Abstract
Background: Abnormalities involving the p16 (also known as cyclin-dependent kinase N2 [CDKN2], p16 [INK4a], or MTS1) and p53 (also known as TP53) tumor suppressor genes are highly prevalent in esophageal adenocarcinomas. Loss of heterozygosity (LOH) at 9p21 and 17p13 chromosomes (locations for p16 and p53 genes, respectively) is frequently observed in the premalignant condition, Barrett’s esophagus. We studied extensively the distribution and heterogeneity of LOH at 9p and 17p chromosomes throughout the Barrett’s segment in patients who have not yet developed esophageal adenocarcinoma. Methods: We evaluated 404 samples from 61 consecutive patients enrolled in the Seattle Barrett’s Esophagus Study from February 1995 through September 1998. All patients had high-grade dysplasia but no diagnosis of cancer. The samples were assayed for LOH at 9p and 17p chromosomes after amplification of genomic DNA by use of polymerase chain reaction and DNA genotyping. The cell fractions were purified by flow cytometry on the basis of DNA content and proliferation-associated antigen labeling. Association between LOH at 9p and LOH at 17p with flow cytometric abnormalities was determined by chi-squared test, and logistic regression models were used to model and test for the extent to which a particular genotype was found in 2-cm intervals. Results and Conclusions: LOH at 9p and 17p chromosomes are highly prevalent somatic genetic lesions in premalignant Barrett’s tissue. LOH at 9p is more common than LOH at 17p in diploid samples and can be detected over greater regions of Barrett’s epithelium. In most patients with high-grade dysplasia, the Barrett’s mucosa contains a mosaic of clones and subclones with different patterns of LOH. Some clones had expanded to involve extensive regions of Barrett’s epithelium. LOH at 9p and 17p chromosomes may be useful biomarkers to stratify patients’ risk of progression to esophageal cancer.
Barrett’s esophagus is a premalignant condition in which the normal squamous epithelium of the esophagus is replaced by metaplastic columnar epithelium (1). Barrett’s esophagus predisposes patients to the development of esophageal adenocarcinoma, a cancer whose incidence has been increasing rapidly in the United States and in regions of Western Europe since the 1970s (2-5). Unfortunately, most esophageal adenocarcinomas are detected when they are advanced and incurable, and more than 90% of patients die of their disease (6).
A systematic protocol of endoscopic surveillance in Barrett’s esophagus can detect esophageal adenocarcinomas when they are early and curable (7,8). Therefore, current recommendations for patients with Barrett’s esophagus include periodic endoscopic biopsy surveillance for early detection of cancer by use of a five-tiered histologic classification of dysplasia and cancer (9). However, the value of endoscopic surveillance has been questioned because multiple studies (10-15) have shown that most patients with Barrett’s esophagus do not progress to cancer. Most patients will not benefit from endoscopic surveillance in terms of increased life expectancy because they will not progress to cancer during their lifetime (14,16,17). Furthermore, the fivetiered histologic classification of dysplasia in Barrett’s esophagus has not been shown to be reproducible in formal, blinded studies (18). High-grade dysplasia is a risk factor for subsequent development of cancer, but the disease in many patients with high-grade dysplasia does not progress and in some it may even regress (19,20). These observations indicate the need for objective, intermediate biomarkers of neoplastic progression in Barrett’s esophagus that can be used alone or in combination with histologic staging to stratify patients’ risk of progressing to cancer. Such diagnostic markers could be used to identify patients at increased risk of progression to cancer so that they could be placed in more frequent surveillance, while patients at lower risk could be counseled, reassured, and undergo less frequent surveillance. In addition, such biomarkers could serve as intermediate end points in clinical and population-based prevention trials.
Barrett’s specialized metaplastic epithelium is hyperproliferative relative to other tissues of the upper gastrointestinal (21-24). Flow-cytometric techniques have been used to identify proliferating epithelial cells, abnormal cell cycle fractions, and cell populations with abnormal DNA contents in Barrett’s esophagus (24-27). These methods are based on cell sorting by either staining DNA alone or dual staining involving proliferation-associated antigen identified by Ki67 antibody and DNA. Prospective studies (28,29) have shown that an increased number of cells with 4N fractions (cells with double the number of chromosomes or an increase in cells in the G2/M fraction of the cell cycle) and/or aneuploidy are risk factors for subsequent neoplastic progression in Barrett’s esophagus. Thus, Ki67 antibody-staining/DNA-content multiparameter flow-cytometric cell sorting can be used to purify populations of cells with 2N, 4N, and aneuploid DNA contents, as well as proliferating cells in the G1 phase of the cell cycle, for subsequent molecular analyses (28,30,31).
Abnormalities involving the p16 (also known as cyclin-dependent kinase N2 [CDKN2], p16 [INK4a], or MTS1) and p53 (also known as TP53) tumor suppressor genes, located on chromosomes 9p21 and 17p13, respectively, are among the most common somatic genetic lesions in human cancers. Loss of heterozygosity (LOH) is the predominant mechanism for inactivating one of the two alleles of each gene, and 9p21 and 17p13 are the two most common regions of LOH in esophageal adenocarcinomas, occurring in approximately 75% and 95% of cases, respectively (31-35). In Barrett’s esophagus, the remaining p16 allele is inactivated in the majority of cases by either CpG island methylation or mutation, and the remaining p53 allele is typically inactivated by mutation (31,36-40). Retrospective investigations (30,31,41) of patients who had already progressed to cancer suggest that p16 and p53 can be inactivated as early events in neoplastic progression before the development of aneuploidy and cancer in Barrett’s esophagus. Furthermore, LOH at 17p is associated with the development of increased 4N fractions that precede the development of aneuploidy in Barrett’s esophagus, probably as a consequence of inactivation of p53’s cell cycle checkpoint functions (28). LOH at 9p and LOH at 17p have also been shown to be precursors of clonal progression in the bladder, lung, and other cancers (42,43). Thus, 9p LOH and 17p LOH are potential candidates for objective molecular markers that can be used in combination with flow-cytometric and histologic staging to stratify patients’ risk of progressing to esophageal adenocarcinoma.
LOH is a common lesion found in most human cancers, but it has been difficult for a variety of reasons to use LOH as a biomarker of neoplastic progression in large clinical or population-based studies. Recently, we developed a method for investigating LOH in large-scale studies using small clinical samples (44). Here, we use this approach to determine the prevalence, distribution, and relationships among LOH at 9p and 17p chromosomes and flow-cytometric abnormalities in flow cytometrically sorted or purified samples from mapped endoscopic biopsy specimens in 61 consecutive patients who had a diagnosis of high-grade dysplasia without cancer in Barrett’s esophagus.
MATERIALS AND METHODS
Patients. We evaluated 61 consecutive patients seen from February 1995 through September 1998 who had a diagnosis of high-grade dysplasia without cancer and who were enrolled in the Seattle Barrett’s Esophagus Study. Sixty of 61 patients had sufficient flow cytometrically purified tissue for LOH analysis. The Seattle Barrett’s Esophagus Study was approved by the Human Subjects Division of the University of Washington (Seattle), with reciprocity from the Fred Hutchinson Cancer Research Center (Seattle, WA). Patients were counseled concerning risks and benefits of endoscopic surveillance for Barrett’s esophagus and informed of potential alternatives, including surgery for high-grade dysplasia.
