Abstract
HCF-1 is a highly conserved and abundant chromatin-associated host cell factor required for transcriptional activation of herpes simplex virus immediate-early genes by the virion protein VP16. HCF-1 exists as a heterodimeric complex of associated N- (HCF-1N) and C- (HCF-1C) terminal subunits that result from proteolytic processing of a precursor protein. We have used small-interfering RNA (siRNA) to inactivate HCF-1 in an array of normal and transformed mammalian cells to identify its cellular functions. Our results show that HCF-1 is a broadly acting regulator of two stages of the cell cycle: exit from mitosis, where it ensures proper cytokinesis, and passage through the G1 phase, where it promotes cell cycle progression. Proteolytic processing is necessary to separate and ensure these two HCF-1 activities, which are performed by separate HCF-1 subunits: the HCF-1N subunit promotes passage through the G1 phase whereas the HCF-1C subunit is involved in proper exit from mitosis. These results suggest that HCF-1 links the regulation of exit from mitosis and the G1 phase of cell growth, possibly to coordinate the reactivation of gene expression after mitosis.
Keywords: cell cycle/herpes simplex virus/mitosis/proteolysis/siRNA silencing
Introduction
Cell proliferation involves a highly regulated series of events in which the cellular genome is replicated faithfully during a synthesis (S) phase and each of the two resulting copies are segregated properly during mitosis (M). These two critical events are normally separated by two growth phases, G1 following mitosis and G2 following S phase. Orderly progression from one phase of the cell cycle to the other is mediated mainly by the regulation of gene expression, and key factors of this process are often transcriptional regulatory proteins.
Here, we elucidate cell cycle functions of the highly conserved and abundant chromatin-associated cell proliferation factor HCF-1, a host-cell factor involved in herpes simplex virus (HSV) immediate-early (IE) gene transcription. In lytic HSV infection, HCF-1 (also known as C1, VCAF and CFF) is incorporated into a multiprotein transcriptional regulatory complex with the viral activator VP16 and the cellular POU-domain transcription factor Oct-1 on HSV IE gene promoters (for a review, see Herr, 1998). Association with VP16 requires the N-terminal 380 amino acids of HCF-1 (LaBoissière et al., 1997; Wilson et al., 1997). This region probably adopts a β-propeller structure and is called the Kelch domain because of its sequence similarity to the Drosophila protein Kelch (Wilson et al., 1997). The HCF-1 Kelch domain is sufficient for VP16-induced complex formation as well as to recruit HCF-1 to chromatin in uninfected cells (Wilson et al., 1997; Wysocka et al., 2001a).
In cells, the majority of HCF-1 exists as a heterodimeric complex of N- (HCF-1N) and C- (HCF-1C) terminal subunits, which results from the proteolytic processing of the 2035 amino acid (HCF-1300) precursor protein and stable association of the resulting subunits (Wilson et al., 1993, 1995; Kristie et al., 1995). This proteolytic cleavage occurs at a series of six 26 amino acid repeats (HCF-1PRO repeats) located near the middle of the HCF-1300 protein (Wilson et al, 1995). HCF-1 processing may involve autocatalytic cleavage at the HCF-1PRO repeats (Vogel and Kristie, 2000a), but the functional significance of HCF-1 processing and subunit association is enigmatic.
HCF-1 is known to play some role in cell proliferation based on studies of the temperature-sensitive baby hamster kidney (BHK) cell line tsBN67 (Goto et al., 1997). In tsBN67 cells, the one X-linked copy of the HCF-1 gene carries a proline to serine substitution at position 134 (P134S) in the HCF-1 Kelch domain. This missense mutation does not grossly alter the stability or processing of HCF-1 at the non-permissive temperature of 40°C (Goto et al., 1997) but it does induce HCF-1 dissociation from chromatin (Wysocka et al., 2001a). This dissociation is followed by a cell proliferation arrest in the G1/G0 phase, 36–48 h after shifting to 40°C (Goto et al., 1997). The cell proliferation arrest is accompanied by a conspicuous multinucleated (primarily binucleated) phenotype in ∼15% of the arrested cells, indicating an associated cytokinesis defect (Reilly and Herr, 2002). Both tsBN67 defects can be rescued by the HCF-1N subunit (Wilson et al., 1997) and are suppressed in naturally arising tsBN67 cell growth revertants or by transformation with the DNA tumor virus SV40 early region, which has suggested that these two defects are connected (Reilly and Herr, 2002; Reilly et al., 2002). The mode of HCF-1 action in these two processes, however, is unknown.
Although providing a window into the involvement of HCF-1 in cell proliferation, tsBN67 cells possess serious limitations including that this cell line was isolated after chemical mutagenesis and may thus contain additional mutations that cooperate with the loss of HCF-1 activity to induce the cell proliferation arrest. Additionally, the function of HCF-1 processing and the HCF-1C subunit have not been elucidated with these cells. Furthermore, if HCF-1 function could be analyzed in different cell lines, it would be possible to determine (i) whether HCF-1 is a key regulator of the cell cycle; (ii) whether its function is restricted to certain cell types; and (iii) whether its function varies between normal and transformed cells.
To address these questions, we have used small-interfering RNA (siRNA) to silence the expression of the HCF-1 gene in normal and transformed cells. Our results indicate that HCF-1 is a universal regulator of cell proliferation and that HCF-1 processing is required to separate and ensure cell growth and cytokinesis functions of HCF-1 provided by the HCF-1N and HCF-1C subunits, respectively.
