was detected as the incorporation of Cy3-dCTP into chromatin DNA. The fluorescence intensities of 40 nuclei were analyzed. Relative average intensities per nucleus were then plotted against time, taking the maximal value as 100%.
Role of Cut5 in the formation of the initiation complex
In budding yeast, Cdc45 and Dpb11/Cut5 play essential roles in the loading of DNA polymerases onto chromatin (see Introduction). However, the precise roles of Dpb11 and Cdc45 in replication initiation have not yet been clarified. We therefore examined the role of Xenopus Cut5 in the loading of DNA polymerases and the mutual dependence of Cut5 and Cdc45 for their chromatin binding. Upon depleting Cut5 and Cdc45 from the extracts, each protein was specifically eliminated while other replication proteins involved in the initiation complex remained in the extracts (Figure 5A). Using these extracts, sperm nuclei were incubated for 40 min and the isolated chromatin fractions were subsequently examined by immunoblotting (Figure 5A, chromatin). After 40 min incubation, DNA replication had been initiated in the mock-depleted extracts and all of the replication proteins so far examined were found to be associated with the chromatin. In the Cut5-depleted extract, ORC and MCM proteins were bound to chromatin at a similar level but neither Cdc45 nor DNA polymerases were detected in the chromatin. In Cdc45-depleted extracts, the binding of DNA polymerases α and ε was suppressed without affecting the formation of pre-RCs. In addition, Cut5 was bound to chromatin at a similar level to that of the mock-depleted extracts. These results show that Cut5 is indispensable for the chromatin binding of Cdc45 and DNA polymerases, but is dispensable for the assembly of pre-RCs, as monitored by the chromatin binding of ORC and MCM proteins.
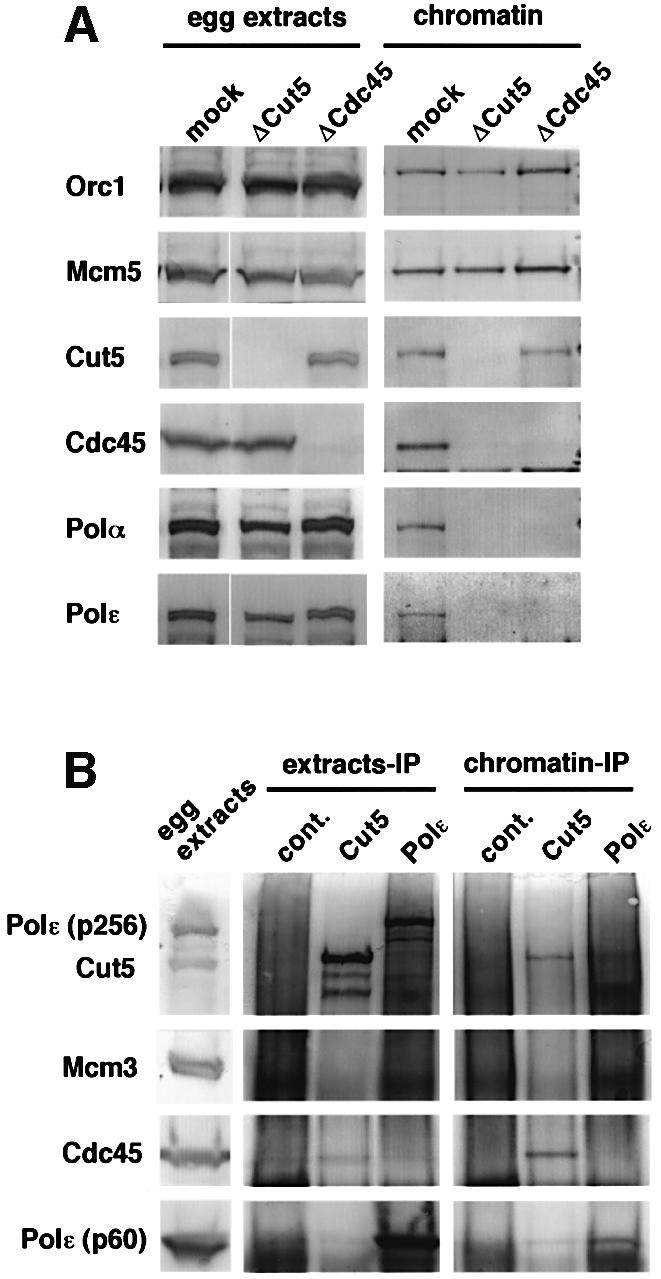
Fig. 5. Requirement of Cut5 for the loading of Cdc45 onto chromatin. (A) Sperm nuclei were incubated in mock-, Cut5- and Cdc45-depleted egg extracts for 40 min at 23°C. For each extract, 1.5 µl depleted extract and the isolated chromatin fractions (chromatin) from 50 µl extracts were resolved with SDS–PAGE and immunoblotted. (B) Co-immunoprecipitation of Cdc45 with Cut5 on replicating chromatin. Sperm nuclei were incubated in S-phase egg extract in the presence of 50 µg/ml aphidicolin for 50 min at 23°C. Chromatin DNA was fragmented by sonication and the fragmented chromatin fractions were clarified by centrifugation. The egg extracts and the fragmented chromatin fractions (chromatin-IP) were immunoprecipitated with antibodies as indicated, then the immunoprecipitates were resolved with SDS–PAGE and immunoblotted.