Biopsy specimen collection. Endoscopic biopsy specimens were obtained, as described previously (7,45). Biopsy specimens were taken at 2-cm intervals in the Barrett’s segment. The cell populations in the biopsy specimen were sorted and purified by flow cytometry. In most instances, purified DNA was available for LOH analysis from tissue taken at sequential 2-cm intervals. We evaluated 404 flow cytometrically purified DNA samples. An average of seven (range, 2-20) flow-purified DNA samples, depending on the Barrett’s segment length, plus a constitutive control were evaluated for each patient.
Flow cytometry. Biopsy specimens were frozen in dimethyl sulfoxide and were stored at –70 °C. Each frozen biopsy specimen was minced, and the homogenates were processed and then sorted by flow cytometry on the basis of cellular DNA content alone or sorted by dual-marker selection involving fluorescent labeling of proliferation-associated antigen by Ki67 antibody and DNA content, as described previously (30,44). Ki67 is an antibody that identifies a proliferation-associated antigen expressed in G1, S, and G2/M but not in G0 phases of the cell cycle (46,47). With the use of Ki67 staining/DNA content flow cytometry, a minimum of two flow cytometrically purified fractions were generated per biopsy specimen, yielding Ki67-negative cells with a 2N DNA content, Ki67-positive cells in G1 phase, cells with a 4N DNA content, or aneuploid fractions. In Fig. 1, these cell cycle fractions will be denoted as 2N (Ki67-negative diploid) and G1 (Ki67-positive diploid). A 4N cell fraction greater than 6% was classified as having an abnormal increased 4N fraction, as described previously (25). The prevalence of abnormalities as determined by flow cytometry in these 61 patients has been reported previously (48). We used these flow cytometric techniques, in addition to LOH at chromosomes 9 and 17, to characterize cell populations that have evolved into apparent clones in vivo as part of the natural disease process. No in vitro culturing or expansion of clones in the laboratory was performed.
Fig. 1.
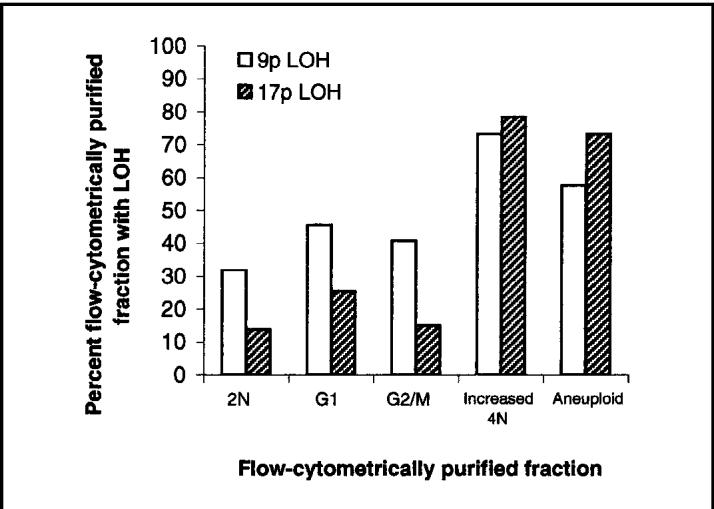
Prevalence of loss of heterozygosity (LOH) at chromosomes 9p and 17p in flow cytometrically sorted cell populations from patient biopsy tissues from Barrett’s epithelium. A total of 404 sorted fractions from 60 patients with a final diagnosis of high-grade dysplasia were evaluated, including 53 2N fractions (diploid G0/G1), 167 G1 fractions (Ki67-positive G1), 108 G2/M fractions (4N DNA content composed of <6% of the cells in the biopsy), 15 increased 4N fractions (4N DNA content composed of >6% of the cells in the biopsy), and 61 aneuploid populations.
Polymerase chain reaction (PCR). DNA was extracted by use of either standard phenol/chloroform or the Puregene DNA Isolation Kit, as recommended by the manufacturer (Gentra Systems, Inc., Minneapolis, MN). Whole genome amplification by use of primer extension preamplification was performed, as described previously (44). Chromosome locus-specific primers labeled with either FAM, TET, or HEX phosphoramidite dyes from Research Genetics (Huntsville, AL) included GATA62F03 (9p23), D9S935 (9p23), D9S925 (9p22.3), D9S932 (9p21.3-22.1), D9S1121 (9p21.3), D9S1118 (9p21), D17S1298 (17p13.3), TP53 (17p13.1) pentanucleotide repeat, TP53 (17p13.1) dinucleotide, D17S1537 (17p13.2), D17S786 (17p13.1), D17S974 (17p12), D17S1303 (17p12), and D17S1288 (17p11). Locus-specific PCR reagents and conditions were described previously (44). PCR products were run on an ABI 373 or an ABI 377 DNA sequencer by use of the internal-size standard Genescan-500 (PE Applied Biosystems, Foster City, CA). After each gel run, lanes were manually tracked and data were visually inspected by use of Genescan and processed by the use of Genotyper software (PE Applied Biosystems). Allele ratios were determined by measuring the fluorescence intensity (peak height) of the smaller (base pair) allele “A” relative to the fluorescent unit intensity of the larger allele “B” (A/B). LOH was determined by assessing the ratio of peak heights in neoplastic tissue samples relative to the ratio in the corresponding normal control. QLoH = (sample allele ratio)/(normal allele ratio). Depending on whether the smaller or larger allele was lost, QLoH could have any value between zero and infinity, with 0.0 being 100% loss of allele A and infinity being 100% loss of allele B. QLoH values less than 0.4 or greater than 2.5 were considered to be clearly indicative of LOH.
Statistical analysis. Exact binomial 95% confidence intervals (CIs) are provided, with observed LOH prevalence rates shown in Table 1. The Pearson chi-squared test was used to test for the association between 9p LOH or 17p LOH with flow-cytometric abnormalities in Table 2. We used the Mantel-Haenszel chi-squared test for stratified tables and the corresponding Mantel-Haenszel common odds ratio estimator (ORMH) to examine the relationship between each LOH and flow cytometric abnormalities, with stratification on presence or absence of the other LOH (49). Statistical analysis for Fig. 1 considers the data as matched pairs of 9p LOH and 17p LOH measurements arising from the flow cytometrically purified samples and uses McNemar’s test (49). Logistic regression models were used to model and test for the extent to which a particular genotype was found in additional 2-cm intervals of the Barrett’s segment given that it was present in at least one 2-cm interval (see Fig. 4). The robust sandwich variance estimator was used to allow for nonindependence of data arising from multiple 2-cm intervals per patient (50). Statistical tests were two-sided.
Table 1.
