Abstract
Figs (Ficus; ca 750 species) and fig wasps (Agaoninae) are obligate mutualists: all figs are pollinated by agaonines that feed exclusively on figs. This extraordinary symbiosis is the most extreme example of specialization in a plant–pollinator interaction and has fuelled much speculation about co-divergence. The hypothesis that pollinator specialization led to the parallel diversification of fig and pollinator lineages (co-divergence) has so far not been tested due to the lack of robust and comprehensive phylogenetic hypotheses for both partners. We produced and combined the most comprehensive molecular phylogenetic trees to date with fossil data to generate independent age estimates for fig and pollinator lineages, using both non-parametric rate smoothing and penalized likelihood dating methods. Molecular dating of ten pairs of interacting lineages provides an unparalleled example of plant–insect co-divergence over a geological time frame spanning at least 60 million years.
Keywords: co-divergence, co-speciation, molecular dating, Ficus, Agaoninae, phylogeny
1. Introduction
Figs have diversified extensively in terrestrial ecosystems throughout the tropics and subtropics. The distinctive fig inflorescence (syconium) is exclusively pollinated by female agaonine wasps that deposit their eggs in some of the flowers (figure 1; Cook & Rasplus 2003). Coevolutionary studies have centred on the specificity and stability of the exchange of pollination services for the rearing of pollinator offspring (Cook & Rasplus 2003; Molbo et al. 2003). Interspecific coevolution involves reciprocal, selected changes in the traits of interacting species, whereas co-divergence can arise purely from the maintenance of a specialized association between two lineages (Page et al. 1996; Page 2003; Percy et al. 2004). Patterns of co-divergence are expected in fig–pollinator relationships owing to extreme host fidelity, and comparisons of fig or fig wasp phylogenies with the classification of the other partner support this possibility (Weiblen 2000; Machado et al. 2001; Jousselin et al. 2003). Molecular phylogenetic trees of figs and their pollinators are suggestive of co-speciation (Herre et al. 1996; Weiblen & Bush 2002), but until now there has been no assessment of temporal congruence, namely whether dates of divergence are correlated between interacting lineages (Page 2003; Percy et al. 2004).
Figure 1.
Fig–wasp symbiosis: the example of Ficus macrophylla Desf. ex. Pers. and its pollinator, Pleistodontes froggatti Mayr. Photograph by JMC, Brisbane, Australia, 2003.
We inferred the phylogeny of 146 diverse Ficus species, representing all major lineages throughout the tropics, based on nuclear ribosomal DNA sequences using both maximum parsimony and Bayesian reconstruction methods. We also obtained an independent estimate of fig wasp phylogeny (Machado et al. 2001). We estimated divergence times for figs and pollinators using independent fossil calibrations for each partner, with non-parametric rate smoothing (NPRS) and penalized-likelihood (PL) dating methods (Sanderson 1997, 2002). We then identified interacting fig and pollinator lineages and compared their respective ages to test for co-divergence. Compared with previous studies (Weiblen 2000; Jousselin et al. 2003), the present paper includes both dense sampling of Ficus species and appropriate outgroups. With a large underlying dataset, we present the first quantitative test of temporal congruence in the fig and pollinator diversification.
2. Material and Methods
(a) Taxon sampling
Total genomic DNA was extracted from 91 taxa of Ficus using CTAB (Doyle & Doyle 1987). In addition, 65 ribosomal internal transcribed spacers (ITS) DNA sequences and 39 external transcribed spacers (ETS) sequences were retrieved from GenBank/EBI (following papers by Weiblen (2000) and Jousselin et al. (2003)), resulting in a total sample of 146 taxa of Ficus. Included material, voucher information, origin and GenBank/EBI accession numbers (AY730059–AY730144 and AY730145–AY730233) are listed in the electronic supplementary material. Matrices have been deposited in Treebase (see http://www.treebase.org). Our sampling covers all fig sections sensu Berg (1989, 2003a–e, 2004), except for three new and small sections in subgenus Sycomorus (Berg 2004); section Hemicardia with three species restricted to the Sino-Himalayan region, section Bosscheria with two species occurring from the Philippines to New Guinea and section Papuasyce with three species from New Guinea to Fiji.