The sequential binding of Cut5 and Cdc45 suggests a possible interaction between these two proteins. In addition, previous studies suggest an interaction between Dpb11/TopBP1 and DNA polymerase ε (Masumoto et al., 2000; Mäkiniemi et al., 2001). We therefore examined physical interactions between Cut5 and other replication proteins. When the egg extracts were immunoprecipitated with anti-Cut5 antibody, we could detect Cut5 and a small amount of Cdc45 in the immunoprecipitates but not other replication proteins, including Mcm3 and DNA polymerase ε (Figure 5B). With antibodies against the 60 kDa subunit of the polymerase, the co-immunoprecipitation of the largest subunit of the polymerase (p256) was detected but not the other proteins, including Cut5. To examine the interaction after the assembly onto chromatin, sperm nuclei were incubated for 50 min in the presence of aphidicolin, an inhibitor of DNA polymerases α, δ and ε, which allowed the maximal binding of Cut5, Cdc45, DNA polymerases α and ε onto chromatin (Mimura et al., 2000). When the fragmented chromatin fractions were then immunoprecipitated with anti-Cut5 antibody, Cdc45 and a very small amount of the 60 kDa subunit of the polymerase were co-precipitated, but not the largest subunit of the polymerase (Figure 5B, chromatin-IP). Using a parallel assay with anti-60 kDa antibody, we could likewise detect only the 60 kDa subunit, suggesting that the largest subunit is rapidly degraded during the isolation. These results show, however, that Cut5 interacts with Cdc45.
S-CDK-dependent and -independent binding of Cut5 to chromatin
To further investigate the role of Cut5, we examined the time-course of the chromatin binding of Cut5 under various conditions using immunofluorescence microscopy (Figure 6A). Before nuclear formation, which was confirmed by the absence of nuclear structures around chromatin DNA under phase contrast microscopy, weak signals of Cut5 were detected on chromatin DNA and focus structures were also observed on some fractions of chromatin (Figure 6A, control 15 min). Only a background signal was detected on DNA from Cut5-depleted extracts (Figure 6A, ΔCut5). The chromatin binding of Cut5 before initiation was not affected by either addition of p21 and roscovitine or geminin, both of which inhibit the replication activity of the extracts completely (Figure 6C). Upon initiation of DNA replication, the chromatin binding of Cut5 was greatly increased but it was almost completely suppressed by addition of p21 and roscovitine or geminin (Figure 6A). These results show that Cut5 binds to chromatin in two distinct modes. The first mode of binding was identified as weak Cut5 signals associated with chromatin even in the absence of S-CDK activity, or in the presence of geminin, which inhibits the formation of pre-RCs through its interaction with Cdt1 (Wohlschlegel et al., 2000; Tada et al., 2001). The second mode of binding was identified as strong signals dependent on pre-RC formation and S-CDK activity.
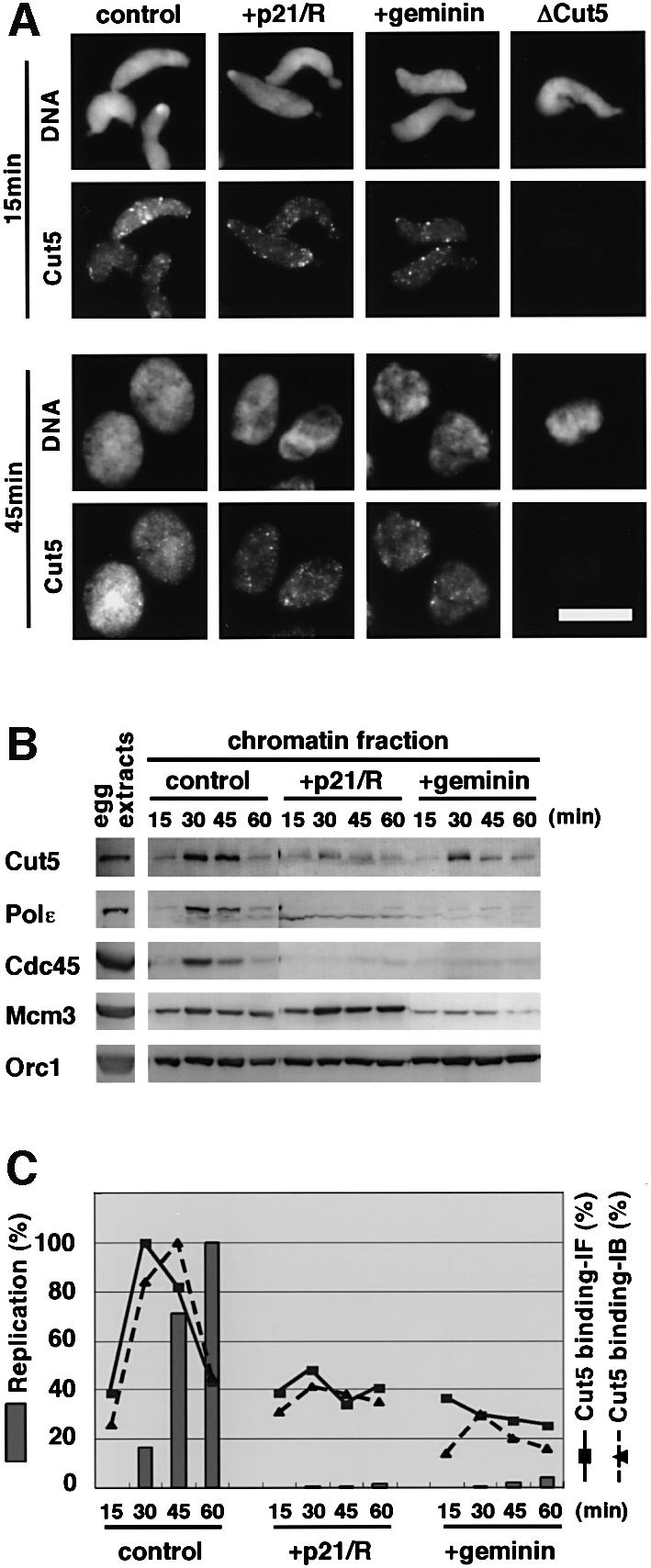
Fig. 6. CDK-dependent and -independent binding of Cut5 to chromatin. (A) Immunofluorescent detection of Cut5 binding. Sperm nuclei were incubated in S-phase egg extracts in the absence (control) or in the presence of 50 µg/ml GST–p21 and 100 µM roscovitine (+p21/rosc.) or 15 µg/ml GST–geminin (+geminin) for the times indicated at 23°C. Cut5-depleted egg extracts (ΔCut5) were used as a negative control of immunofluorescence. Samples were treated with 0.25% NP-40 and then fixed with 3.7% formaldehyde. Nuclear localization of Cut5 was visualized with rabbit anti-Cut5 antibody followed by Alexa 488-labelled anti-rabbit IgG. DNA was visualized with Hoechst 33258. Scale bar 10 µm. (B) Time-course of chromatin binding of various replication proteins. Sperm nuclei were incubated in 50 µl of the extracts for the times indicated in the absence (control) or presence of p21 and roscovitine (+p21/rosc.) or 15 µg/ml GST–geminin (+geminin). One microliter of egg extract and the isolated chromatin fractions were resolved by SDS–PAGE, then immunoblotted with antibodies against various replication proteins. (C) Time-courses of DNA replication and chromatin binding of Cut5. Fluorescence images captured as in (A) and the immunoblotted bands in (B) were quantified in order to estimate the amounts of chromatin-bound Cut5. Replication activity was detected as the incorporation of Cy3-dCTP into DNA. The fluorescence intensities of 40 nuclei were analyzed using NIH Image software. Relative average intensities per nucleus were then plotted against time, taking the maximal value as 100% (Replication and Cut5 binding-IF). For immunoblotting, the intensities of immunoblotted bands of Cut5 were normalized against those of Orc1, a loading control for chromatin fractions, and ratios of the amounts of Cut5 to Orc1 were then plotted against time, taking the maximal value as 100% (Cut5 binding-IB).