Prevalence of 9p and 17p LOH in 60 patients with a maximum diagnosis of high-grade dysplasia*
| No. with LOH/No. studied (% [95% CI]) | |
|---|---|
| 60 total patients, any LOH on 9p or 17p | 44/60 (73 [60% — 84%]) |
| Patients informative for at least one locus | |
| Prevalence of 9p LOH | 35/59 (59 [46%—72%]) |
| Prevalence of 17p LOH | 34/57 (60 [46%—72%]) |
| 56 patients in whom both 9p and 17p loci could be evaluated | |
| Patients with neither | 13/56 (23 [13%—36%]) |
| Patients with 9p LOH only | 9/56 (16 [8%—28%]) |
| Patients with 17p LOH only | 9/56 (16 [8%—28%]) |
| Patients with both 17p LOH and 9p LOH | 25/56 (45 [31%—59%]) |
LOH = loss of heterozygosity; CI = confidence interval.
Table 2.
Prevalence of 9p LOH and 17p LOH in patients with and without flow-cytometric abnormalities*
| Patients with increased 4N and/or aneuploidy | Patients with normal flow-cytometric characteristic (diploid only) | ||||||
|---|---|---|---|---|---|---|---|
| 9p het | 9p LOH | Total | 9p het | 9p LOH | Total | ||
| 17p het | 1 (3)† | 2 (6) | 3 (9) | 17p het | 12 (52) | 7 (30) | 19 (83) |
| 17pLOH | 8 (24) | 22 (67) | 30 (91) | 17p LOH | 1 (4) | 3 (13) | 4 (17) |
| Total | 9 (27) | 24 (73) | 33‡ | Total | 13 (57) | 10 (43) | 23‡ |
LOH = loss of heterozygosity; het = heterozygous; increased 4N refers to samples with more than 6% of cells containing 4N DNA.
Numbers of patients (% of total), rounding of totals is done after adding the nonrounded percentages).
Includes only the 56 patients who were informative for markers on both 9p and 17p.
Fig. 4.
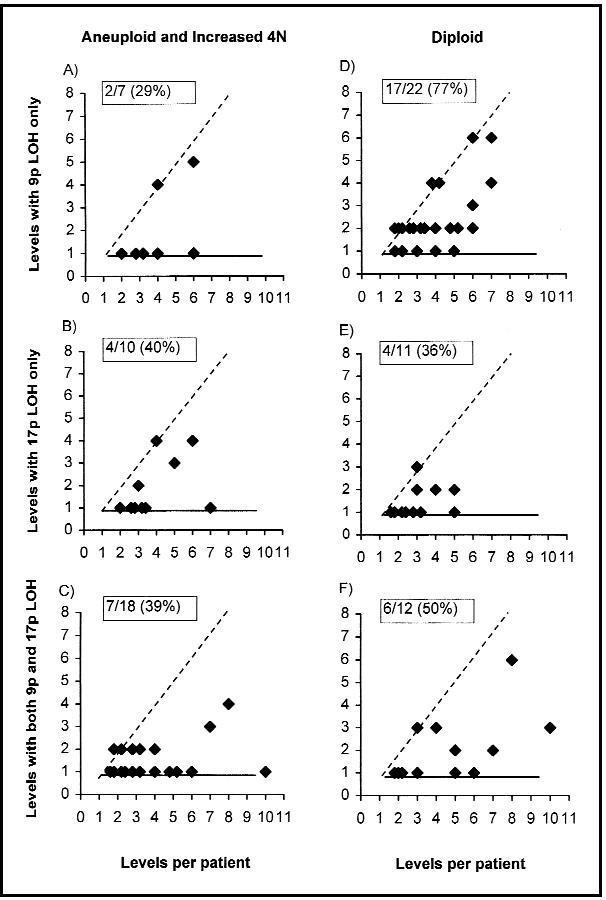
Expansion of loss of heterozygosity (LOH) at 9p and 17p chromosomes with and without flow-cytometric abnormalities. The levels with LOH (y-axis) in each flow-cytometric category (increased 4N and/or aneuploid [panels A– < C] or diploid [panels D– < F]) were plotted against the number of (2-cm) levels evaluated for each patient (x-axis). The number of 2-cm levels evaluated varies relative to the Barrett’s segment length. Samples with 9p LOH only are plotted on panels A and D, 17p LOH are plotted only on panels B and E, and both 9p LOH and 17p LOH are plotted together on panels C and F. Points on the dashed diagonal line have undergone complete expansion within the Barrett’s segment, points on the horizontal have not undergone expansion, and clones in between have undergone incomplete expansion. The percent of clones with expansion is given in the upper left corner of each graph.
RESULTS
Prevalence of 9p LOH and 17p LOH in Patients With High-Grade Dysplasia
We determined the prevalence of LOH at chromosomal regions encompassing p16 and p53 genes by use of six and eight short-tandem repeats (STRs) on chromosomes 9p and 17p, respectively (Table 1). A high prevalence of LOH was detected; 44 (73%) of 60 patients (95% CI = 60%-84%) had LOH at one or more loci on 9p, 17p, or both chromosomes.
Association of Aneuploidy and Increased 4N With 9p LOH and 17p LOH
Aneuploid cell populations and increased 4N fractions in esophageal biopsy specimens have been shown to identify a subset of patients at increased risk of neoplastic progression in Barrett’s esophagus (28,29). Because we flow cytometrically purified diploid (2N), diploid Ki67-positive cell fraction in G1 phase, 4N, and aneuploid fractions for LOH analysis, we were able to evaluate relationships among ploidy, cell cycle abnormalities, and LOH involving 9p and 17p chromosomes throughout the Barrett’s segment of each patient. Fifty-six patients were informative for one or more loci at or flanking both the p16 and p53 genes. Thirty-three of the 56 patients had increased 4N fractions and/or aneuploidy, and 23 had only diploid samples (Table 2) (48). In patients with increased 4N and/or aneuploidy, 30 (91%) of 33 (95% CI = 76%-98%) had 17p LOH compared with only four (17%) of 23 (95% CI = 5%-39%) who had diploid samples only (P<.001; odds ratio [OR] = 48; 95% CI = 10-225). 9p LOH was also more common in patients with increased 4N and/or aneuploidy; 24 (73%) of 33 (95% CI = 54%-87%) had 9p LOH compared with 10 (43%) of 23 (95% CI = 23%-66%) with diploid samples only (P = .03; OR = 3.5; 95% CI = 1.1-10.5). We recognize that there may be a biologic basis for the association between 9p LOH and 17p LOH and thus took into account the possible confounding effects by looking at the association between flow-cytometric abnormalities and each LOH, with stratification on the other LOH. The association between patients with 17p LOH and flow-cytometric abnormalities remains equally strong with or without adjusting for a confounding effect of 9p LOH (ORMH = 40; 95% CI = 8.0-200; P<.001). However, when accounting for 17p LOH status, there is no evidence for an association between patients with 9p LOH and aneuploidy and/or increased 4N fractions (ORMH = 1.7; 95% CI = 0.32-8.9; P = .54).