Previous studies (Weiblen 2000; Jousselin et al. 2003) have shown that ITS and ETS sequences of Ficus are so divergent from those of other genera (e.g. Morus, Broussonetia, Brosimum and Artocarpus) in family Moraceae, that no satisfactory alignment could be performed. As a result, rooting the tree of Ficus has been problematic and Ficus section Pharmacosycea has often been used as the root based on preliminary analyses of 11 rbcL sequences (Herre et al. 1996) and indications from morphology (Berg 1989). Other molecular studies based on various plastid regions (Herre et al. 1996; Sytsma et al. 2002; Datwyler & Weiblen 2004) have, however, shown that the tribe Castilleae is more closely related to Ficus than any of the previously attempted outgroups. We collected DNA sequences of Poulsenia, Castilla, Sparattosyce and Antiaropsis in Castilleae, finding them to be alignable with Ficus and therefore suitable as outgroups.
For the phylogenetic analyses of the pollinating wasps, we used the dataset generated by Machado et al. (2001). This matrix includes mitochondrial cytochrome oxidase subunit I (cox1=COI) gene sequences of 816 nucleotides for 36 of the associated pollinator species, representing 15 out of 20 agaonine genera.
(b) PCR amplification and DNA sequencing
A total of 1230 aligned nucleotide positions across the ITS and ETS (Baldwin et al. 1995; Baldwin & Markos 1998) were amplified using primers 17SE and 26SE (Sun et al. 1994) and Hel1 and 18S ETS (Baldwin & Markos 1998), respectively. Standard automated sequencing protocols (Jousselin et al. 2003) were used except that DMSO was added to all reactions. Six taxa were only sequenced for ITS, because ETS could not be amplified (see electronic supplementary material).
(c) Phylogenetic reconstructions
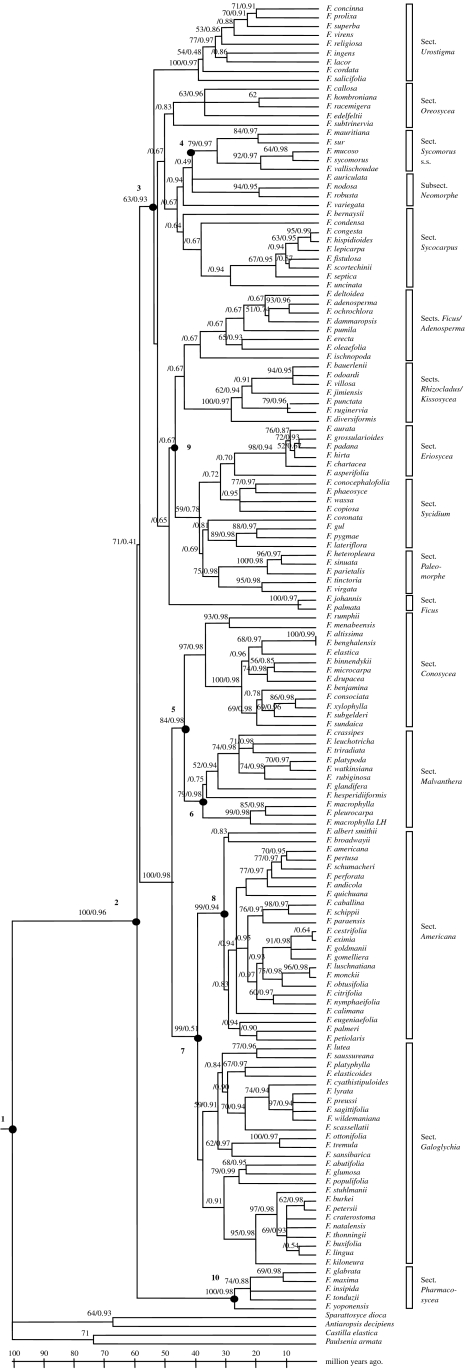
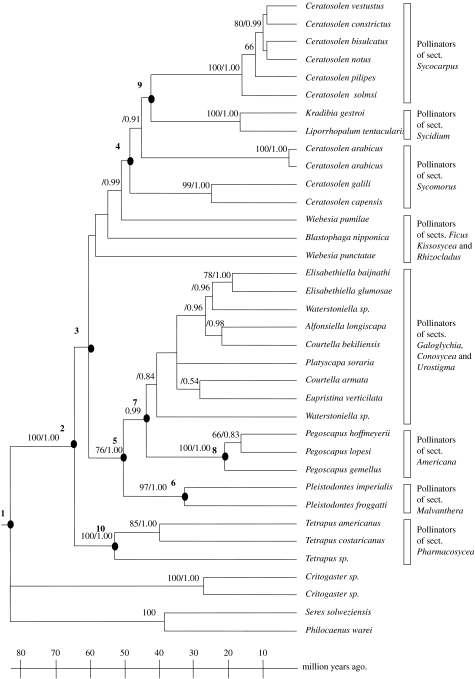
The large number of Ficus sequences analysed prevented us from using maximum-likelihood (ML) searches, so we adopted the following strategy. We performed 500 heuristic search replicates of Subtree-Pruning and Regrafting using maximum-parsimony (MP) criterion as implemented in PAUP* 4.0b10 (Swofford 2002). We then optimized ML branch lengths on one of the trees from the MP analysis using the save trees option in paup and the most suited HKY85+γ model of molecular evolution, with all parameters estimated from the data (figure 2). The phylogenetic tree of wasps, containing far fewer taxa (36), was reconstructed directly under the ML optimality criterion using five Tree Bisection-Reconnection replicates in PAUP* 4.0b10 (Swofford 2002; figure 3).