Results
HCF-1 is depleted efficiently and specifically by siRNA
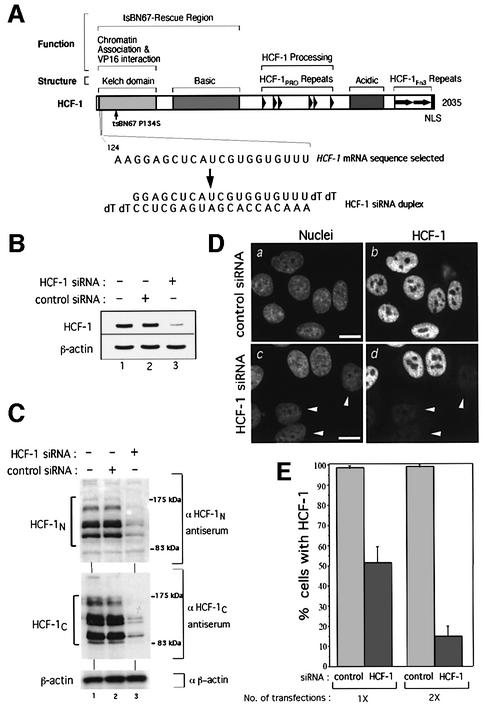
To determine the function of mammalian HCF-1, we used the siRNA strategy (Elbashir et al., 2001). Figure 1A shows a schematic of HCF-1, along with the HCF-1 mRNA sequence and corresponding HCF-1 siRNA duplex designed to disrupt HCF-1 synthesis. Successful loss of HCF-1 mRNA 3 days after two rounds of HCF-1 siRNA treatment but not a parallel treatment with an irrelevant luciferase mRNA-specific siRNA (control) is shown in Figure 1B, using a semi-quantitative RT–PCR analysis. In contrast, β-actin mRNA levels were unaffected (compare lane 3 with lanes 1 and 2, Figure 1B). Furthermore, immunoblot analysis showed that the HCF-1N and HCF-1C subunits were also significantly depleted by the same treatment (compare lane 3 with lanes 1 and 2, Figure 1C), whereas the β-actin protein was not affected (bottom panel). Thus, the HCF-1 siRNA induced the disappearance of both HCF-1 subunits by triggering the degradation of the HCF-1 mRNA. As expected, each strand of the HCF-1 siRNA alone did not affect HCF-1 expression (data not shown).
Fig. 1. Human HCF-1 is depleted efficiently and specifically by siRNA. (A) Schematic structure of HCF-1 (Wysocka et al., 2001a). Above are shown (i) structural or sequence elements and (ii) functional regions. The position of the tsBN67 P134S missense mutation is indicated. The selected HCF-1 coding region RNA sequence (nucleotides 124–144; Wilson et al., 1993) and the corresponding HCF-1 siRNA duplex are indicated below. (B) HCF-1 (top) and β-actin (bottom) mRNA levels in untreated HeLa cells (lane 1) or 3 days after treatment with either control (lane 2) or HCF-1 (lane 3) siRNA. (C) Immunoblot analysis of HCF-1N (top), HCF-1C (middle) and β-actin (bottom) proteins of untreated HeLa cells (lanes 1) or 3 days after treatment with control (lanes 2) or HCF-1 (lanes 3) siRNA. (D) Control (a and b) or HCF-1 (c and d) siRNA-treated cells were subjected to analysis after paraformaldehyde fixation by nuclear DAPI (a and c) and αHCF-1N (b and d) staining. Arrowheads point to cells exhibiting HCF-1 depletion. Scale bar: 5 µm. (E) Percentage of HCF-1-positive cells after one or two transfections with control or HCF-1 siRNA as determined by immunofluorescence. The percentage of HCF-1-positive nuclei was obtained by counting 200 cells per sample in each of three independent experiments.
To evaluate the efficacy of siRNA-induced HCF-1 depletion, HeLa cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) to identify nuclei, and with αHCF-1N antiserum to identify HCF-1-positive cells 3 days after siRNA treatment, as shown in Figure 1D. Whereas all of the untreated cells were positive for HCF-1 (compare a and b), many (but not all) of the HCF-1 siRNA-treated cells showed a sharp decrease in HCF-1 staining (compare c and d, arrowheads). One transfection led to 50% HCF-1-negative cells after 3 days, whereas two transfections separated by 12 h led to 85% HCF-1-negative cells, as shown in Figure 1E. These results indicate that the selected HCF-1 siRNA depletes HeLa cell HCF-1 protein with the efficiency influenced by the number of transfections. Below, we performed two rounds of siRNA treatment to deplete HCF-1 effectively.
HCF-1 depletion prevents progression through S phase
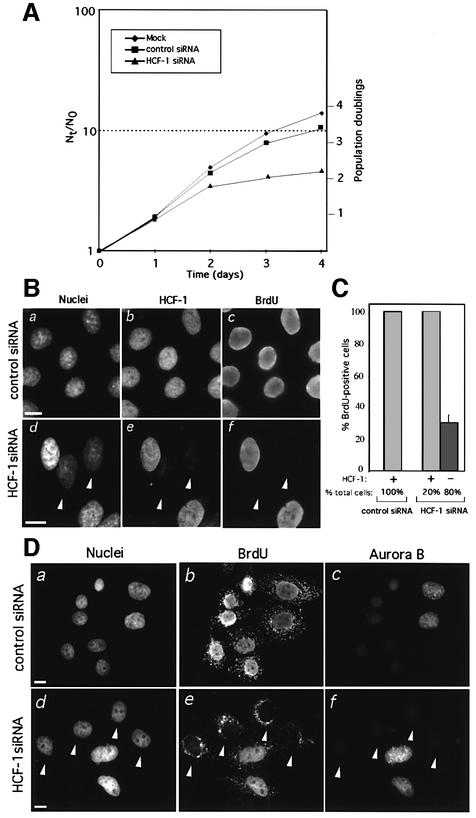
Owing to the P134S missense mutation, tsBN67 cells display a temperature-sensitive cell proliferation arrest (Goto et al., 1997) but retain normal HCF-1 processing and stable HCF-1N/P134S and HCF-1C subunits. The cellular phenotype resulting from depletion of both HCF-1 subunits is not known, however. To examine such a phenotype, we compared the proliferation rates of HeLa cells either untreated or treated with control or HCF-1 siRNAs as shown in Figure 2A. HCF-1 siRNA-transfected cells displayed a decrease in cell proliferation 2 days after transfection, whereas control siRNA-transfected cells continued to proliferate at nearly the same rate as mock-treated cells. Thus, as with temperature treatment of tsBN67 cells, depletion of HCF-1 by siRNA in HeLa cells is accompanied by changes in the rates of cell proliferation.