We determined the presence or absence of 9p LOH and 17p LOH in 404 flow cytometrically purified samples from all patients, separated into 2N (diploid), Ki67-positive diploid G1, normal 4N (G2/M), increased 4N (>6% of the cells with 4N in the cell cycle), and aneuploid cell populations (Fig. 1). 9p LOH was detected in 42% of normal-sorted diploid fractions (2N, G1, and normal 4N G2/M fractions) in contrast to 17p LOH, which was detected in only 20% of the same samples [P<.001]). 9p LOH and 17p LOH were detected in 73% and 78% of increased 4N fractions, respectively. 9p LOH was detected in 58% of aneuploid populations, and 17p LOH was detected in 73% of aneuploid populations. The difference between 9p LOH and 17p LOH in increased 4N and aneuploid populations was not statistically significant (P = .10). We re-evaluated these results using only patients with LOH and excluding patients without LOH and found similar results to those in which all patients were analyzed together.
Ordering LOH at Chromosomes 9p and 17p
We used the method of clonal ordering (comparing genetic changes within a clone two by two to determine whether the events are dependent or independent and whether they occur in a specific order relative to each other) to determine the relative order of occurrence of the two LOH events in 21 patients who had both 9p LOH and 17p LOH in a single clonal population (Table 3) (30,31,41,51). For LOH on one chromosome arm to be classified as occurring “before” the other, a patient must have at least one sample with both 9p LOH and 17p LOH as well as a separate sample with LOH on only one of the arms, and the losses must have the same STR patterns. 17p LOH was rarely detected before 9p LOH in one (5%) of 21 patients, but 9p LOH was detected before 17p LOH in 11 (52%) of 21. The two abnormalities were always detected together in nine (43%) of 21 patients (i.e., all samples had lost the same allele and loci on each chromosome arm). In an additional four patients, 9p LOH and 17p LOH were detected in mutually exclusive clones, none of which had both 9p LOH and 17p LOH in the same sample. In 19 other patients, we detected either 9p LOH or 17p LOH but not both. We detected 9p LOH without 17p LOH in 10 patients and 17p LOH without 9p LOH in nine patients, supporting the concept that either abnormality can occur in the absence of the other.
Table 3.
Ordering of loss of heterozygosity (LOH) events in patients with 9p LOH, 17p LOH, or both
| Patients with an order* | No. with order |
|---|---|
| 17p before 9p | 1/21 (5%) |
| 9p before 17p | 11/21 (52%) |
| 9p and 17p together | 9/21 (43%) |
| Other patients with 9p LOH and/or 17p LOH | |
| 9p and 17p in mutually exclusive clones | 4 patients |
| Either 9p LOH or 17p LOH but not both | 19 patients |
Of 44 patients with 9p LOH, 17p LOH, or both, an order could be determined in 21 patients who had clones with both 9p LOH and 17p LOH detected in the same DNA sample.
Genetic Clonal Diversity in Premalignant Tissue
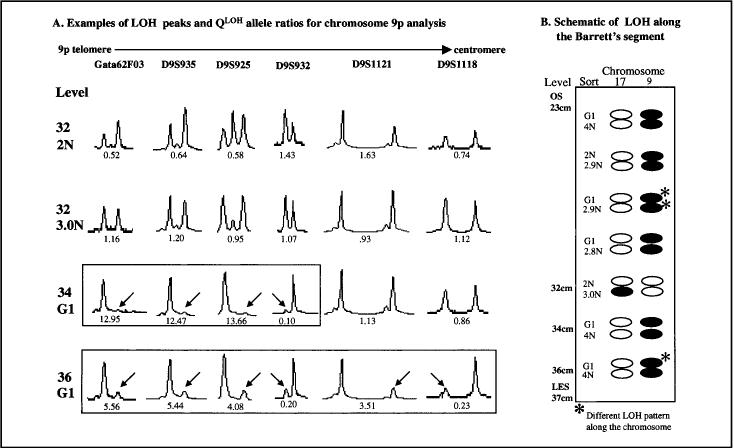
We investigated clonal diversity in premalignant tissue by assessing 9p LOH and 17p LOH systematically throughout the Barrett’s segment in all patients. The ploidy, lost allele (i.e., smaller or larger allele in base-pair size), and patterns of loss along each chromosome as well as between the two chromosome arms (9p and 17p) were used to define clonal populations and to determine the number of distinct clones that were present in the evaluated segment (Table 4). Forty-four patients had LOH at one or more STRs. In 14 (32%) of the 44 patients, we found evidence for only a single clone (e.g., Fig. 2, A; case No. 252). In these 14 cases, all abnormal samples had the same ploidy and LOH pattern for both chromosomes. However, for the majority of patients, we found evidence that the Barrett’s segment contained a mosaic of clones and subclones with different patterns of LOH and ploidies. For example, 13 (30%) of the 44 patients had the same LOH pattern in both the diploid and the aneuploid populations (e.g., Fig. 2, B; case No. 916). In these patients, LOH could have developed in diploid progenitor cells before the evolution of aneuploid progeny. Four (9%) of the 44 patients had mutually exclusive clones, with 9p LOH in one subset of samples and 17p LOH in others but never both 9p LOH and 17p LOH in the same sample (e.g., Fig. 2, D and E; case Nos. 368 and 790, respectively). These clones arose either independently of each other or from a common progenitor that had one or more abnormalities not being assessed in this study. Seven (16%) of the 44 patients had alternate alleles lost in different samples (e.g., Fig. 2, F; case No. 306). In these cases, one or more samples had LOH of the larger allele, whereas other samples from the same patient had LOH of the smaller allele. We also found evidence for clonal evolution of LOH in 13 (30%) of 44 patients, as evidenced by the accumulation of loss on different chromosomes in different samples or the expansion of the region of loss along the length of the chromosome. For example, in Fig. 2, C (case No. 994), the 1.8N population had both 9p LOH and 17p LOH, whereas other samples had only 9p LOH, suggesting that a diploid clone with 9p LOH was the progenitor for the 1.8N aneuploid progeny with both 9p LOH and 17p LOH. Expansion of loss along the length of the chromosome was identified in three patients (e.g., Fig. 3). In this patient, the sorted G1 fraction at the 34-cm level had 9p LOH involving STR polymorphic markers GATA62F03, D9S935, D9S925, and D9S932 with retention of heterozygosity at D9S1121 and D9S1118, consistent with a terminal deletion of 9p, but the sorted G1 fraction at the 36-cm level had 9p LOH involving all STRs evaluated, indicating the evolution of additional losses.
Table 4.
Prevalence of clonal heterogeneity in premalignant tissue
| Clonal heterogeneity | No. of patients* (%) |
|---|---|
| Single clone | 14 (32) |
| Ploidy change, same LOH pattern† | 13 (30) |
| Mutually exclusive clones‡ | 4 (9) |
| Alternate alleles lost§ | 7 (16) |
| Change in LOH pattern, accumulation of loss∥ | 13 (30) |
Some patients had loss of heterozygosity (LOH) patterns in multiple categories. Percent is calculated from 44 total patients.
Patients with the same alleles and loci lost on each chromosome in all samples with LOH.
Patients have both 9p LOH and 17p LOH but never both in the same samples.
Patients with one clone that has loss of the smaller allele and a second clone that has loss of the larger allele.