Figure 2.
Ultrametric tree of Ficus (ITS+ETS). MP tree saved under estimated ML conditions and made ultrametric with NPRS; bootstrap percentages/posterior probabilities above branches.
Figure 3.
Ultrametric ML tree of fig pollinating Agonine wasps (cox1); bootstrap percentages/posterior probabilities above branches.
Support was assessed by both Bayesian analyses and bootstrap re-sampling. Bayesian analysis was performed using MrBayes 2.01 (Hulsenbeck & Ronquist 2001). We used an HKY85 model of evolution (lset NST=2 RATES=equal). The analysis was performed with 1 000 000 generations on four Monte Carlo Markov chains with equal rates and trees sampled every 10 generations (mcmc NGEN=1 000 000, PRINTFREQ=100, SAMPLEFREQ=10, NCHAINS=4). We plotted generation number against the likelihood scores to locate the ‘burn in’. The first 10 000 trees of low-posterior probability were deleted, and all remaining trees were imported into PAUP* 4.0b10 (Swofford 2002). A 50% majority rule consensus tree was produced to yield the posterior probabilities of clades. A total of 500 bootstrap replicates with TBR swapping, equal weighting and a limit of one random addition sequence per bootstrap replicate was performed.
(d) Dating phylogenies
We estimated divergence times for figs and pollinators using independent fossil calibrations for each partner and both NPRS (as implemented in TreeEdit 1.0; Sanderson 1997; Rambaut & Charleston 2001) as well as PL (in r8s 2.0; Sanderson 2002, 2003) to account for deviations from the assumption of a molecular clock. Confidence intervals for ages were calculated by reapplying NPRS to 100 bootstrapped matrices. Sixty million years (Myr) old fossilized achenes assigned to Ficus were used to constrain the minimum age of the fig radiation (Collinson 1989). We calibrated the wasp phylogeny using fossil Pegoscapus from Dominican amber, constraining the genus to be at least 21 Myr old (Machado et al. 2001).
The fig wasp symbiosis is regarded as extremely species specific. Although deviations from one-to-one species specificity are known (Molbo et al. 2003), associations between pollinator genera and Ficus sections are often congruent based on previous phylogenetic analyses (Herre et al. 1996; Weiblen 2000; Machado et al. 2001; Jousselin et al. 2003). We identified ten ecologically associated lineages of figs and pollinators and compared their ages (see figures 2–4 and electronic supplementary material). These 10 pairs represent possible co-cladogenetic events across the phylogeny at higher taxonomic levels for which the corresponding clades of figs and wasps were both resolved. A tanglegram generated with Treemap (Page 1995) showing the 10 nodes is provided in the electronic supplementary material. A plot of the age of wasp lineages against the corresponding fig lineages was constructed and regression analysis was performed following Percy et al. (2004; figure 4). To evaluate whether the correlation could be due to chance alone, sums of squares of perpendicular offsets from a perfect linear regression (slope=1) were compared to 10 000 randomized sets of 10 pairs of ages drawn from the phylogenetic trees of figs and wasps.
Figure 4.