Fig. 2. HCF-1 depletion prevents G1 progression. (A) Proliferation rates of untreated HeLa cells or after treatment with either control or HCF-1 siRNA. Cells were seeded at a density of 8 × 104 cells per well in 6-well plates the day before transfection. At the indicated times after the first of two transfections, cells were harvested and counted from duplicate plates, and the results averaged. N0, the number of cells at time 0; Nt, number of cells at each specified time point. (B) Long-term BrdU incorporation assay of HCF-1 or control siRNA-treated HeLa cells. Nuclei were identified by DAPI (a and d), HCF-1 by αHCF-1N (b and e), and BrdU by αBrdU (c and f) staining, after ethanol fixation. Arrowheads identify HCF-1-negative cells, which lack BrdU incorporation. (C) Quantification of the long-term BrdU incorporation assay of HCF-1-positive and negative HeLa cells after treatment with either control or HCF-1 siRNA. These percentages were determined by counting 200 cell per sample in each of three independent experiments. (D) Detection of Aurora-B in HeLa cells treated with either control (a–c) or HCF-1 (d–f) siRNA. (a and d) nuclear DAPI staining; (b and e) BrdU staining; (c and f) Aurora-B staining. Cells were fixed with paraformaldehyde, for which BrdU staining labels cytoplasmic mitochondria. White arrowheads identify BrdU- and Aurora-B-negative cells. Scale bar: 5 µm.
To characterize the proliferative status of individual HCF-1-depleted cells, we identified S phase-competent cells with a long-term bromodeoxyuridine (BrdU) incorporation assay, as shown in Figure 2B. Transfected cells were grown for 2 days and then incubated with BrdU for a further 24 h to ensure that all proliferating cells are labeled as they pass through S phase. Cells subsequently were stained (i) with DAPI to identify nuclei; (ii) with αHCF-1N antiserum to distinguish the HCF-1-positive and -negative cells; and (iii) with αBrdU antiserum to identify proliferating cells. Proliferating siRNA control-treated cells all contained HCF-1 and incorporated BrdU (Figure 2B, compare a–c). In contrast, siRNA-induced HCF-1- depleted cells failed to incorporate BrdU (arrowheads), whereas accompanying HCF-1-positive cells incorporated BrdU (compare positive and negatively stained cells in e and f). As shown in Figure 2C, quantification of BrdU-positive and negative cells shows that all of the HCF-1-positive cells in both the control (100% of the total cells) and HCF-1 siRNA-transfected (20% of total cells) samples incorporated BrdU, indicating that this assay can effectively measure S phase-competent cells. In the HCF-1 siRNA-treated sample, however, of the 80% of the cells that were HCF-1 negative, 70% failed to incorporate BrdU (Figure 2C), showing an evident albeit incomplete defect in entering or progressing through S phase in the absence of HCF-1.
HCF-1-depleted cells are arrested in the G1 phase
To determine if individual arrested HCF-1-depleted cells are in a specific phase of the cell cycle, we took advantage of the differential cell cycle phase-specific staining of Aurora-B kinase in HeLa cells (Crosio et al., 2002): by immunofluorescence, Aurora-B kinase is not detected in G1/S and displays a punctate nuclear distribution in G2. We used the G2-specific Aurora-B staining pattern to distinguish G1 and G2 phase cells. As shown in Figure 2D, all control siRNA-treated cells incorporated BrdU (panel b) and showed either background non-Aurora-B G1/S staining with generally smaller nuclei, or G2 punctate nuclear Aurora-B staining with generally larger nuclei (compare six G1/S phase cells on the left and two G2 cells on the right in a–c). In HCF-1 siRNA-treated cells, BrdU-negative cells appeared (arrowheads, panel e). These cells correspond to HCF-1-depleted cells (see Figure 2B and C). All of these BrdU-negative cells also exhibit the G1/S phase-specific absence of Aurora-B staining (panel f). Because these cells failed to enter S phase at any time during the long-term BrdU labeling, these results suggest that HCF-1 depletion leads to a G1 phase arrest.
Inactivation of the pRb protein family is not sufficient to rescue the cell cycle arrest induced by HCF-1 depletion
The G1 phase arrest induced by the depletion of HCF-1 in HeLa cells is reminiscent of the temperature-induced arrest of tsBN67 cells. The HCF-1 siRNA HeLa cell proliferation arrest is intriguing because inactivation of the pRb protein family in tsBN67 cells by the SV40 early region (SV40e) can suppress the effect of the P134S HCF-1 mutation (Reilly et al., 2002), and in HeLa cells the pRb protein family is already inactivated by the human papillomavirus E7 protein (Masters, 2002). These results suggest that the effect of the P134S mutation is less severe than that of HCF-1 depletion. To test this hypothesis, we asked whether HCF-1 depletion in tsBN67SV40e cells disrupts their proliferation.
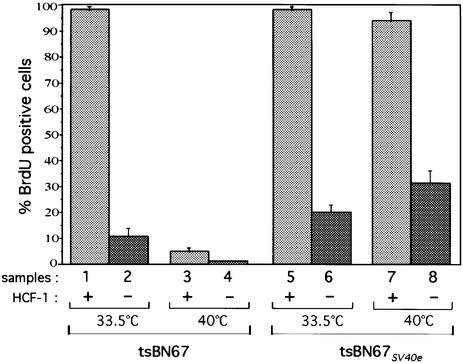
After hamster HCF-1 siRNA transfection, cells were grown for 3 days at permissive (33.5°C) or non-permissive (40°C) temperature, which was sufficient to deplete HCF-1 as determined by immunoblot analysis (data not shown), and then incubated in the presence of BrdU for 24 h. As shown in Figure 3, consistent with previous results (Goto et al., 1997; Reilly et al., 2002), HCF-1-positive tsBN67 cells incorporated BrdU at 33.5°C but not at 40°C (compare samples 1 and 3), whereas the HCF-1-positive tsBN67SV40e cells incorporated BrdU at both temperatures (compare samples 5 and 7). In contrast, HCF-1 depletion induced a cell cycle arrest in tsBN67 cells at both 33.5 and 40°C (compare samples 2 and 4). Interestingly, the HCF-1 depletion also led to the same arrest of tsBN67SV40e cells (samples 6 and 8). These results indicate that the mutant P134S HCF-1 protein retains one or more activities that can cooperate with the SV40e region to promote cell proliferation. Interestingly, HCF-1-depleted tsBN67SV40e cells display an elevated number of BrdU-positive cells at 33.5 and 40°C as compared with the parental tsBN67 cells (compare samples 2, 4, 6 and 8); indeed the level of BrdU-positive tsBN67SV40e cells is very similar to that in HCF-1-depleted HeLa cells (compare Figures 2C and 3), suggesting that inactivation of the pRb protein family still bypasses some cell proliferation requirements for HCF-1 in HCF-1-depleted cells.