Patients have clones with different LOH patterns along the chromosome or clones that have evolved to contain LOH on both 9p and 17p chromosomes.
Fig. 2.
Loss of heterozygosity (LOH) along the Barrett’s epithelial segment in individual patients. Six examples of patients with different LOH patterns. OS (ora serrata) and LES (lower esophageal sphincter) are endoscopic features used to define the region of Barrett’s within the esophagus. cm = location along the esophagus in centimeters from the incisors; sort = type of sample sorted (either Ki67-positive G1, Ki67-negative 2N, 4N [normal G2/M], Inc 4N [increased 4N], or aneuploid); open circles = no LOH; filled circles = LOH; and hatched circles = alternate allele lost. A) Case No. 252, single clone; B) case No. 916, change in ploidy; C) case No. 994, accumulation of loss; D) case No. 368 and E) case No. 790, mutually exclusive clones; and F) case No. 306, alternate allele patterns. Asterisk indicates a different LOH pattern along the same chromosome, i.e., different extent of loss.
Fig. 3.
Genotyping by use of fluorescent staining of markers for loss of heterozygosity (LOH) analysis. See Fig. 2 legend for the definitions of OS and LES. Fluorescent traces (graphic) for LOH results for all chromosome 9 loci in case No. 336 are shown. A) The QLoH ratio (see “Materials and Methods” section) is given below the peaks. LOH is indicated by an arrow pointing to the lost allele. B) Schematic of all LOH results along the Barrett’s epithelial segment. An extra allele was found in the diploid and aneuploid populations from level 32, but additional alleles were not used as markers of clonality in this study. Asterisk indicates different LOH pattern along the same chromosomes, as indicated by boxed patterns. Open circles = no LOH; filled circles = LOH.
Distribution of Clones With LOH in the Barrett’s Segment
The extent to which a clone with a specific somatic genetic abnormality can expand within Barrett’s mucosa may influence the risk of progression by increasing the size of the population in which subsequent genetic events can occur. Furthermore, the extent of clonal expansion may also determine whether a given biopsy protocol can detect the abnormality and, therefore, its utility as an intermediate marker of progression. We assessed the extent to which different clones expanded in the Barrett’s segment (Fig. 4). Some clones showed little expansion, being detected at only a single 2-cm level of the Barrett’s segment, whereas others expanded to involve the entire evaluated Barrett’s segment length. Many clones showed incomplete expansion and were detected at multiple 2-cm intervals of the Barrett’s segment, even though they did not involve the entire segment. Because the length of the Barrett’s segment differs among patients, the number of levels evaluated for each patient varied, ranging from 2 to 20 cm per patient (average, 6 cm per patient). Some diploid clones with LOH can undergo complete expansion to involve the entire evaluated Barrett’s segment. Clones expanding to multiple 2-cm intervals in the Barrett’s segment were detected in 17 (77%) of 22 diploid clones with 9p LOH only, in four (36%) of 11 diploid clones with 17p LOH only (P = .05), and in six (50%) of 12 diploid clones with both 9p LOH and 17p LOH (P = .13). Aneuploid and increased 4N populations with 9p LOH, 17p LOH, or both could also expand over variable Barrett’s segment lengths from 2 to 20 cm. Aneuploid or increased 4N clones showed expansion in 29%, 40%, and 39% of the clones with 9p LOH, 17p LOH, or both, respectively. Diploid clones with 9p LOH only were typically detected over a greater extent of Barrett’s epithelium than aneuploid and increased 4N populations (P = .02) and diploid populations with either 17p LOH only or both 9p LOH and 17p LOH (P = .05).
DISCUSSION
LOH at chromosomes 9p and 17p, targeting p16 and p53 genes, respectively, are among the most common genetic lesions in esophageal adenocarcinomas. It is likely that inactivation of these pathways by LOH is part of the pathogenesis of these cancers because 9p and 17p LOH arise at a similar stage of progression as mutations in p53 and p16 genes and p16 CpG island methylation (31,35,36,39,41,52-56). Retrospective studies (30,31,36,41,52) of patients who had developed an esophageal adenocarcinoma have shown that 9p LOH and 17p LOH can develop in premalignant Barrett’s epithelium before aneuploidy and cancer. However, it has previously been difficult to prospectively investigate LOH in large numbers of small clinical samples from human premalignant conditions in vivo (53-55). Here, we demonstrate that our biopsy protocol, combined with fluorescence-based PCR and LOH analysis of flow cytometrically sorted human samples, can be used to investigate the biologic behavior of clones with 9p LOH, 17p LOH, or both in the premalignant epithelium of patients who have not developed esophageal adenocarcinoma. Our results extend earlier investigations by demonstrating that 9p LOH and 17p LOH are highly prevalent somatic genetic abnormalities in patients with high-grade dysplasia and no diagnosis of cancer in Barrett’s esophagus. Clones with 9p LOH and/or 17p LOH can undergo variable expansion within the Barrett’s segment and evolve aneuploid and increased 4N fractions without progressing to cancer. Our results indicate a surprising degree of clonal heterogeneity in premalignant Barrett’s epithelium consistent with a complex pattern of evolution of neoplastic cell lineages rather than a simple linear pathway of progression (41,57).
High-grade dysplasia is a risk factor for progression to cancer in Barrett’s esophagus, but prospective studies (19,20) have shown that many patients do not progress to cancer. We have found striking somatic genetic heterogeneity in the Barrett’s mucosa of patients with high-grade dysplasia. For example, 23% of the evaluable patients had no detectable LOH, 16% had only 9p LOH, 16% had only 17p LOH, and 45% had both. Although prospective validation will be essential, it may be that somatic genetic assessment of 9p LOH and 17p LOH can identify low- and high-risk patients. For example, the fraction of patients with both LOH events is comparable to the proportion of patients previously diagnosed with high-grade dysplasia in the Seattle Barrett’s Esophagus Study whose disease progressed (Reid BJ: unpublished results).
Our results extend our understanding of the biologic behavior of clones with 9p LOH, 17p LOH, or both in human neoplastic progression in vivo. A previous study (41) in patients whose disease had already progressed to cancer suggested that there was no obligate order of 9p LOH and 17p LOH during neoplastic progression in Barrett’s esophagus. Our results clearly indicate that 9p LOH or 17p LOH can each arise without the other in premalignant Barrett’s epithelium. 9p LOH, however, was more frequently detected before 17p LOH in patients in whom an order could be determined. The observation that 9p LOH can be detected before other genetic abnormalities during progression is consistent with those in other cancers in which it has been suggested that 9p LOH is an early event (42,58-60). Clones with 17p LOH were more likely to be associated with increased 4N fractions or aneuploid cell populations, probably as a consequence of loss of cell cycle checkpoint functions of p53 with subsequent genetic instability (28,61,62). In contrast, diploid clones with 9p LOH were rarely associated with aneuploidy or increased 4N fractions in the absence of 17p LOH and tended to spread to large regions of esophageal mucosa, possibly as a consequence of inactivation of p16’s G1/S-phase- or senescence-related functions (63,64). Our observations might result from clonal expansion of diploid cells with 9p LOH, creating a field in which 17p LOH could develop as a later event. An alternative hypothesis is that inactivation of p53 alone is not as permissive for clonal expansion, thereby limiting detection of clones with only 17p LOH.