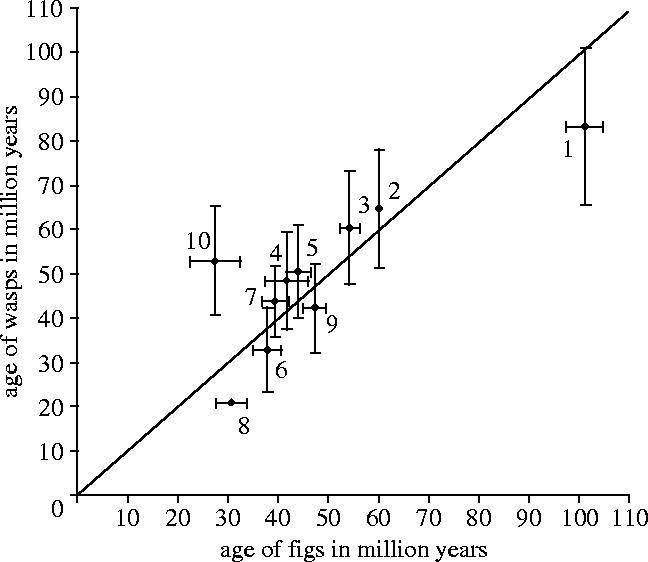
Temporal congruence of fig lineages and their associated pollinator wasp lineages based on independent, fossil calibrated molecular phylogenetic topologies. (1) Ficus/Agaoninae root nodes (101.24±3.71/83.33±17.61 Myr), (2) Ficus/Agaoninae crown groups (constrained to 60 Myr (Collinson 1989)/64.69±13.30 Myr), (3) Dioecious Ficus/associated pollinator stem lineages (54.34±1.84/60.49±12.76 Myr), (4) Sycomorus s.s./associated Ceratosolen stem lineages (41.70±4.41/48.52±10.94), (5) Malvanthera/Pleistodontes stem lineages (44.08±2.39/50.54±10.45 Myr), (6) Malvanthera/Pleistodontes crown groups (37.84±2.73/32.78±9.47 Myr), (7) Americana/Pegoscapus stem lineages (39.47±2.68/43.71±7.87 Myr), (8) Americana/Pegoscapus crown groups (30.67±2.98 Myr/constrained to 21 Myr; Machado et al. 2001), (9) Sycidium/Kradibia plus Lipporhopalum stem lineages (47.28±2.28/42.25±9.95 Myr), (10) Pharmacosycea/Tetrapus crown lineages (27.47±5.03/52.89±12.28 Myr).
3. Results
(a) Phylogeny of fig species
The combined dataset, including 146 taxa of Ficus and four outgroups, represents by far the most comprehensive phylogenetic study of figs to date. Out of 1354 aligned nucleotides, 478 characters were parsimony informative. The MP analysis of the combined dataset generated 74 most parsimonious trees of 2010 steps with a consistency index of 0.52 and a retention index of 0.83. One of the trees is shown in figure 2 (MP and ML trees have been deposited in Treebase, see §2).
Our results are generally consistent with previous phylogenetic studies of figs (Weiblen 2000; Jousselin et al. 2003), with one notable exception. Figure 2 indicates that subgenus Sycomorus (Berg 2004) is not monophyletic due to the nesting of section Adenosperma with subgenus Ficus. We attribute this finding to error in phylogeny estimation owing to a lack of non-parametric bootstrap support and morphological evidence for the monophyly of subgenus Sycomorus (Berg 1989; Weiblen 2000). The inclusion of outgroups in the ITS/ETS analyses indicated that Ficus is monophyletic and strongly supported (100% Bootstrap support, BS, 0.96 Bayesian posterior probability, PP). The earliest diverging lineage is section Pharmacosycea (100% BS, PP=0.98), the sister group to the rest of the figs (71% BS, PP=0.41). Although weakly supported, this finding is consistent with morphology (Berg 1989) and a preliminary analysis of chloroplast DNA sequences (Herre et al. 1996).
The remainder of the figs split into two major groups, one including a well-supported subgenus Urostigma (100% BS, PP=0.98), but excluding section Urostigma itself. Within subgenus Urostigma, the well supported neotropical section Americana (99% BS, PP=0.94) may be sister to the African section Galoglychia (99% BS, PP=0.51), although section Galoglychia is not supported by BS>50% and is paraphyletic with respect to section Americana in 35 of the 74 most parsimonious trees. Subgenus Urostigma also shows the Australasian section Malvanthera (79% BS, PP=0.98) and the mainly Asian section Conosycea (97% BS, PP=0.98) as sister clades (84% BS, PP=0.98). Ficus elastica, which has traditionally been placed in a section of its own (Stilpnophyllum; Berg 1989; Jousselin et al. 2003), or more recently with members of section Malvanthera Corner (Stilpnophyllum s.l.; Berg & Corner 2005) is here well embedded within section Conosycea. Ficus rumphii of the Asian section Leucogyne (two species; Berg 1989) also appears to be embedded in section Conosycea, where it groups with the Madagascan Ficus menabeensis.
The other major lineage (63% BS, PP=0.93) is weakly supported by bootstrapping and resolution is poor. Section Urostigma (100% BS, PP=0.97) is an early diverging lineage within this clade, which also includes sections Oreosycea (BS<50%, PP=0.83) and Sycomorus (79% BS, PP=0.97) plus all of the dioecious figs. Subgenus Synoecia (Berg 2003a,d) including the two root climbing sections Kissosycea and Rhizocladus is strongly supported (BS 100%, PP=0.97).