To compare the two modes of Cut5 binding more quantitatively, the amounts of Cut5 bound to chromatin were estimated based on immunofluorescence intensities (Figure 6C). In the control egg extract, the amount of Cut5 binding at 30 min had increased ∼3-fold from the initial level (15 min) and it then decreased back to the initial level at 60 min, at which point replication was almost complete. In contrast, the amount of Cut5 remained at the initial level throughout the incubation with p21 and roscovitine or geminin. Similar results were also obtained from an immunoblotting analysis of chromatin fractions (Figure 6B, Cut5) and the amounts of chromatin-bound Cut5 estimated from the blotting data fit well with those from the immunofluorescence data (Figure 6C). In accordance with the observation of DNA replication activity by immunofluorescence, both DNA polymerase ε and Cdc45 bound to chromatin had returned to nearly basal levels at 60 min, when replication was almost complete. In the presence of geminin, the chromatin binding of Mcm3 was inhibited but Orc1 and Cut5 were not affected (Figure 6B). These results show that the low level of Cut5 binding is independent of S-CDK and pre-RC formation.
Thus, Cut5 binds to chromatin in both S-CDK- independent and S-CDK-dependent modes but it is not known which mode is required for DNA replication. We first examined whether or not S-CDK-independent binding of Cut5 plays any role in DNA replication. In order to restrict Cut5 to its first, S-CDK-independent, binding mode, sperm nuclei were incubated in egg extract with p21 and roscovitine for 15 min and the chromatin fractions were then isolated from the extract. The isolated chromatin fractions were transferred to and incubated in mock- and Cut5-depleted egg extracts, and DNA replication activities were measured (Figure 7A). As shown in the figure, both extracts supported DNA replication to a similar level but Cut5 binding remained at a constant low level in the Cut5-depleted extract (Figure 7A, compare left and middle). As a negative control, sperm nuclei incubated in the Cut5-depleted extract throughout showed only marginal replication activity and the level of Cut5 binding was as low as 20% of the control level of CDK-independent binding (Figure 7A, right), indicating that Cut5 was almost completely depleted from this extract. These results show that the quantity of Cut5 bound in the absence of S-CDK is sufficient to support full DNA replication.
Fig. 7. Requirement of chromatin binding of Cut5 for DNA replication. (A) Sperm nuclei were pre-incubated in S-phase egg extract in the presence of 50 µg/ml p21 and 100 µM roscovitine (p21/R) for 15 min at 23°C. The chromatin fractions were then isolated and incubated in mock- (Cont.+p21/R→mock) and Cut5-depleted (Cont.+p21/R→ ΔCut5) egg extracts at 23°C. At the times indicated, which include 15 min pre-incubation, samples were treated with NP-40, fixed with formaldehyde, and the amount of Cut5 bound to chromatin and the incorporation of Cy3-dCTP were analyzed as described in the legend for Figure 6. As a negative control, sperm nuclei were incubated in the Cut5-depleted egg extract throughout the time-course (ΔCut5). (B) S-CDK-dependent binding of Cut5 to chromatin in the absence of Cdc45. Sperm nuclei were incubated in mock- and Cdc45-depleted egg extracts with or without p21/roscovitine at 23°C. After incubation for the times indicated, samples were analyzed as in (A).
Cut5 binding to chromatin increased only after the action of S-CDK (Figure 6). Since S-CDK activity is required for the binding of Cdc45 to chromatin, a question remains as to whether S-CDK-dependent binding of Cut5 requires Cdc45 or not. We therefore examined the binding of Cut5 in the absence and presence of Cdc45. With mock-depleted extracts, Cut5 binding increased in an S-CDK-dependent manner (Figure 7B, compare mock and mock+ p21/R). Similar results were obtained with Cdc45-depleted extracts (Figure 7B, ΔCdc45 and ΔCdc45+p21/R). The Cut5 binding in Cdc45-depleted extracts increased with incubation time and reached a higher level than in the mock-depleted extracts (Figure 7B, mock and ΔCdc45). These results show that Cdc45 is dispensable for the S-CDK-dependent binding of Cut5. Taken together, the modes of Cut5 binding to chromatin are summarized as follows. Initially, small but significant amounts of Cut5 bind to chromatin before the activation of S-CDK and this is sufficient for DNA replication. The majority of Cut5 binding occurs after the activation of S-CDK, independently of Cdc45 and DNA replication, and this increased amount of binding is dispensable for DNA replication.