Fig. 3. Inactivation of the pRb protein family is not sufficient to rescue the cell cycle arrest induced by HCF-1 depletion. Quantification of long-term BrdU incorporation in HCF-1-positive (odd numbered samples) and negative (even numbered samples) tsBN67 (samples 1–4) and tsBN67SV40e (samples 5–8) cells after treatment with hamster-specific HCF-1 siRNA. The percentages were obtained by counting 200 cells per sample in each of three independent experiments. Control siRNA-treated tsBN67 and tsBN67SV40e cells displayed the same BrdU incorporation pattern as the HCF-1-positive HCF-1 siRNA-transfected cells shown in the figure (data not shown).
HCF-1 depletion causes a cell cycle arrest in normal as well as in oncogenically transformed cells
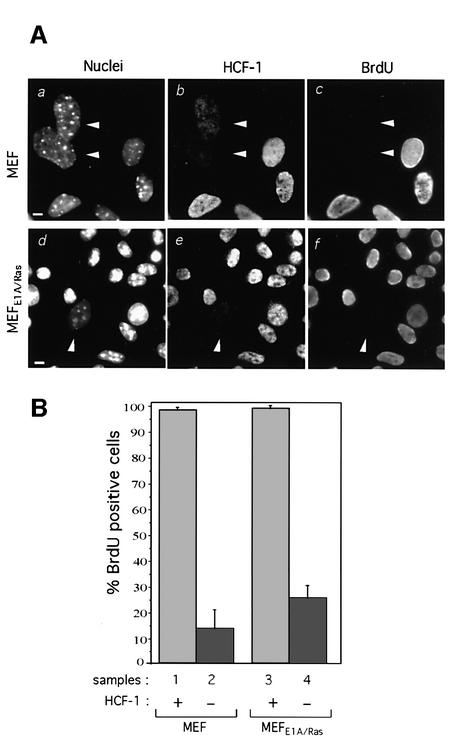
The ability to disrupt S phase entry by siRNA in both HeLa and tsBN67 cells—cell lines from different species, tissues and oncogenic transformation state—indicates that the role(s) of HCF-1 in cell proliferation is not restricted to specific cells. Because the genetic and growth properties of HeLa and tsBN67 cells are altered, we asked whether HCF-1 depletion causes cell proliferation defects in normal cells. We therefore treated mouse embryo fibroblasts (MEFs) with mouse-specific HCF-1 siRNA and analyzed them by the long-term BrdU incorporation assay. By performing immunofluorescence on individual cells, we were able to detect 5–10% HCF-1-depleted MEFs after HCF-1-siRNA treatment. As shown in Figure 4A (a–c), all HCF-1-positive MEFs incorporated BrdU, whereas HCF-1-depleted cells (arrowheads) did not, indicating that loss of HCF-1 in normal cells also induces a cell cycle arrest. [For unexplained reasons, HCF-1-depleted MEFs often displayed enlarged nuclei (see panel a).] Quantification (Figure 4B, samples 1 and 2) shows that essentially all of the HCF-1-positive cells but <15% of the HCF-1-depleted cells entered S phase, suggesting that HCF-1 is important for cell cycle progression in normal cells.
Fig. 4. HCF-1 depletion causes a cell proliferation arrest in primary and oncogenically transformed cells. (A) Long-term BrdU incorporation by MEF (a–c) and MEFE1A/Ras (d–f) cells after treatment with mouse HCF-1 siRNA. Nuclear DAPI (a and d), αHCF-1C (b and e) and αBrdU (c and f) staining are shown. Arrowheads point to HCF-1 depleted cells, which are negative for BrdU incorporation. (B) Quantification of BrdU-positive cells in HCF-1 siRNA-treated MEF (samples 1 and 2) and MEFE1A/Ras (samples 3 and 4) cells. The percentage of cells incorporating BrdU was obtained by counting 200 cells per sample in each of three independent experiments. Scale bar: 5 µm.
To determine whether defined oncogenic cellular transformation could relieve the requirement for HCF-1 function, we tested the effects of siRNA-induced HCF-1 depletion in low-passage adenovirus E1A and oncogenic Ras V12-transformed MEFs (MEFE1A/Ras). As shown in Figure 4A (d–f), even though the efficiency of HCF-1 depletion was low, we observe in individual MEFE1A/Ras cells that these potent oncogenes do not relieve the requirement for HCF-1, although quantification shows that, as with tsBN67SV40e and tsBN67 cells, there is some induction of S phase entry compared with the wild-type MEFs (25% versus 13%, samples 2 and 4, Figure 4B). These results show that HCF-1 is a broad, if not universal, key promoter of cell growth and proliferation in mammalian cells.
Silent mutations in the HCF-1 mRNA sequence targeted by the HCF-1 siRNA protect from siRNA-induced silencing
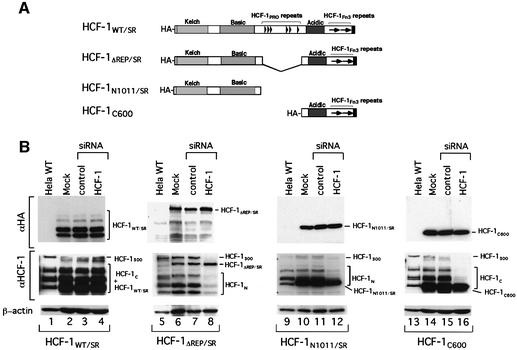
To identify the regions of HCF-1 required to promote cell proliferation, a set of four recombinant flu hemagglutinin (HA) epitope-tagged siRNA-resistant (SR) HCF-1 molecules were stably expressed in HeLa cells. To prevent destruction of the recombinant HCF-1 mRNAs containing the sequence targeted by the HCF-1 siRNA, we used the strategy of Lassus et al. (2002): nine silent mutations were introduced into the HCF-1 sequence corresponding to the siRNA duplex, creating siRNA-resistant HCF-1 (HCF-1SR) proteins. As expected, these silent mutations did not affect rescue of the tsBN67 cell proliferation defect (data not shown). Figure 5A shows the structure of the four HA-HCF-1SR proteins: (i) HCF-1WT/SR representing the wild-type protein; (ii) HCF-1ΔREP/SR lacking the HCF-1PRO repeats and deficient in HCF-1 proteolytic processing; (iii) HCF-1N1011/SR representing the HCF-1N subunit; and (iv) the naturally siRNA-resistant HCF-1C600 representing the HCF-1C subunit.