(b) Dating the divergence of figs and their pollinating wasps
Figure 4 illustrates the respective ages plotted with standard errors for 10 interacting fig and pollinator lineages (linear regression through the origin with r=0.968 not significantly different from r=1, t=0.165, p=0.386). Ages estimated from trees dated with NPRS and PL methods were highly correlated (R2=0.897 and 0.960 for figs and wasps, respectively), although PL provided slightly younger ages depending on smoothing parameters. NPRS ages are shown in figure 4. Comparing 10 000 random sets of 10 pairs of ages taken from both phylogenetic trees showed that the pattern observed in figure 4 is highly significant (p=0.002). The standard errors for the dates in the fig phylogeny were narrower than the intervals obtained for the wasp phylogeny, reflecting the denser taxon sampling and better resolution in the fig phylogenetic tree.
4. Conclusion
We have produced the most comprehensive phylogeny of figs to date and this supports the idea that section Pharmacosycea is the oldest section in the genus. With a dense taxon sampling, our phylogenetic analyses support the monophyly of most fig sections, especially within the monoecious subgenera Urostigma and Pharmacosycea. With over 750 species, Ficus is a large genus, and more detailed studies of phylogenetic patterns and evolutionary processes in the fig–wasp interaction should focus on smaller, more manageable subsets of species such as sections of the genus. Knowing the monophyly of a group of figs is a prerequisite for evaluating possible co-speciation in the mutualism. Not all clades are well supported in our analysis and future molecular systematic work should focus on the relationship between monoecious and dioecious figs (particularly the relationships of sections Urostigma and Oreosycea) and the sectional classification of dioecious figs.
Two previous molecular studies have estimated the date of origin of figs and their pollinating wasp. Machado et al. (2001) obtained an age interval of 75–100 Myr for the crown group of the wasps, a date that is older than available fossil evidence of Ficus by at least 15 Myr. More recently, Datwyler & Weiblen (2004) used three calibration points to date their phylogenetic tree of Moraceae based on ndhF sequences of over 80 taxa representing 33 genera. They obtained an estimate of 83 Myr for the root node of Ficus. We obtained confidence intervals of 98–105 Myr for the age of the root node of Ficus and 66–101 for the age of the root node of the wasps. The crown group of Ficus was constrained to 60 Myr by a fossil achene, and for the crown group of the wasps we obtained a confidence interval of 51–78 Myr (figure 4). Our results confirm previously published dates suggesting a time frame of 60–100 Myr ago for the origin of the fig–wasp association. However, the use of fossils for dating yields minimum age estimates, because the fossil record may not coincide with the earliest appearance. Confirmation of these dates could be given by analysing whether they are compatible with biogeographic scenarios for Ficus (see Machado et al. 2001; Zerega et al. in press). If the age estimates we have obtained are correct, this could imply long distance oceanic dispersal being an important process explaining the present distribution of Ficus. For instance, the south American section Pharmacosycea would have separated from the rest of the figs only 60 million years ago (node 2 on figure 2), which post-dates the separation of South America from Africa (about 90–100 Myr ago) during the break up of Gondwana. Likewise, the American section Americana and the African section Galoglychia would have separated around 40–50 million years ago (node 7 on figure 2).
Phylogenetic double-dating has so far only been used to evaluate co-speciation between parasitic psyllids (Hemiptera) and their hosts in the genistoid legumes (Genisteae; Percy et al. 2004). The authors found that all but one of the putative co-speciation events were in fact asynchronous, indicating that the psyllids colonized hosts that had already diversified rather than co-speciating contemporaneously with their hosts. By comparison, the fig-pollinating wasp system exhibits strong evidence for co-diversification in at least 10 interacting lineages.
Coevolution between mutualistic partners and between hosts and parasites is a long-held hypothesis, but the prevalence of coevolution between interacting taxa is unknown, largely because only a small number of associations have been studied in sufficient detail to document long-term coevolution.
The best-documented case is that between pocket gophers (Geomyidae) and their chewing lice (Phthiraptera; see Hafner et al. 2003 and references therein). Independent phylogenies of host and parasite lineages, based on sequences of the mitochondrial cox1 gene, show significant congruence both at high-taxonomic levels and within genera. Although lice may be transmitted horizontally between individuals, such dispersal relies on host-to-host contact, which is almost exclusively intraspecific among gophers. Other biological aspects, such as hair diameter, may also restrict the suitability of other potential host species for dispersing lice. Consequently, Hafner and co-workers suggest that the pattern of co-cladogenesis results primarily from lack of opportunity to colonize new host species.