Determination of the order of reaction of S-CDK, Cut5 and Cdc45 in the initiation of DNA replication
Both Cut5 and S-CDK are required for the initiation of DNA replication, and S-CDK-independent binding of Cut5 is fully responsible for this initiation. This raises two possibilities as to the function of Cut5: one is that both Cut5-dependent and S-CDK-dependent processes are separately required for initiation, the other is that Cut5 binding is required for the action of S-CDK in initiation. In order to distinguish these two possibilities, we examined whether or not S-CDK could act in the absence of Cut5. For this purpose, we first incubated sperm nuclei in Cut5-depleted extracts in the absence of CDK inhibitors for 40 min. Since the addition of CDK inhibitors after 40 min incubation could only slightly inhibit the replication activity of mock-depleted extracts (data not shown, see also Strausfeld et al., 1994), it is reasonable to assume that S-CDK function has been almost executed after 40 min incubation. Next, the nuclear fractions were isolated and transferred to egg extracts in the presence or absence of S-CDK inhibitors, and DNA replication activity was examined (Figure 8A). In the absence of the inhibitors, we could detect as much DNA replication activity as was observed in the control extracts (data not shown), and the chromatin association of Cut5 also increased with time (Figure 8A, ΔCut5→Cont.). On the other hand, in the presence of CDK inhibitors during the incubation, DNA replication was reduced to as little as 20% and Cut5 binding to <50% of the levels observed without the inhibitors (compare Figure 8A, ΔCut5→Cont. and ΔCut5→Cont.+p21/R). A similarly low level of association of Cut5 was observed when the inhibitors were included in the extracts throughout the experiment (Figure 8A, ΔCut5+p21/R→Cont.+p21/R). These results show that S-CDK activity is still required even after pre-incubating chromatin in Cut5-depleted extracts in the presence of CDK activity.
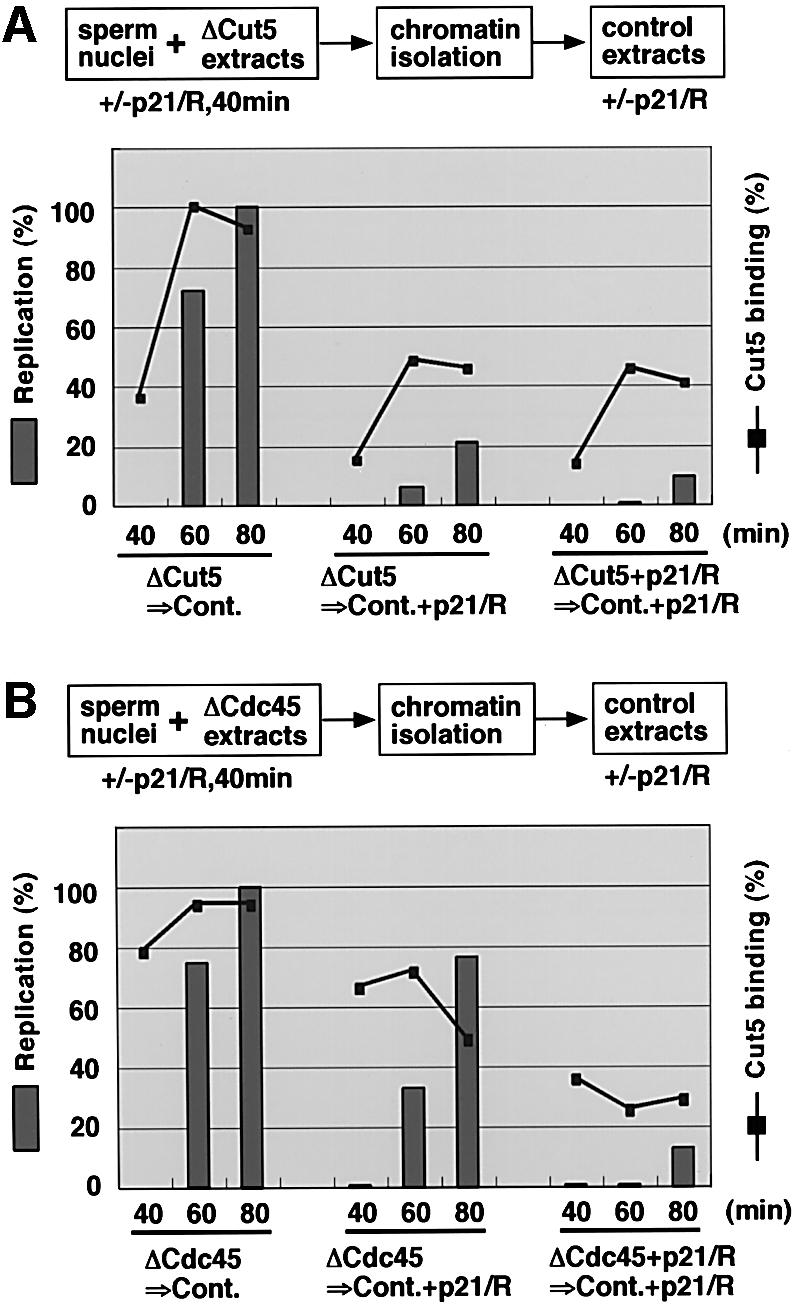
Fig. 8. Requirement of Cut5 and Cdc45 for the action of S-CDK. (A) Sperm nuclei were pre-incubated in Cut5-depleted egg extracts in the presence or absence of 50 µg/ml p21 and 100 µM roscovitine (p21/R) for 40 min at 23°C. The chromatin fractions were isolated and incubated in egg extracts in the absence (Cont.) or presence of p21/ roscovitine (Cont.+p21/R) at 23°C. After incubation for the times indicated, which include 40 min pre-incubation time, samples were treated with NP-40 and fixed with formaldehyde. The amount of Cut5 bound to chromatin and the incorporation of Cy3-dCTP were analyzed as described in the legend for Figure 6. (B) Similar experiments to those in (A) were performed, using Cdc45-depleted extracts instead of Cut5-depleted extracts.