Fig. 5. Silent mutations in the HCF-1 mRNA sequence targeted by the HCF-1 siRNA duplex prevent ectopic HCF-1 depletion by siRNA. (A) Schematic structures of the four siRNA-resistant (SR) HCF-1 recombinant proteins. (B) Immunoblot analysis of untreated wild-type HeLa cells (lanes 1, 5, 9 and 13) or HeLa cells synthesizing the different HCF-1SR recombinant proteins either untreated (lanes 2, 6, 10 and 14) or treated with control (lanes 3, 7, 11 and 15) or HCF-1 (lanes 4, 8, 12 and 16) siRNA. HCF-1300, HCF-1 precursor. The HCF-1SR proteins were visualized alone (αHA tag antiserum, top panel) or together with endogenous HCF-1 proteins (middle panel) using the αHCF-1N (lanes 5–12) or αHCF-1C (lanes 1–4 and 13–16–) antisera. β-actin, loading control (bottom panel).
Each of these four HeLa cell lines was subjected to siRNA treatment and analyzed by immunoblot as shown in Figure 5B. The αHA antiserum showed that each of the HCF-1SR proteins is resistant to siRNA treatment (top panels, compare lanes 2–4, 6–8, 10–12 and 14–16), whereas HCF-1 antisera showed that the endogenous HCF-1 subunits disappeared after HCF-1 siRNA treatment for the HCF-1ΔREP/SR (middle panel, lanes 7 and 8), HCF-1N1011/SR (lanes 11 and 12) and HCF-1C600 (lanes 15 and 16) cell lines; for the HCF-1WT/SR sample, it was not possible to ascertain the loss of endogenous HCF-1 owing to the co-migration of the HCF-1WT/SR subunits.
The HCF-1N subunit is necessary and sufficient to promote G1 phase progression and S phase entry
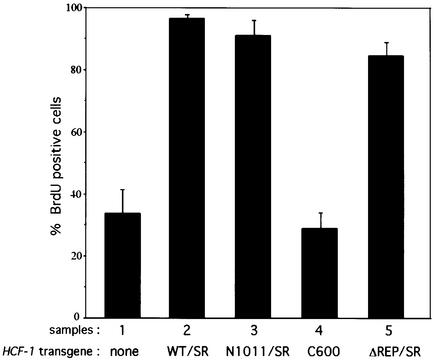
We next examined the ability of the different HA-HCF-1SR proteins to rescue the S phase entry defect in endogenous HCF-1-depleted cells, by performing the long-term BrdU incorporation assay as shown in Figure 6. The control siRNA had no effect on BrdU incorporation in any of the cell lines (data not shown). Owing to the inability to identify HCF-1-depleted cells with the HCF-1WT/SR and HCF-1ΔREP/SR cell lines, the quantification in Figure 6 includes all cells irrespective of their HCF-1 depletion status. Thus, ∼35% of the wild-type HeLa cells were S phase competent (sample 1), which represents the background in this assay. Expression of the HCF-1WT/SR protein restored the ability of the cells to incorporate BrdU, confirming that the S phase defect is indeed caused by HCF-1 depletion (sample 2). HCF-1 processing is not required to rescue the S phase defect because the HCF-1ΔREP/SR protein restored BrdU incorporation to near wild-type levels (compare samples 2 and 5).
Fig. 6. The HCF-1N subunit is necessary and sufficient to promote G1 progression and entry into S phase. Quantification of the long-term BrdU incorporation in parental HeLa cells (sample 1) or HeLA cells synthesizing HCF-1SR proteins (samples 2–5) performed as described in Materials and methods is shown. The percentage of cells incorporating BrdU was obtained by counting 200 cells per sample in each of three independent experiments.
In previous experiments with tsBN67 cells, the HCF-1N1011 but not the HCF-1C600 construct could rescue cell proliferation at non-permissive temperature (Wilson et al., 1997). In these experiments, however, it was not possible to determine whether the HCF-1N subunit alone is sufficient to rescue cell cycle progression because the P134S mutant protein in tsBN67 cells is synthesized at near wild-type levels and processed normally. By using siRNA, however, we can deplete the HCF-1N and HCF-1C subunits simultaneously (see Figure 1C) and address the roles of the two HCF-1 subunits independently. Interestingly, expression of the HCF-1N subunit alone (i.e. HCF-1N1011/SR), but not the HCF-1C subunit (i.e. HCF-1C600), was sufficient to restore BrdU incorporation to wild-type levels (compare samples 3 and 4). Thus, the HCF-1N subunit is necessary and sufficient to promote G1 phase progression and S-phase entry.
HCF-1-depleted cells arrest with a population of multinucleated cells
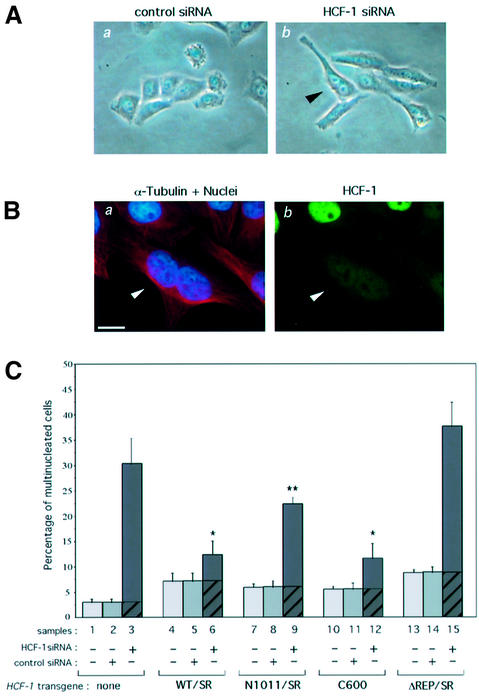
As mentioned above, temperature-arrested tsBN67 cells display a multinucleated, primarily binucleated, phenotype in ∼15% of arrested cells (Reilly and Herr, 2002). Three days after transfection, we examined the siRNA-treated HeLa cells for a similar phenotype, as shown in Figure 7. Like wild-type HeLa cells, the control siRNA-treated cells were clustered and displayed a compact shape with few multinucleated cells (Figure 7A, a). In contrast, the HCF-1 siRNA-treated HeLa cells displayed elongated shapes and many multinucleated, predominantly binucleated, cells (e.g. see arrowhead in b), indicating that, as with cell growth, the loss of HCF-1 function in different cell types can lead to a multinucleation defect, probably arising from improper cytokinesis.