Another classic system is the obligate pollination mutualism between the yucca (Agavaceae) and the yucca moth (Lepidoptera; Pellmyr 2003), but no analysis of parallel cladogenesis has yet been conducted due to the lack of phylogenetic estimates for the host plants.
In addition, an obligate pollination mutualism between Glochidion trees (Phyllantaceae) and Epicephala moths (Gracillariidae) was recently described (Kato et al. 2003). Several different methods for investigating the level of co-cladogenesis between phylogenies indicated that there is a greater degree of correlation between the Glochidion and Epicephala phylogenetic trees than expected in a random association (Kawakita et al. 2004). Coevolution with pollinators has also been suggested in Phyllanthus, another genus in Phyllantaceae (Kawakita & Kato 2004).
Likewise for palms, a diversity of insect pollination mutualisms have been described (Henderson 1986), but not yet studied in a phylogenetic framework.
All of these systems show deviations from perfect phylogenetic congruence, which could be due to host-shifting, independent speciation and/or extinction events, and error associated with phylogeny estimation. A number of studies have provided evidence that various hemipteran insect taxa, such as mealybugs (Baumann & Baumann 2005), white flies (Thao & Baumann 2004) and their primary bacterial endosymbionts, share phylogenetic histories. These systems tend to show perfect congruence, but this is consistent with a single infection of the hosts with an ancestor of the endosymbionts followed by vertical transmission. Other studies have simply failed to demonstrate coevolution between associated partners. For instance, Desdevises et al. (2002) found that host–parasite associations between Sparidae (Teleostei) fishes and their parasites of the genus Lamellodiscus (Monogenea) were due more to ecological factors than to coevolutionary processes.
Molecular dating showed that the yucca–yucca moth association arose at least 40 million years ago (Pellmyr 2003) and long-term co-divergence was recently reported for Simian foamy RNA viruses and old World primates (Switzer et al. 2005). The phylogenetic trees were remarkably congruent in both branching order and divergence times over 30 million years, strongly supporting co-speciation in this host–parasite system.
The strength of the relationship between the independently inferred ages of closely associated fig and pollinator lineages in the present study provides the most compelling evidence to date for long-term co-divergence in this now classical mutualism during at least the past 60 million years. Having established a scenario of parallel diversification of fig and wasp lineages, future studies should focus on the extent of co-speciation in the fig–wasp symbiosis based on manageable monophyletic groups of figs in comparison with the associated pollinators as exemplified by Weiblen & Bush (2002) for Ficus subgenus Sycomorus and the pollinating wasps of the genus Ceratosolen. Another promising line of investigation would be to examine whether the dates obtained in the present study are compatible with biogeographic scenarios and dates obtained for other groups and what implications these dates have for explaining the present distribution of figs and fig pollinators.
Acknowledgments
We thank K. Turk and E. Yektaei-Karin for their help with DNA sequencing, and Mark Chase and two anonymous reviewers for their valuable comments on the manuscript. We also thank The Forest Herbarium, Royal Forest Department, Thailand (BKF), C. C. Berg (BG), F. Billiet (BR), F. Forest and E. van Jaarsfeld (NBG), H. V. Hansen (C), B. Chantarasuwan, E. Jousselin, K. Oyama, R. Samuel and H. T. Simonsen for providing fig material. This work was partially funded by the Danish Carlsberg Foundation (NR), the Johannes Fog's Foundation (NR), the NERC (UK) (JC) and the European Commission (OIF to NR and VS).
Footnotes
Present address: Department of Ecology and Evolution, University of Lausanne, 1015 Lausanne, Switzerland.