Both S-CDK and Cut5 are required for the loading of Cdc45 but it is not known whether S-CDK function is required simultaneously with Cdc45 or not. In order to clarify this point, sperm nuclei were incubated in Cdc45-depleted extracts for 40 min. The chromatin fractions were then isolated, transferred to egg extracts in the presence or absence of the CDK inhibitors, and DNA replication activities were measured (Figure 8B). In the control experiment without the inhibitors, we found high activity of DNA replication and high levels of Cut5 binding to chromatin during the incubation (Figure 8B, ΔCdc45→ Cont.). Surprisingly, even in the presence of the inhibitors, we could detect DNA replication activity as high as 70% of control levels (Figure 8B, ΔCdc45→Cont. and ΔCdc45→Cont.+p21/R). However, DNA replication was almost completely inhibited when CDK activity was inhibited throughout the experiment (Figure 8B, ΔCdc45+p21/R→Cont.+p21/R). These results show that S-CDK could act in the absence of Cdc45. In other words, Cdc45 functions after the action of S-CDK.
Discussion
In this paper, we have identified Xenopus Cut5 and characterized its function using the cell-free replication system of Xenopus egg extracts. Major features of the present findings are as follows. First, we found that Cut5 is required for the loading of Cdc45 onto chromatin, which eventually leads to the loading of replicative DNA polymerases, but not for the elongation stages of DNA replication. Secondly, we found that Cut5 binds to chromatin in two distinct modes—one is S-CDK-independent and the other S-CDK-dependent—and that the quantity of Cut5 bound in the former mode is sufficient to support full replication. Thirdly, we found that S-CDK acts after the binding of Cut5 and before the binding of Cdc45 to chromatin. These results demonstrate that Cut5 plays a crucial role in S-CDK-dependent formation of the pre-initiation complex of DNA replication.
Essential role of Cut5 in the initiation of DNA replication
Previous studies have shown that Cut5 homologs, including SpCut5, ScDpb11 and HsTopBP1, are implicated in DNA replication (Saka and Yanagida, 1993; Araki et al., 1995; Mäkiniemi et al., 2001). In addition, a recent report on the budding yeast dpb11-1 mutant suggests that Dpb11 plays an important role in the loading of polymerases onto replication origins (Masumoto et al., 2000). Here, we found that Xenopus Cut5 is essential for DNA replication in Xenopus egg extracts (Figure 3), and that it is required for the loading of polymerases α and ε (Figure 5A). These results confirm the universal role of Cut5 in replication initiation. We also found that Cut5 is required for the binding of Cdc45 to chromatin, while Cdc45 is not required for Cut5 binding. Since Cdc45 is required for polymerase loading (Mimura and Takisawa, 1998; Aparicio et al., 1999; Mimura et al., 2000), these results suggest the sequential binding of Cut5, Cdc45 and DNA polymerases onto chromatin. In contrast, a chromatin immunoprecipitation assay with budding yeast suggested that Cdc45 associates with some early origins during G1 phase (Aparicio et al., 1999) and that Dpb11 becomes associated with origins only at the onset of S-phase (Masumoto et al., 2000), thus supporting the idea that Dpb11 functions after the binding of Cdc45. However, the amount of Cdc45 bound to chromatin increases only at the onset of S-phase (Aparicio et al., 1999; Zou and Stillman, 2000) and Dpb11 is required for the chromatin binding of Cdc45 (Y.-S.Tak and H.Araki, personal communication). This would suggest that Cut5 is required for functional chromatin binding of Cdc45. In accordance with this notion, Xenopus Cut5 binds to chromatin before nuclear formation and in the absence of S-CDK activity, while the binding of Cdc45 is dependent on both these events (Mimura and Takisawa, 1998).
A recent study suggests that Mcm10 is also required for the binding of Cdc45 to chromatin in a nucleus-free system derived from Xenopus egg extracts (Wohlschlegel et al., 2002). In this system, the recruitment of Mcm10 onto chromatin is independent of S-CDK and Cdc7 activities but it is dependent on MCM proteins and occurs only after the addition of nucleoplasmic extract, which may mimic the nuclear formation in our system. It is not known whether Cut5 binding is dependent on Mcm10 or not but since it is apparently independent of pre-RC formation, it is reasonable to assume that Cut5 binding is independent of Mcm10. The genetic interactions between Dpb11 and Mcm10 (Kawasaki et al., 2000) may suggest that Cut5/Dpb11 is required for the loading of Cdc45 in cooperation with Mcm10.
Physical interaction of Dpb11/TopBP1 with DNA polymerase ε suggests that Dpb11/TopBP1 has a direct role in the loading of the polymerase onto unwound origins (Masumoto et al., 2000; Mäkiniemi et al., 2001). We found that only a small amount of the 60 kDa subunit of polymerase ε could be co-immunoprecipitated with Xenopus Cut5 in fragmented chromatin fractions, and that the co-immunoprecipitation of the polymerase with Cut5 could not be detected using the egg extracts (Figure 5B). These results suggest that a physical interaction between Cut5 and DNA polymerase ε is weak and may be transient. Actually, a significant interaction between Dpb11 and the polymerase was observed only after the chemical crosslinking of the proteins bound to chromatin fractions (Masumoto et al., 2000). More importantly, we found the co-immunoprecipitation of Cdc45 and Cut5 from egg extracts as well as chromatin fractions (Figure 5B). Such a physical interaction between Cut5 and Cdc45 suggests that Cut5 plays a direct role in the loading of Cdc45 onto chromatin.