Fig. 7. HCF-1 depletion causes a cytokinesis defect that is rescued by the HCF-1C subunit. (A) Representative phase-contrast images of HeLa cells are shown after treatment with control (a) or HCF-1 (b) siRNA. The black arrowhead points to an elongated and binucleated cell. (B) Immunofluorescence analysis of a binucleated cell after HCF-1 siRNA treatment. (a) α-tubulin (red) and DAPI (blue) staining; (b) αHCF-1N (green) staining. Arrowhead, HCF-1-depleted binucleated cell. Scale bar: 5 µm. (C) Quantification of multinucleated cells of parental HeLa cells (samples 1–3) or HeLa cells synthesizing the HCF-1SR proteins (samples 4–15), either untreated (samples 1, 4, 7, 10 and 13), or treated with control (samples 2, 5, 8, 11, and 14) or HCF-1 (samples 3, 6, 9, 12, and 15) siRNA. Multinucleated cells were identified by immunofluorescence using DAPI, and αHA and α-tubulin antisera. The percentage of multinucleated cells was obtained by counting 200 cells per sample in each of three independent experiments. P-value of <0.02 (*) or <0.06 (**) as determined by Student’s t-test.
Fluorescent staining with (i) DAPI to identify the nuclei, (ii) αHCF-1C antiserum to detect HCF-1 and (iii) α-tubulin antiserum to determine the cell borders revealed that the multinucleated cells were depleted of HCF-1 (Figure 7B, see arrowhead), indicating that the multinucleation phenotype is indeed due to the inactivation of HCF-1.
The HCF-1C subunit ensures proper cytokinesis
Using the same α-tubulin–DAPI fluorescence assay as in Figure 7B, we quantitated the levels of multinucleation in the siRNA-treated cells 3 days after transfection. Analysis of the parental HeLa cells shows that ∼3% of untreated or control siRNA-treated cells are multinucleated (compare samples 1 and 2 in Figure 7C), whereas 25–35% of the HCF-1 siRNA-treated cells were multinucleated (sample 3). Thus, loss of HCF-1 function in different cell types (HeLa versus BHK) and by different mechanisms (HCF-1 siRNA versus HCF-1 temperature sensitivity) leads to a pronounced defect in mitosis, probably cytokinesis, in many but not all cells.
To determine which regions of HCF-1 are involved in cytokinesis, we assayed the ability of the HCF-1SR proteins to rescue HCF-1 siRNA-induced multinucleation. Before any siRNA treatment, the percentage of multinucleated cells present in the different populations of HeLa cells varied between 5 and 8%, and this variation was not affected by the transfection of control siRNA (Figure 7C, compare samples 4 and 5, 7 and 8, 10 and 11, and 13 and 14); this variability could result from the overexpression of the various recombinant HCF-1 proteins and represents the multinucleation background for each sample (see the hatched portion of the bar in samples 3, 6, 9, 12 and 15).
The HCF-1WT/SR cells displayed a pronounced decrease in the level of HCF-1 siRNA-induced multinucleation: the HCF-1 siRNA treatment induced multinucleation in 6% of the HCF-1WT/SR cells as opposed to 27% in the parental HeLa cells (compare the solid portions of samples 3 and 6). Thus, the wild-type HCF-1 siRNA-resistant construct can rescue much but not all of the multinucleation defect, indicating that the cytokinesis defect is indeed caused by loss of HCF-1 function.
HCF-1 siRNA treatment led to induction of multinucleation in 18% of the HCF-1N1011/SR cells (Figure 7C, sample 9), indicating that the HCF-1N subunit cannot independently rescue the cytokinesis defect effectively. In contrast, the HCF-1C subunit alone suppressed the multinucleation phenotype (sample 12). These results suggest that the roles of HCF-1 in cell growth and cell division, rather than connected as previously suggested (Reilly and Herr, 2002; Reilly et al., 2002), are largely segregated into the different HCF-1 subunits: the HCF-1N subunit promotes cell growth and the HCF-1C subunit ensures proper cell division.
Proteolytic processing of HCF-1 is important for ensuring proper cytokinesis
To date, the role of HCF-1 processing has been enigmatic; it is not required to rescue tsBN67 (Wilson et al., 1997) nor HeLa cell HCF-1 siRNA-induced (Figure 6) cell growth defects. Inactivation of HCF-1 processing in the HCF-1ΔREP/SR construct results, however, in failure to suppress the HeLa cell HCF-1 siRNA-induced multinucleation phenotype (Figure 7C, sample 15). Thus, surprisingly, proteolytic processing of HCF-1 separates the cell growth and mitotic functions of the HCF-1N and HCF-1C subunits, respectively, and is required for the HCF-1C subunit to function in cytokinesis.
The HCF-1N subunit can promote G1 phase progression and S phase entry after improper cytokinesis
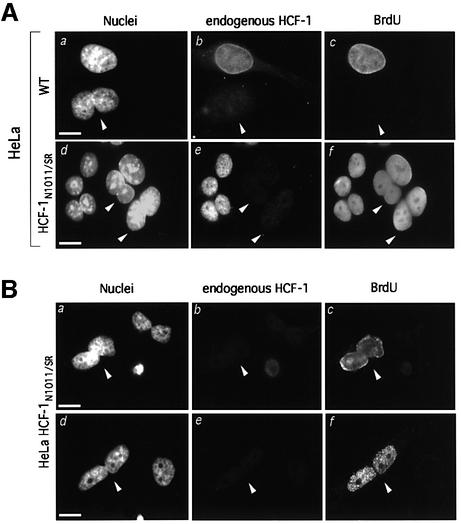
The HCF-1 siRNA-induced cell proliferation arrest occurs in cells with either a single nucleus or multiple nuclei, as illustrated for a binucleated cell in Figure 8A (a–c). Because HCF-1N1011/SR cells display wild-type HCF-1 siRNA-resistant cell growth (Figure 6) but defective cytokinesis (Figure 7), we asked whether these cells can enter S phase after improper cytokinesis. Figure 8A (d–f) shows that binucleated HCF-1N1011/SR cells incorporate BrdU in a long-term labeling assay (see arrowheads).