Supplementary Material
References
- Baldwin B.G, Markos S. Phylogenetic utility of the external transcribed spacer (ETS) of 18S–26S rDNA: congruence of ETS and ITS trees of Calycadenia (Compositae) Mol. Phylogenet. Evol. 1998;10:449–463. doi: 10.1006/mpev.1998.0545. 10.1006/mpev.1998.0545 [DOI] [PubMed] [Google Scholar]
- Baldwin B.G, Sanderson M.J, Porter J.M, Wojciechowski M.F, Campbell C.S, Donoghue M.J. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann. Mo. Bot. Gard. 1995;82:247–277. [Google Scholar]
- Baumann L, Baumann P. Cospeciation between the primary endosymbionts of mealybugs and their hosts. Curr. Microbiol. 2005;50:84–87. doi: 10.1007/s00284-004-4437-x. 10.1007/s00284-004-4437-x [DOI] [PubMed] [Google Scholar]
- Berg C.C. Classification and distribution of Ficus. Experientia. 1989;45:605–611. 10.1007/BF01975677 [Google Scholar]
- Berg C.C. Flora Malesiana precursor for the treatment of Moraceae 1: the main subdivison of Ficus: the subgenera. Blumea. 2003a;48:167–178. [Google Scholar]
- Berg C.C. Flora Malesiana precursor for the treatment of Moraceae 2: Ficus subgenus Pharmacosycea section Oreosycea. Blumea. 2003b;48:289–301. [Google Scholar]
- Berg C.C. Flora Malesiana precursor for the treatment of Moraceae 3: Ficus subgenus Ficus. Blumea. 2003c;48:529–550. [Google Scholar]
- Berg C.C. Flora Malesiana precursor for the treatment of Moraceae 4: Ficus subgenus Synoecia. Blumea. 2003d;48:551–571. [Google Scholar]
- Berg C.C. Flora Malesiana precursor for the treatment of Moraceae 5: Ficus subgenus Sycidium. Blumea. 2003e;48:573–597. [Google Scholar]
- Berg C.C. Flora Malesiana precursor for the treatment of Moraceae 6: Ficus subgenus sycomorus. Blumea. 2004;49:155–200. [Google Scholar]
- Berg, C. C. & Corner, E. J. H. 2005 Moraceae. Flora Malesiana. Ser. I, vol. 17, part 2.
- Collinson M.E. The fossil history of the Moraceae, Urticaceae (including Cecropiaceae), and Cannabaceae. In: Crane P, Blackmore S, editors. Evolution, systematics, and fossil history of the hamamelidae: higher hamamelidae vol. 2. (Systematics Association Special Volumes) Clarendon Press; Oxford, UK: 1989. pp. 319–339. [Google Scholar]
- Cook J.M, Rasplus J.-Y. Mutualists with attitude: coevolving fig wasps and figs. Trends Ecol. Evol. 2003;18:241–248. 10.1016/S0169-5347(03)00062-4 [Google Scholar]
- Datwyler S.L, Weiblen G.D. On the origin of the fig: phylogenetic relationships of Moraceae from ndhF sequences. Am. J. Bot. 2004;91:767–777. doi: 10.3732/ajb.91.5.767. [DOI] [PubMed] [Google Scholar]
- Desdevises Y, Morand S, Jousson O, Legendre P. Coevolution between Lamellodiscus (Monogenea: Diplectanidae) and Sparidae (Teleostei): the study of a complex host–parasite system. Evolution. 2002;56:2459–2471. doi: 10.1111/j.0014-3820.2002.tb00171.x. [DOI] [PubMed] [Google Scholar]
- Doyle J.J, Doyle J.L. A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- Hafner M.S, Demastes J.W, Spradling T.A, Reed D.L. Cophylogeny between pocket gophers and chewing lice. In: Page R.D.M, editor. Tangled trees: phylogeny, cospeciation and coevolution. The University of Chicago Press; Chicago, IL: 2003. pp. 195–220. [Google Scholar]
- Henderson A. A review of pollination studies in the Palmae. Bot. Rev. 1986;52:221–259. [Google Scholar]
- Herre E.A, Machado C.A, Bermingham E, Nason J.D, Windsor D.M, McCafferty S.S, Van Houten W, Bachmann K. Molecular phylogenies of figs and their pollinator wasps. J. Biogeogr. 1996;23:521–530. [Google Scholar]
- Hulsenbeck J.P, Ronquist F. Mr Bayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Jousselin E, Rasplus J.-Y, Kjellberg F. Convergence and coevolution in a mutualism: evidence from a molecular phylogeny of Ficus. Evolution. 2003;57:1255–1269. doi: 10.1554/02-445. [DOI] [PubMed] [Google Scholar]
- Kato M, Takimura A, Kawakita A. An obligate pollination mutualism and reciprocal diversification in the tree genus Glochidion (Euphorbiaceae) Proc. Natl Acad. Sci. USA. 2003;100:5264–5267. doi: 10.1073/pnas.0837153100. 10.1073/pnas.0837153100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakita A, Kato M. Evolution of obligate pollination mutualism in New Caledonian Phyllanthus (Euphorbiaceae) Am. J. Bot. 2004;91:410–415. doi: 10.3732/ajb.91.3.410. [DOI] [PubMed] [Google Scholar]