Two modes of Cut5 binding to chromatin
Here we found that Cut5 binds to chromatin in both an S-CDK-dependent and an S-CDK-independent manner (Figure 6). To obtain further insights into the function of Cut5, we estimated the amount of Cut5 bound to chromatin independently of S-CDK. The concentration of Cut5 in the extracts was estimated as 5 ng/µl based on immunoblotting (Figure 3A), and the amount of Cut5 on chromatin (control 15 min) from 50 µl of the egg extracts (4000 nuclei/µl) was calculated to be roughly equal to the amount in 0.3 µl of the extracts (Figure 6B). Given that the Xenopus genome contains 2.9 × 109 bp of DNA (Wohlschlegel et al., 2002), Cut5 binds to chromatin at one molecule/∼104 bp, which is close to the average replicon size (∼10 kb) in Xenopus egg extract (Walter and Newport, 1997; Blow et al., 2001). This result may infer that S-CDK-independent binding is dependent on the ORC. Since recombinant TopBP1 binds to DNA in vitro (Yamane and Tsuruo, 1999), it is now important to clarify whether Cut5 also has the ability to bind to chromatin independently of ORC.
The S-CDK-dependent binding of Cut5 is dispensable for replication activity (Figure 7A) and does not require Cdc45 (Figure 7B), showing that it does not depend on origin firing events, such as the loading of DNA polymerases. Previous studies have shown that Cut5 has dual functions, one for DNA replication and the other for the S-phase checkpoint (Saka and Yanagida, 1993; Saka et al., 1994; Araki et al., 1995; Wang and Elledge, 1999). It is tempting to speculate that the CDK-dependent Cut5 binding may be required for the checkpoint function. Since a defect in Cut5 leads to premature mitosis without DNA replication (Saka and Yanagida, 1993; Araki et al., 1995), Cut5 may monitor entry into S-phase and send signals to prevent premature mitosis.
Sequential action of Cut5, S-CDK and Cdc45 in the formation of pre-ICs
Two kinase activities, DDK and S-CDK, are required for the initiation of DNA replication (Takisawa et al., 2000; Bell and Dutta, 2002). In the present study, we did not examine the role of DDK in the chromatin binding of Cut5. However, DDK associates with chromatin at about the time of nuclear assembly (Jares and Blow, 2000), suggesting that S-CDK-independent binding does not require DDK. Since DDK acts before S-CDK in the Xenopus system (Jares and Blow, 2000; Walter, 2000), our results suggest that DDK and S-CDK both act sequentially after the binding of Cut5. Our results further show that Cut5 but not Cdc45 is required for S-CDK to function in initiation (Figure 8). Since the S-CDK-independent binding of Cut5 is sufficient for initiation (Figure 7A), it is reasonable to assume that Cut5 binds first, then S-CDK functions and finally Cdc45 joins in the formation of pre-ICs. This scenario suggests that the target of S-CDK may be Cut5 itself. Cut5 homologs in metazoans have a conserved CDK target site (see Figure 1), and Xenopus Cut5 is phosphorylated by cyclin A/Cdk2 in vitro (our unpublished observation). However, the mutant Xenopus protein obtained by converting threonine into alanine in the CDK site (amino acid 814) still supported the replication activity of Cut5-depleted extracts (Supple mentary data). In addition, yeast Cut5/Dpb11 does not contain such a CDK target site and no evidence has been obtained for its phosphorylation. Recent studies with yeast demonstrate that Sld2 is a novel target of S-CDK, and that S-CDK-dependent phosphorylation of Sld2/Drc1 positively regulates DNA replication through the association of Sld2/Drc1 with Dpb11/Cut5 (Masumoto et al., 2002; Noguchi et al., 2002). A homolog of Sld2/Drc1 has not been identified in other organisms but the present data suggest that such a homolog is also functioning in higher eukaryotes. Further study of the function of Cut5 will clarify the molecular mechanisms for the action of S-CDK in pre-RC activation in higher eukaryotes.
Materials and methods
Cloning and sequencing of Xenopus Cut5
To clone Xenopus Cut5, we obtained a partial sequence of a potential Xenopus Cut5 (accession No. AW635109) from the EST database by homology searching with human TopBP1. The sequence was amplified from Xenopus oocyte cDNA by PCR using 5′-primer (ATTATACACCAGCAAAGGAATC) and 3′-primer (ATGTTGGTCACAGCACTCAG). The amplified PCR fragment was used as a probe to screen a λZAPII-derived cDNA library of Xenopus oocyte mRNA. From 5 × 105 plaques, we isolated four positive clones for Xenopus Cut5 with insert sizes of ∼1.5 kb, 2 kb, 3 kb and 4.7 kb. The largest clone, of 4.7 kb, was sequenced with an automatic DNA sequencer (ABI, 377). The sequence thus determined encoded a protein lacking the N-terminus so the 5′ region was further expanded using the 5′RACE system (Gibco BRL) from total Xenopus oocyte RNA. The full-length Xenopus Cut5 gene (accession No. AB091779) consisted of 5078 bp containing a 5′UTR of 103 bp, an ORF of 4542 bp and a 3′UTR of 433 bp.
Protein expression and antibody production
A full-length ORF of Xenopus Cut5 was prepared as follows. The plasmid (pBluescript) carrying the cDNA of 4.7 kb (corresponding to amino acids 86–1513) was digested with EcoRI and XhoI and a 4.7 kb fragment was subcloned into 6× His-tag vector pFastBac HTb (Gibco BRL). The plasmid obtained was further digested with NcoI and ApaI and the digested plasmid containing the ORF, corresponding to amino acids 310–1513, was used for the following ligation reaction. The N-terminal coding region (corresponding to amino acids 1–309) was amplified by RT–PCR from total Xenopus oocyte RNA using 5′primer (TACCATGGCTTCGAGTGAAAACG) and 3′primer (AAGGGCCCGACTATTTGGTTTGC). The PCR product was digested with NcoI and ApaI, whose recognition sites were included in the primer ends (indicated by underlining). The digested PCR product and the digested plasmid were ligated to construct the full-length ORF (amino acids 1–1513). A recombinant baculovirus 6× His–Xenopus Cut5 was obtained using BAC-TO-BAC HT recombinant baculovirus expression system (Gibco BRL). The recombinant protein was expressed in Sf9 cells and purified on a Ni-NTA column (Qiagen) following the instructions of the supplier. Polyclonal rabbit antiserum was raised against the recombinant protein (Hokudo Inc., Japan) and further affinity-purified with the recombinant protein immobilized on Affi-Gel 10 (Bio-Rad).