Fig. 8. The HCF-1N subunit can promote G1 phase progression and S phase entry after improper cytokinesis. (A) Immunofluorescence analysis of the long-term BrdU incorporation assay of multinucleated cells is shown for parental (a–c) and HCF-1N1011/SR (d–f) HeLa cells. (a and d) DAPI staining; (b and e) αHCF-1C staining for endogenous HCF-1; (c and f) αBrdU staining. White arrowheads identify HCF-1-depleted binucleated cells. (B) Immunofluorescence analysis of short-term BrdU incorporation in HCF-1 siRNA-treated HCF-1N1011/SR cells. (a and d) DAPI staining; (b and e) αHCF-1C staining for endogenous HCF-1; (c and f) αBrdU staining. Arrowheads identify BrdU-positive, HCF-1-depleted binucleated cells. Scale bar: 5 µm.
To determine whether these labeled multinucleated HCF-1N1011/SR cells indeed result from S phase entry (as opposed to improper mitosis after being labeled in S phase), we performed a short BrdU incorporation assay (1 h). As shown in Figure 8B, in this short-term assay, multinucleated HCF-1-depleted BrdU-positive cells appeared (arrowheads), suggesting that the HCF-1N subunit can promote S phase entry even after improper cell division owing to the loss of HCF-1C subunit function. The short-term BrdU staining patterns in Figure 8B (c and f) differ (diffuse and punctate, respectively), indicating labeling during different stages of S phase (O’Keefe et al., 1992), but the patterns are similar in both nuclei of the individual cells. These results suggest that, within a binucleated HCF-1N1011/SR cell, both nuclei appear to progress through S phase synchronously. These results illustrate how the role of the HCF-1N subunit in cell growth is independent of HCF-1C subunit function.
Discussion
We have used siRNA to inactivate HCF-1 in cells of multiple mammalian species, and in normal and transformed cells to unravel its cellular function. Our results show that HCF-1 is a broadly acting regulator of two stages of the mammalian cell cycle: passage through the G1 phase and exit from mitosis. These two activities of HCF-1 are performed by separate HCF-1 subunits. The HCF-1N subunit promotes cell growth and passage through the G1 phase, whereas the HCF-1C subunit ensures proper exit from mitosis. HCF-1 proteolytic processing is necessary for HCF-1C subunit function in cytokinesis. These results reveal the varied and broad roles of HCF-1 in the mammalian cell cycle and show that, in addition to its important role in HSV replication, HCF-1 possesses important cell proliferation regulatory functions in uninfected cells.
A limitation of the siRNA approach is that gene inactivation is restricted to the transfected cells and even all of the transfected cells do not respond to the siRNA treatment. We therefore used multiple single-cell assays to determine the effects of HCF-1 loss in individual cells, including (i) detection of HCF-1-depleted cells; (ii) long-term BrdU incorporation to assay entry and progression through S phase; (iii) Aurora-B localization combined with BrdU incorporation to differentiate G1, S and G2 phase cells; and (iv) α-tubulin and DAPI staining to detect multinucleated cells. These assays allowed us to study the role of HCF-1 in both cell growth and division. To show that the effect of the HCF-1 siRNA was due to the loss of HCF-1 function, we created siRNA-resistant wild-type HCF-1 molecules in HeLa cells and rescued the wild-type phenotype. Use of mutant siRNA-resistant HCF-1 molecules resulted in the identification of the different roles of the HCF-1N and HCF-1C subunits, and HCF-1 processing in human cell cycle progression.
Role of HCF-1 proteolysis in promoting proper cytokinesis
An unusual feature of HCF-1 is its proteolytic processing, which occurs stochastically at six centrally located 26 amino acid repeats, resulting in stable N- and C-terminal subunits, the large majority of which remain associated with one another after proteolysis (Wilson et al., 1995). Although HCF-1 proteolytic processing has been conserved in Drosophila (Mahajan et al., 2003), albeit not Caenorhabditis elegans (Wysocka et al., 2001b), its role in HCF-1 function has been a mystery because of the absence of any cellular assay to study this process. The only previous cellular model for HCF-1 function has been the tsBN67 hamster cell line in which processing of the mutant HCF-1 occurs normally (Goto et al., 1997) and processing of wild-type HCF-1 is not required to rescue HCF-1 function (Wilson et al., 1997). The inability of the HCF-1ΔREP molecule to rescue the multinucleated phenotype indicates that HCF-1 processing is important for the cytokinesis functions of the HCF-1C subunit. Alternatively, the HCF-1PRO repeat or flanking sequences that are deleted in the HCF-1ΔREP construct play a role in cytokinesis. We do not favor this hypothesis because, in contrast to the rest of the protein, the sequences flanking the HCF-1PRO repeats are not evolutionarily conserved (Goto et al., 1997; Kristie, 1997) and therefore probably do not possess important functional activities. We suggest that the proteolytic processing is important to permit the dissociation of the HCF-1 subunits at a specific phase of the cell cycle, perhaps mitosis, as a part of performing their function in the cell. In this manner, HCF-1 proteolytic processing separates cell growth and mitotic functions of HCF-1 and ensures their proper execution.
The proteolytic processing properties of HCF-1 are similar to those recently described for the mixed lineage leukemia (MLL, also known as acute lymphoblastic leukemia or ALL) oncoprotein (Nakamura et al., 2002; Yokoyama et al., 2002; Hsieh et al., 2003). MLL is a 3968 amino acid protein that is processed at two sites to create stable subunits that remain associated with one another. Interestingly, much like the results described here, the MLL N- and C-terminal subunits possess separate negative and positive transcriptional activities, respectively. In this regard, we note that the HCF-1C subunit possesses a well-defined transcriptional activation domain (Luciano and Wilson, 2002) and the HCF-1N subunit possesses both repressive and activating properties (Vogel and Kristie, 2000b; Piluso et al., 2002).
HCF-1N functions to promote G1 phase progression
The siRNA-induced inactivation of HCF-1 causes a G1 phase cell cycle arrest in human cells that is reminiscent of the HCF-1 P134S temperature-induced G1/G0 arrest in hamster tsBN67 cells (Goto et al., 1997). Owing, however, to the stability of the P134S HCF-1 protein in tsBN67 cells, it was not possible to determine all the regions of HCF-1 that are necessary and sufficient to promote cell proliferation (Wilson et al., 1997). By using siRNA, we depleted both of the HCF-1N and HCF-1C subunits simultaneously and showed, by use of siRNA-resistant molecules, that the HCF-1N subunit alone is sufficient to promote G1 phase progression as assayed by S phase entry. Thus, the HCF-1N subunit apparently is sufficient to rescue cell growth.