We have shown previously that, in patients whose disease had already progressed to cancer, the Barrett’s epithelium surrounding the malignancy contained multiple aneuploid cell populations and neoplastic cell lineages with multiple molecular abnormalities (31,41,51,56,65). Our present results extend these observations by demonstrating that the metaplastic epithelium of patients who have not yet developed cancer can consist of a mosaic of clones and subclones. Some mosaics appear to develop from a common progenitor that evolves progeny with additional LOH events or changes in ploidy (Fig. 2, C). However, a minority of mosaics may also develop from neoplastic cell lineages that arise independently (Fig. 2, D and E). In these cases, some clones have 9p LOH, whereas others have 17p LOH, none have both, or patients have neoplastic cell lineages with loss of alternate 9p or 17p alleles (Fig. 2, F). It is possible in these cases that an unknown common progenitor evolved bifurcating neoplastic cell lineages leading to apparently independent pathways. These apparently independent clones are consistent with other studies, suggesting possible multifocal clonal evolution of human neoplasms (59,66-68).
Although LOH is a common lesion found in the vast majority of human cancers and precancerous lesions, it has been difficult to use LOH as a biomarker in large-scale clinical or population-based studies. We recently described a method for high-throughput analysis of LOH in small, clinical samples (44). Here, we demonstrate that this method can be applied to a human premalignant condition to detect the presence, extent, and biologic characteristics of clones with 9p LOH, 17p LOH, or both. Our results suggest a model of clonal evolution during neoplastic progression in Barrett’s esophagus. Inactivation of one allele of p16 and p53, as assessed by 9p and 17p LOH, is a common genetic alteration in premalignant Barrett’s epithelium and can develop before flow-cytometric biomarkers of progression, such as increased 4N fractions and aneuploidy. Diploid populations with 9p LOH spread to involve variable regions of esophageal mucosa, sometimes involving the entire Barrett’s segment. Although either 9p LOH or 17p LOH can occur without the other, 9p LOH tends to be detected before 17p LOH when the two abnormalities can be ordered relative to each other in single patients. Progenitor clones can either arise independently or evolve bifurcations that lead to divergent progeny and extensive clonal heterogeneity within premalignant tissue. Most patients with aneuploidy and increased 4N fractions have both 9p LOH and 17p LOH, and it seems likely that the acquisition of 9p LOH and 17p LOH predisposes to the evolution of aneuploid cell populations and other genetic abnormalities that culminate in the development of cancer. Prospective follow-up will be required to determine which, if any, of the abnormal clones with 9p LOH and/or 17p LOH will undergo further progression. Our findings indicate that, while human neoplastic progression is complex, the techniques and protocols used in this study can allow future studies that will assess the usefulness of LOH as a biomarker of progression.
Acknowledgments
We thank Michael Barrett, Thomas Paulson, and David Wong (Fred Hutchinson Cancer Research Center, Seattle, WA) for their critical review of this manuscript.
Footnotes
Supported by Public Health Service grants R01CA61202, R01CA78828, and R01CA72030 from the National Cancer Institute, National Institutes of Health, Department of Health and Human Services.
REFERENCES
- (1).Phillips RW, Wong RK. Barrett’s esophagus. Natural history, incidence, etiology, and complications. Gastroenterol Clin North Am. 1991;20:791–816. [PubMed] [Google Scholar]
- (2).Reid BJ. Barrett’s esophagus and esophageal adenocarcinoma. Gastroenterol Clin North Am. 1991;20:817–34. [PubMed] [Google Scholar]
- (3).Blot WJ, Devesa SS, Kneller RW, Fraumeni JF., Jr. Rising incidence of adenocarcinoma of the esophagus and gastric cardia. JAMA. 1991;265:1287–9. [PubMed] [Google Scholar]
- (4).Powell J, McConkey CC. The rising trend in oesophageal adenocarcinoma and gastric cardia. Eur J Cancer Prev. 1992;1:265–9. doi: 10.1097/00008469-199204000-00008. [DOI] [PubMed] [Google Scholar]
- (5).Devesa SS, Blot WJ, Fraumeni JF., Jr. Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer. 1998;83:2049–53. [PubMed] [Google Scholar]
- (6).Farrow DC, Vaughan TL. Determinants of survival following the diagnosis of esophageal adenocarcinoma (United States) Cancer Causes Control. 1996;7:322–7. doi: 10.1007/BF00052937. [DOI] [PubMed] [Google Scholar]
- (7).Levine DS, Haggitt RC, Blount PL, Rabinovitch PS, Rusch VW, Reid BJ. An endoscopic biopsy protocol can differentiate high-grade dysplasia from early adenocarcinoma in Barrett’s esophagus. Gastroenterology. 1993;105:40–50. doi: 10.1016/0016-5085(93)90008-z. [DOI] [PubMed] [Google Scholar]
- (8).Peters JH, Clark GW, Ireland AP, Chandrasoma P, Smyrk TC, DeMeester TR. Outcome of adenocarcinoma arising in Barrett’s esophagus in endoscopically surveyed and nonsurveyed patients. J Thorac Cardiovasc Surg. 1994;108:813–21. discussion 821-2. [PubMed] [Google Scholar]
- (9).Sampliner RE. Practice guidelines on the diagnosis, surveillance, and therapy of Barrett’s esophagus. The Practice Parameters Committee of the American College of Gastroenterology. Am J Gastroenterol. 1998;93:1028–32. doi: 10.1111/j.1572-0241.1998.00362.x. [DOI] [PubMed] [Google Scholar]
- (10).Spechler SJ, Robbins AH, Rubins HB, Vincent ME, Heeren T, Doos WG, et al. Adenocarcinoma and Barrett’s esophagus. An overrated risk. Gastroenterology. 1984;87:927–33. [PubMed] [Google Scholar]
- (11).Cameron AJ, Ott BJ, Payne WS. The incidence of adenocarcinoma in columnar-lined (Barrett’s) esophagus. N Engl J Med. 1985;313:857–9. doi: 10.1056/NEJM198510033131404. [DOI] [PubMed] [Google Scholar]
- (12).Hameeteman W, Tytgat GN, Houthoff HJ, van den Tweel JG. Barrett’s esophagus: development of dysplasia and adenocarcinoma. Gastroenterology. 1989;96:1249–56. doi: 10.1016/s0016-5085(89)80011-3. [DOI] [PubMed] [Google Scholar]