Complementation in yeast mutant strains
A full-length coding region of Xenopus Cut5 (corresponding to amino acids 1–1513) and an N-terminal coding region of Xenopus Cut5 (corresponding to amino acids 1–761) were cloned into the pREP expression vector (Maundrell, 1993) at the NdeI and SalI sites for S.pombe, and into the pKT10 expression vector (Tanaka et al., 1990) at the KpnI and XhoI sites for S.cerevisiae. S.pombe cut5-T401 and S.cerevisiae dpb11-1 mutants were transformed with these vectors and transformants were plated at restrictive, semi-permissive or permissive temperature.
Preparation of Xenopus egg extract and chromatin fractions
S-phase egg extract and demembranated sperm nuclei were prepared as described (Kubota and Takisawa, 1993). Aphidicolin was added to the egg extracts at a concentration of 40 µg/ml to inhibit DNA polymerases. GST–p21 (Mimura and Takisawa, 1998) and roscovitine (Sigma) were added to the extracts at 50 µg/ml and 100 µM, respectively, to inhibit S-CDK. GST–geminin (Mimura et al., 2000) was added at 15 µg/ml to inhibit the licensing reaction. To isolate chromatin fractions, sperm nuclei were incubated in 50–100 µl of the extracts (4000 nuclei/1 µl extracts) for appropriate times at 23°C. The samples were diluted with 10 vol of EB (100 mM KCl, 2.5 mM MgCl2 and 50 mM HEPES–KOH pH 7.5) containing 0.25% NP-40, then centrifuged through a 10% sucrose layer at 10 000 g and the pellets were washed with EB. The pellets were treated as follows. For immunoblotting, the chromatin-associated proteins were extracted with SDS–PAGE sample buffer and separated from DNA by passage through a 0.45 µm filter. Quantification of the immunoblots was performed with NIH Image software. For immunoprecipitation, the pellets were resuspended in 1 vol of EB with protease inhibitors (1 µg/ml each of leupeptin, pepstatin and aprotinin). The suspensions were then sonicated with a sonicator (U50; IKA Labotechnik) until DNA was sheared into fragments with an average size of 700 bp and the soluble fractions were recoverd by centrifugation at 10 000 g. For the chromatin transfer experiments, the chromatin fractions were prepared as described above except that NP-40 was omitted from the buffers, a 20% sucrose layer was used and the isolated chromatin fractions were resuspended in egg extracts.
Assay for DNA replication activity
The replication activities of the egg extracts were measured as the incorporation of [α-32P]dCTP or Cy3-dCTP into sperm DNA. [α-32P]dCTP incorporation was carried out as described (Mimura and Takisawa, 1998) except that the autoradiography was quantified by Image Gauge software (Fuji Film). For Cy3-dCTP incorporation, the method was as described below in fluorescence microscopy.
Immunodepletion and immunoprecipitation
Immunodepletion of Xenopus proteins was carried out as described (Mimura and Takisawa, 1998) except that rProtein A Sepharose Fast Flow (Amersham Biosciences) was used instead of Affi-Prep protein A matrix (Bio-Rad). The immunoprecipitations of egg extracts and fragmented chromatin fractions were performed as described (Mimura and Takisawa, 1998; Mimura et al., 2000).
Fluorescence microscopy
Sperm nuclei (4000/µl) were incubated in 10 µl of the egg extract containing 10 µM Cy3-dCTP (Amersham Biosciences) for appropriate times at 23°C. The samples were prepared as described (Mimura et al., 2000) and fluorescence images were captured with the OpenLab imaging program (Improvision) and analyzed with NIH Image software (Kubota et al., 1997) to estimate the amounts of chromatin-bound Cut5 and the replication activities of the egg extracts.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Hiroyuki Araki and Hisao Masukata for complementation analysis of Xenopus Cut5 with yeast mutants, Shou Waga for antibody against the p60 subunit of Xenopus Pol ε, Yon-Soo Tak and Hiroyuki Araki for providing information before publication, Taro S.Masuda and Catherine Merrick for critical reading of the manuscript. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas (A) from the Ministry of Education, Science, Sports and Culture, Japan.