- Kawakita A, Takimura A, Terachi T, Sota T, Kato M. Cospeciation analysis of an obligate pollination mutualism: have Glochidion trees (Euphorbiaceae) and pollinating Epicephala moths (Gracillariidae) diversified in parallel? Evolution. 2004;58:2201–2214. doi: 10.1111/j.0014-3820.2004.tb01598.x. [DOI] [PubMed] [Google Scholar]
- Machado C.A, Jousselin E, Kjellberg F, Compton S.G, Herre E.A. Phylogenetic relationships, historical biogeography and character evolution of fig-pollinating wasps. Proc. R. Soc. B. 2001;268:685–694. doi: 10.1098/rspb.2000.1418. 10.1098/rspb.2000.1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molbo D, Machado C.A, Sevenster J.G, Keller L, Herre E.A. Cryptic species of fig-pollinating wasps: implications for the evolution of the fig–wasp mutualism, sex allocation, and precision of adaptation. Proc. Natl Acad. Sci. USA. 2003;100:5867–5872. doi: 10.1073/pnas.0930903100. 10.1073/pnas.0930903100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R. D. M. 1995 TREEMAP program. Availability: http://taxonomy.zoology.gla.ac.uk/rod/treemap.html
- Page R.D.M. Introduction. In: Page R.D.M, editor. Tangled trees: phylogeny, cospeciation and coevolution. The University of Chicago Press; Chicago, IL: 2003. pp. 1–21. [Google Scholar]
- Page R.D.M, Clayton D.H, Patterson A.M. Lice and cospeciation: a response to Barker. Int. J. Parasitol. 1996;26:213–218. doi: 10.1016/0020-7519(95)00115-8. 10.1016/0020-7519(95)00115-8 [DOI] [PubMed] [Google Scholar]
- Pellmyr O. Yuccas, yucca moths, and coevolution: a review. Ann. Mo. Bot. Gard. 2003;90:35–55. [Google Scholar]
- Percy D.M, Page R.D.M, Cronk Q.C.B. Plant–insect interactions: double-dating associated insect and plant lineages reveals asynchronous radiations. Syst. Biol. 2004;53:120–127. doi: 10.1080/10635150490264996. 10.1080/10635150490264996 [DOI] [PubMed] [Google Scholar]
- Rambaut A, Charleston M. University of Oxford; UK: 2001. TreeEdit: phylogenetic tree editor, version 1. [Google Scholar]
- Sanderson M.J. A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol. Biol. Evol. 1997;14:1218–1231. [Google Scholar]
- Sanderson M.J. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Sanderson M.J. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. 10.1093/bioinformatics/19.2.301 [DOI] [PubMed] [Google Scholar]
- Sun Y, Skinner D.Z, Liang G.H, Hulbert S.H. Phylogenetic analysis of Sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theor. Appl. Genet. 1994;89:26–32. doi: 10.1007/BF00226978. 10.1007/BF00226978 [DOI] [PubMed] [Google Scholar]
- Switzer W.M, et al. Ancient co-speciation of simian foamy viruses and primates. Nature. 2005;434:376–380. doi: 10.1038/nature03341. 10.1038/nature03341 [DOI] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, MA: 2002. PAUP*: phylogenetic methods using parsimony (*and other methods), version 4. [Google Scholar]
- Sytsma K.J, Morawetz J, Pires J.C, Nepokroeff M, Conti E, Zijhra M, Hall J.C, Chase M.W. Urticalean rosids: circumscription, rosid ancestry, and phylogenetics based on rbcL, trnL-F and ndhF sequences. Am. J. Bot. 2002;89:1531–1546. doi: 10.3732/ajb.89.9.1531. [DOI] [PubMed] [Google Scholar]
- Thao M.L, Baumann P. Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts. Appl. Environ. Microbiol. 2004;70:3401–3406. doi: 10.1128/AEM.70.6.3401-3406.2004. 10.1128/AEM.70.6.3401-3406.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiblen G.D. Phylogenetic relationships of functionally dioecious Ficus (Moraceae) based on ribosomal DNA sequences and morphology. Am. J. Bot. 2000;87:1342–1357. [PubMed] [Google Scholar]
- Weiblen G.D, Bush G.L. Speciation in fig pollinators and parasites. Mol. Ecol. 2002;11:1573–1578. doi: 10.1046/j.1365-294x.2002.01529.x. 10.1046/j.1365-294X.2002.01529.x [DOI] [PubMed] [Google Scholar]
- Zerega, N. J. C., Clement, W. L., Datwyler, S. L. & Weiblen, G. D. In press. Biogeography and divergence times in the mulberry family (Moraceae). Mol. Phyl. Evol. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.