- (13).Miros M, Kerlin P, Walker N. Only patients with dysplasia progress to adenocarcinoma in Barrett’s oesophagus. Gut. 1991;32:1441–6. doi: 10.1136/gut.32.12.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).van der Burgh A, Dees J, Hop WC, van Blankenstein M. Oesophageal cancer is an uncommon cause of death in patients with Barrett’s oesophagus. Gut. 1996;39:5–8. doi: 10.1136/gut.39.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Drewitz DJ, Sampliner RE, Garewal HS. The incidence of adenocarcinoma in Barrett’s esophagus: a prospective study of 170 patients followed 4.8 years. Am J Gastroenterol. 1997;92:212–5. [PubMed] [Google Scholar]
- (16).Provenzale D, Kemp JA, Arora S, Wong JB. A guide for surveillance of patients with Barrett’s esophagus. Am J Gastroenterol. 1994;89:670–80. [PubMed] [Google Scholar]
- (17).Grimm I, Shaheen N, Bozymski EM. Surveillance for Barrett’s esophagus: are we saving lives. Gastroenterology. 1997;112:661–2. doi: 10.1053/gast.1997.v112.agast970661. [DOI] [PubMed] [Google Scholar]
- (18).Reid BJ, Haggitt RC, Rubin CE, Roth G, Surawicz CM, Van Belle G, et al. Observer variation in the diagnosis of dysplasia in Barrett’s esophagus. Hum Pathol. 1988;19:166–78. doi: 10.1016/s0046-8177(88)80344-7. [DOI] [PubMed] [Google Scholar]
- (19).Levine DS, Haggitt CR, Irvine S, Reid BJ. Natural history of high-grade dysplasia in Barrett’s esophagus [abstract] Gastroenterology. 1996;110:A550. [Google Scholar]
- (20).Schnell T, Sontag SJ, Chejfec G, Kurucar C, O’Connell S, Levine G, et al. High grade dysplasia still is not an indication for surgery in patients with Barrett’s esophagus: an update [abstract] Gastroenterology. 1998;114:A280. [Google Scholar]
- (21).Herbst JJ, Berenson MM, McCloskey DW, Wiser WC. Cell proliferation in esophageal columnar epithelium (Barrett’s esophagus) Gastroenterology. 1978;75:683–7. [PubMed] [Google Scholar]
- (22).Pellish LJ, Hermos JA, Eastwood GL. Cell proliferation in three types of Barrett’s epithelium. Gut. 1980;21:26–31. doi: 10.1136/gut.21.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Gray MR, Hall PA, Nash J, Ansari B, Lane DP, Kingsnorth AN. Epithelial proliferation in Barrett’s esophagus by proliferating cell nuclear antigen immunolocalization. Gastroenterology. 1992;103:1769–76. doi: 10.1016/0016-5085(92)91433-5. [DOI] [PubMed] [Google Scholar]
- (24).Reid BJ, Sanchez CA, Blount PL, Levine DS. Barrett’s esophagus: cell cycle abnormalities in advancing stages of neoplastic progression. Gastroenterology. 1993;105:119–29. doi: 10.1016/0016-5085(93)90017-7. [DOI] [PubMed] [Google Scholar]
- (25).Reid BJ, Haggitt RC, Rubin CE, Rabinovitch PS. Barrett’s esophagus. Correlation between flow cytometry and histology in detection of patients at risk for adenocarcinoma. Gastroenterology. 1987;93:1–11. [PubMed] [Google Scholar]
- (26).Fennerty MB, Sampliner RE, Way D, Riddell R, Steinbronn K, Garewal HS. Discordance between flow cytometric abnormalities and dysplasia in Barrett’s esophagus. Gastroenterology. 1989;97:815–20. doi: 10.1016/0016-5085(89)91483-2. [DOI] [PubMed] [Google Scholar]
- (27).Robaszkiewicz M, Reid BJ, Volant A, Cauvin JM, Rabinovitch PS, Gouerou H. Flow-cytometric DNA content analysis of esophageal squamous cell carcinomas. Gastroenterology. 1991;101:1588–93. doi: 10.1016/0016-5085(91)90396-3. [DOI] [PubMed] [Google Scholar]
- (28).Galipeau PC, Cowan DS, Sanchez CA, Barrett MT, Emond MJ, Levine DS, et al. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett’s esophagus. Proc Natl Acad Sci U S A. 1996;93:7081–4. doi: 10.1073/pnas.93.14.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Reid BJ, Blount PL, Rubin CE, Levine DS, Haggitt RC, Rabinovitch PS. Flow-cytometric and histological progression to malignancy in Barrett’s esophagus: prospective endoscopic surveillance of a cohort. Gastroenterology. 1992;102:1212–9. [PubMed] [Google Scholar]
- (30).Blount PL, Galipeau PC, Sanchez CA, Neshat K, Levine DS, Yin J, et al. 17p allelic losses in diploid cells of patients with Barrett’s esophagus who develop aneuploidy. Cancer Res. 1994;54:2292–5. [PubMed] [Google Scholar]
- (31).Barrett MT, Sanchez CA, Galipeau PC, Neshat K, Emond M, Reid BJ. Allelic loss of 9p21 and mutation of the CDKN2/p16 gene develop as early lesions during neoplastic progression in Barrett’s esophagus. Oncogene. 1996;13:1867–73. [PubMed] [Google Scholar]
- (32).Blount PL, Ramel S, Raskind WH, Haggitt RC, Sanchez CA, Dean PJ, et al. 17p allelic deletions and p53 protein overexpression in Barrett’s adenocarcinoma. Cancer Res. 1991;51:5482–6. [PubMed] [Google Scholar]
- (33).Hammoud ZT, Kaleem Z, Cooper JD, Sundaresan RS, Patterson GA, Goodfellow PJ. Allelotype analysis of esophageal adenocarcinomas: evidence for the involvement of sequences on the long arm of chromosome 4. Cancer Res. 1996;56:4499–502. [PubMed] [Google Scholar]
- (34).Barrett MT, Galipeau PC, Sanchez CA, Emond MJ, Reid BJ. Determination of the frequency of loss of heterozygosity in esophageal adenocarcinoma by cell sorting, whole genome amplification and microsatellite polymorphisms. Oncogene. 1996;12:1873–8. [PubMed] [Google Scholar]
- (35).Dolan K, Garde J, Gosney J, Sissons M, Wright T, Kingsnorth AN, et al. Allelotype analysis of oesophageal adenocarcinoma: loss of heterozygosity occurs at multiple sites. Br J Cancer. 1998;78:950–7. doi: 10.1038/bjc.1998.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Hamelin R, Flejou JF, Muzeau F, Potet F, Laurent-Puig P, Fekete F, et al. TP53 gene mutations and p53 protein immunoreactivity in malignant and premalignant Barrett’s esophagus. Gastroenterology. 1994;107:1012–8. doi: 10.1016/0016-5085(94)90225-9. [DOI] [PubMed] [Google Scholar]
- (37).Neshat K, Sanchez CA, Galipeau PC, Blount PL, Levine DS, Joslyn G, et al. p53 mutations in Barrett’s adenocarcinoma and high-grade dysplasia. Gastroenterology. 1994;106:1589–95. doi: 10.1016/0016-5085(94)90415-4. [DOI] [PubMed] [Google Scholar]
- (38).Gleeson CM, Sloan JM, McGuigan JA, Ritchie AJ, Russell SE. Base transitions at CpG dinucleotides in the p53 gene are common in esophageal adenocarcinoma. Cancer Res. 1995;55:3406–11. [PubMed] [Google Scholar]