References
- Aparicio O.M., Stout,A.M. and Bell,S.P. (1999) Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc. Natl Acad. Sci. USA, 96, 9130–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H., Leem,S.H., Phongdara,A. and Sugino,A. (1995) Dpb11, which interacts with DNA polymerase II (ε) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc. Natl Acad. Sci. USA, 92, 11791–11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.P. and Dutta,A. (2002) DNA replication in eukaryotic cells. Annu. Rev. Biochem., 71, 333–374. [DOI] [PubMed] [Google Scholar]
- Blow J.J. and Laskey,R.A. (1986) Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell, 47, 577–587. [DOI] [PubMed] [Google Scholar]
- Blow J.J., Gillespie,P.J., Francis,D. and Jackson,D.A. (2001) Replication origins in Xenopus egg extract are 5–15 kilobases apart and are activated in clusters that fire at different times. J. Cell Biol., 152, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares P. and Blow,J.J. (2000) Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev., 14, 1528–1540. [PMC free article] [PubMed] [Google Scholar]
- Jiang W., McDonald,D., Hope,T.J. and Hunter,T. (1999) Mammalian Cdc7–Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J., 18, 5703–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y., Masumoto,H., Sugino,A. and Araki,H. (1998) Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol. Cell. Biol., 18, 6102–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki Y., Hiraga,S. and Sugino,A. (2000) Interactions between Mcm10p and other replication factors are required for proper initiation and elongation of chromosomal DNA replication in Saccharomyces cerevisiae. Genes Cells, 5, 975–989. [DOI] [PubMed] [Google Scholar]
- Kubota Y. and Takisawa,H. (1993) Determination of initiation of DNA replication before and after nuclear formation in Xenopus egg cell-free extracts. J. Cell Biol., 123, 1321–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y., Mimura,S., Nishimoto,S., Masuda,T., Nojima,H. and Takisawa,H. (1997) Licensing of DNA replication by a multi-protein complex of MCM/P1 proteins in Xenopus eggs. EMBO J., 16, 3320–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkiniemi M., Hillukkala,T., Tuusa,J., Reini,K., Vaara,M., Huang,D., Pospiech,H., Majuri,I., Westerling,T., Makela,T.P. and Syvaoja,J.E. (2001) BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J. Biol. Chem., 276, 30399–30406. [DOI] [PubMed] [Google Scholar]
- Masumoto H., Sugino,A. and Araki,H. (2000) Dpb11 controls the association between DNA polymerases α and ε and the autonomously replicating sequence region of budding yeast. Mol. Cell. Biol., 20, 2809–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H., Muramatsu,S., Kamimura,Y. and Araki,H. (2002) S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature, 415, 651–655. [DOI] [PubMed] [Google Scholar]
- Maundrell K. (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene, 123, 127–130. [DOI] [PubMed] [Google Scholar]
- Mimura S. and Takisawa,H. (1998) Xenopus Cdc45-dependent loading of DNA polymerase α onto chromatin under the control of S-phase Cdk. EMBO J., 17, 5699–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S., Masuda,T., Matsui,T. and Takisawa,H. (2000) Central role for Cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells, 5, 439–452. [DOI] [PubMed] [Google Scholar]
- Nishitani H. and Lygerou,Z. (2002) Control of DNA replication licensing in a cell cycle. Genes Cells, 7, 523–534. [DOI] [PubMed] [Google Scholar]
- Nguyen V.Q., Co,C. and Li,J.J. (2001) Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature, 411, 1068–1073. [DOI] [PubMed] [Google Scholar]
- Noguchi E., Shanahan,P., Noguchi,C. and Russell,P. (2002) CDK phosphorylation of Drc1 regulates DNA replication in fission yeast. Curr. Biol., 12, 599–605. [DOI] [PubMed] [Google Scholar]
- Saka Y. and Yanagida,M. (1993) Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+. Cell, 74, 383–393. [DOI] [PubMed] [Google Scholar]
- Saka Y., Fantes,P., Sutani,T., McInerny,C., Creanor,J. and Yanagida,M. (1994) Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J., 13, 5319–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld U.P., Howell,M., Rempel,R., Maller,J.L., Hunt,T. and Blow,J.J. (1994) Cip1 blocks the initiation of DNA replication in Xenopus extracts by inhibition of cyclin-dependent kinases. Curr. Biol., 4, 876–883. [DOI] [PubMed] [Google Scholar]
- Tada S., Li,A., Maiorano,D., Mechali,M. and Blow,J.J. (2001) Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol., 3, 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takisawa H., Mimura,S. and Kubota,Y. (2000) Eukaryotic DNA replication: from pre-replication complex to initiation complex. Curr. Opin. Cell Biol., 12, 690–696. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku,M., Tamanoi,F., Kaziro,Y., Matsumoto,K. and Toh-e,A. (1990) IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol. Cell. Biol., 10, 4303–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T. and Nasmyth,K. (1998) Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J., 17, 5182–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga S., Masuda,T., Takisawa,H. and Sugino,A. (2001) DNA polymerase ε is required for coordinated and efficient chromosomal DNA replication in Xenopus egg extracts. Proc. Natl Acad. Sci. USA, 98, 4978–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J.C. (2000) Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J. Biol. Chem., 275, 39773–39778. [DOI] [PubMed] [Google Scholar]
- Walter J. and Newport,J.W. (1997) Regulation of replicon size in Xenopus egg extracts. Science, 275, 993–995. [DOI] [PubMed] [Google Scholar]
- Walter J. and Newport,J. (2000) Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase α. Mol. Cell, 5, 617–627. [DOI] [PubMed] [Google Scholar]
- Wang H. and Elledge, (1999) DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 96, 3824–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel J.A., Dwyer,B.T., Dhar,S.K., Cvetic,C., Walter,J.C. and Dutta,A. (2000) Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science, 290, 2309–2312. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel J.A., Dhar,S.K., Prokhorova,T.A., Dutta,A. and Walter,J.C. (2002) Xenopus Mcm10 binds to origins of DNA replication after Mcm2-7 and stimulates origin binding of Cdc45. Mol. Cell, 9, 233–240. [DOI] [PubMed] [Google Scholar]
- Yamamoto R.R., Axton,J.M., Yamamoto,Y., Saunders,R.D., Glover,D.M., Henderson,D.S. (2000) The Drosophila mus101 gene, which links DNA repair, replication and condensation of heterochromatin in mitosis, encodes a protein with seven BRCA1 C-terminus domains. Genetics, 156, 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K. and Tsuruo,T. (1999) Conserved BRCT regions of TopBP1 and of the tumor suppressor BRCA1 bind strand breaks and termini of DNA. Oncogene, 18, 5194–5203. [DOI] [PubMed] [Google Scholar]
- Yamane K., Kawabata,M. and Tsuruo,T. (1997) A DNA-topoisomerase-II-binding protein with eight repeating regions similar to DNA-repair enzymes and to a cell-cycle regulator. Eur. J. Biochem., 250, 794–799. [DOI] [PubMed] [Google Scholar]
- Yamane K., Wu,X. and Chen,J. (2002) A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol. Cell. Biol., 22, 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L. and Stillman,B. (1998) Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science, 280, 593–596. [DOI] [PubMed] [Google Scholar]
- Zou L. and Stillman,B. (2000) Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol. Cell. Biol., 20, 3086–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]