- (39).Wong DJ, Barrett MT, Stoger R, Emond MJ, Reid BJ. p16INK4a promoter is hypermethylated at a high frequency in esophageal adenocarcinomas. Cancer Res. 1997;57:2619–22. [PubMed] [Google Scholar]
- (40).Klump B, Hsieh CJ, Holzmann K, Gregor M, Porschen R. Hypermethylation of the CDKN2/p16 promoter during neoplastic progression in Barrett’s esophagus. Gastroenterology. 1998;115:1381–6. doi: 10.1016/s0016-5085(98)70016-2. [DOI] [PubMed] [Google Scholar]
- (41).Barrett MT, Sanchez CA, Prevo LJ, Wong DJ, Galipeau PC, Paulson TG, et al. Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Genet. 1999;22:106–9. doi: 10.1038/8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Sidransky D, Frost P, Von Eschenbach A, Oyasu R, Preisinger AC, Vogelstein B. Clonal origin of bladder cancer. N Engl J Med. 1992;326:737–40. doi: 10.1056/NEJM199203123261104. [DOI] [PubMed] [Google Scholar]
- (43).Kishimoto Y, Sugio K, Hung JY, Virmani AK, McIntire DD, Minna JD, et al. Allele-specific loss in chromosome 9p loci in preneoplastic lesions accompanying non-small-cell lung cancers. J Natl Cancer Inst. 1995;87:1224–9. doi: 10.1093/jnci/87.16.1224. [DOI] [PubMed] [Google Scholar]
- (44).Paulson TG, Galipeau PC, Reid BJ. Loss of heterozygosity analysis using whole genome amplification, cell sorting and fluorescence-based PCR. Genome Res. 1999;9:482–91. [PMC free article] [PubMed] [Google Scholar]
- (45).Reid BJ, Weinstein WM, Lewin KJ, Haggitt RC, VanDeventer G, DenBesten L, et al. Endoscopic biopsy can detect high-grade dysplasia or early adenocarcinoma in Barrett’s esophagus without grossly recognizable neoplastic lesions. Gastroenterology. 1988;94:81–90. doi: 10.1016/0016-5085(88)90613-0. [DOI] [PubMed] [Google Scholar]
- (46).Gerdes J, Schwab U, Lemke H, Stein H. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer. 1983;31:13–20. doi: 10.1002/ijc.2910310104. [DOI] [PubMed] [Google Scholar]
- (47).Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710–5. [PubMed] [Google Scholar]
- (48).Prevo LJ, Sanchez CA, Galipeau PC, Reid BJ. p53-Mutant clones and field effects in Barrett’s esophagus. Cancer Res. 1999;59:4784–7. [PubMed] [Google Scholar]
- (49).Fleiss JL. Statistical methods for rates and proportions. John Wiley & Sons; New York (NY): 1981. [Google Scholar]
- (50).Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–30. [PubMed] [Google Scholar]
- (51).Blount PL, Meltzer SJ, Yin J, Huang Y, Krasna MJ, Reid BJ. Clonal ordering of 17p and 5q allelic losses in Barrett dysplasia and adenocarcinoma. Proc Natl Acad Sci U S A. 1993;90:3221–5. doi: 10.1073/pnas.90.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Gleeson CM, Sloan JM, McManus DT, Maxwell P, Arthur K, McGuigan JA, et al. Comparison of p53 and DNA content abnormalities in adenocarcinoma of the oesophagus and gastric cardia. Br J Cancer. 1998;77:277–86. doi: 10.1038/bjc.1998.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Tarmin L, Yin J, Zhou X, Suzuki H, Jiang HY, Rhyu MG, et al. Frequent loss of heterozygosity on chromosome 9 in adenocarcinoma and squamous cell carcinoma of the esophagus. Cancer Res. 1994;54:6094–6. [PubMed] [Google Scholar]
- (54).Zhou X, Tarmin L, Yin J, Jiang HY, Suzuki H, Rhyu MG, et al. The MTS1 gene is frequently mutated in primary human esophageal tumors. Oncogene. 1994;9:3737–41. [PubMed] [Google Scholar]
- (55).Muzeau F, Flejou JF, Thomas G, Hamelin R. Loss of heterozygosity on chromosome 9 and p16 (MTS1, CDKN2) gene mutations in esophageal cancers. Int J Cancer. 1997;72:27–30. doi: 10.1002/(sici)1097-0215(19970703)72:1<27::aid-ijc3>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- (56).Neshat K, Sanchez CA, Galipeau PC, Cowan DS, Ramel S, Levine DS, et al. Barrett’s esophagus: a model of human neoplastic progression. Cold Spring Harb Symp Quant Biol. 1994;59:577–83. doi: 10.1101/sqb.1994.059.01.065. [DOI] [PubMed] [Google Scholar]
- (57).Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- (58).An HX, Niederacher D, Picard F, van Roeyen C, Bender HG, Beckmann MW. Frequent allele loss on 9p21-22 defines a smallest common region in the vicinity of the CDKN2 gene in sporadic breast cancer. Genes Chromosomes Cancer. 1996;17:14–20. doi: 10.1002/(SICI)1098-2264(199609)17:1<14::AID-GCC3>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- (59).Fujii H, Marsh C, Cairns P, Sidransky D, Gabrielson E. Genetic divergence in the clonal evolution of breast cancer. Cancer Res. 1996;56:1493–7. [PubMed] [Google Scholar]
- (60).Takahashi T, Habuchi T, Kakehi Y, Mitsumori K, Akao T, Terachi T, et al. Clonal and chronological genetic analysis of multifocal cancers of the bladder and upper urinary tract. Cancer Res. 1998;58:5835–41. [PubMed] [Google Scholar]
- (61).Carder P, Wyllie AH, Purdie CA, Morris RG, White S, Piris J, et al. Stabilised p53 facilitates aneuploid clonal divergence in colorectal cancer. Oncogene. 1993;8:1397–401. [PubMed] [Google Scholar]
- (62).Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–72. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- (63).Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–7. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- (64).Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol. 1996;16:859–67. doi: 10.1128/mcb.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Rabinovitch PS, Reid BJ, Haggitt RC, Norwood TH, Rubin CE. Progression to cancer in Barrett’s esophagus is associated with genomic instability. Lab Invest. 1989;60:65–71. [PubMed] [Google Scholar]
- (66).Oda T, Tsuda H, Scarpa A, Sakamoto M, Hirohashi S. Mutation pattern of the p53 gene as a diagnostic marker for multiple hepatocellular carcinoma. Cancer Res. 1992;52:3674–8. [PubMed] [Google Scholar]
- (67).Chung KY, Mukhopadhyay T, Kim J, Casson A, Ro JY, Goepfert H, et al. Discordant p53 gene mutations in primary head and neck cancers and corresponding second primary cancers of the upper aerodigestive tract. Cancer Res. 1993;53:1676–83. [PubMed] [Google Scholar]
- (68).Sozzi G, Miozzo M, Pastorino U, Pilotti S, Donghi R, Giarola M, et al. Genetic evidence for an independent origin of multiple preneoplastic and neoplastic lung lesions. Cancer Res. 1995;55:135–40. [PubMed] [Google Scholar]