Because the HCF-1N subunit interacts with and modulates the transcriptional activity of many transcription factors such as GABP, LZIP and Miz-1 (Vogel and Kristie, 2000b; Luciano and Wilson, 2002; Piluso et al., 2002) and associates selectively with the Sin3 histone deacetylase and Set1/Ash2 histone methyltransferase (Wysocka et al., 2003), HCF-1 probably promotes progression through G1 by regulating gene transcription. Although the relative significance of these various interactions remains to be unraveled, the role of HCF-1 in Miz-1 function is particularly interesting. Miz-1 can inhibit G1 phase progression, possibly by activating the promoter for the cyclin-dependent kinase inhibitor p15INK4b, which in turn inhibits pRb inactivation by phosphorylation and thus S phase entry. HCF-1 can interfere with Miz-1 p15INK4b activation, suggesting that it can indirectly promote S phase entry by permitting phosphorylation and inactivation of pRb (Piluso et al., 2002). Consistent with this model, inactivation of pRb family members by the SV40 large T antigen and adenovirus oncoproteins can rescue the cell cycle defect in tsBN67 cells (Reilly et al., 2002).
This model, however, probably does not explain all of HCF-1’s roles in promoting G1 progression because, although pRb is inactivated in HeLa, tsBN67SV40e and MEFE1A/Ras cells, our results show that the depletion of HCF-1 in these cells still causes a defect in cell growth, albeit less severe than in normal cells (Figures 2–4). These results indicate that HCF-1 is probably involved in multiple regulatory pathways. Consistent with this hypothesis, in addition to a role in transcription, HCF-1 recently has been implicated in pre-mRNA spliceosome assembly by interacting with snRNPs (Ajuh et al., 2002).
HCF-1C functions to ensure proper cytokinesis
An important finding of our studies is that the role of HCF-1 in mitosis, specifically cytokinesis, is played by the HCF-1C subunit (Figure 7). How might the HCF-1C subunit ensure proper exit from mitosis? Interestingly, the HCF-1C subunit associates with protein phosphatase 1 (PP1; Ajuh et al., 2000). There are three closely related PP1 isoforms, α, β and γ, encoded by three different genes (for a review, see Cohen, 2002). PP1γ, at least, plays a role in cytokinesis (Cheng et al., 2000). Although it is not known which PP1 proteins associate with HCF-1 and which PP1 targets are specified by HCF-1 in vivo, it is tempting to imagine that the HCF-1C subunit modulates the activity of one or more PP1 isoforms, especially PP1γ, during mitosis to ensure proper cytokinesis.
If the HCF-1C subunit is responsible for ensuring proper cytokinesis, then why do tsBN67 cells, which contain wild-type HCF-1C subunits, display this mitotic defect (Reilly and Herr, 2002)? Perhaps, the temperature-sensitive P134S HCF-1N subunit has a dominant inhibitory effect on HCF-1C function in tsBN67 cells, resulting in the multinucleation phenotype even in the presence of the wild-type HCF-1C subunit. Additionally, the HCF-1N subunit may play a role in cytokinesis as evidenced by the partial rescue of the multinucleation phenotype by the HCF-1N1011/SR construct (Figure 7C). We note that even our wild-type HCF-1WT/SR cDNA construct does not completely rescue the multinucleation defect; this may be due to the absence of an HCF-1 variant (called HCF-1Δ382–450) produced by alternative pre-mRNA splicing in which the resulting HCF-1N/Δ382–450 and HCF-1C subunits fail to associate (Wilson et al., 1995).
Does HCF-1 regulate the transition from mitosis to the G1 or G0 phase of the cell cycle?
Of the many steps in the orderly progression through the cell cycle, the regulation of the M/G1 phase transition is perhaps the least well understood. Many very important events that regulate cell fate occur at this point in the cell cycle: it is here that the gene expression program of the cell is re-established following the dramatic chromosomal condensation events of mitosis. Furthermore, it is at this stage that cells generally make the decision to exit from the cell cycle into the G0 phase and differentiate. Our current understanding of the roles of HCF-1 in cell proliferation suggests that it plays multiple roles in the M/G1 phase transition, specifically cytokinesis and early G1 cell cycle progression. Through proteolytic processing and the generation of a heterodimeric complex of HCF-1N and HCF-1C subunits with distinct functions, HCF-1 is apparently able to link the regulation of exit from mitosis and the G1 phase of cell growth. If true, HCF-1 could also play important roles in the cellular decision to enter the G0 phase and initiate specific gene expression cascades involved in cell differentiation. Such activities of HCF-1 in this critical transition of the cell cycle could explain why HCF-1 is an attractive target of the HSV VP16 regulator to control the lytic or latent outcome of HSV infection in proliferating and non-proliferating cells.
Materials and methods
Cell culture
HeLa cells, MEFs and tsBN67 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS). MEFs (Serrano et al., 1997) were provided by E.De Stanchina and S.Lowe (Cold Spring Harbor Laboratory).
Immunoblot analysis
For immunoblot analysis, cells washed with phosphate-buffered saline (PBS) were lysed in SDS Laemmli sample buffer and, after clarification by centrifugation, equal amounts of protein were resolved by SDS–PAGE, transferred to a nitrocellulose membrane (Schleicher & Schuell) and probed with either polyclonal rabbit antisera to the HCF-1N (αN18; Goto et al. 1997) or HCF-1C (αH12; Wilson et al., 1993) subunits, or monoclonal antibodies to the HA tag (12CA5; see Wilson et al., 1993) or β-actin (Sigma), followed by appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies. The immunoreactive bands were detected by chemiluminescence as described (Pierce).
Mutagenesis and generation of cell lines
A human neomycin-resistant expression vector with the genomic β-actin promoter and exon 1, intron 1 and the β-actin ATG initiation codon in exon 2 (pHBA-BGH; Miller and Rizzino, 1996) was modified to encode the HA epitope tag (SYPYDVPDYASLGGPS) downstream of the ATG in a SalI site, creating pAPN. The HA epitope tag sequence is followed by a SpeI–BamHI–HindIII polylinker. XbaI–BamHI cDNA fragments encoding the wild-type human HCF-1, the HCF-1N1011 subunit (residues 2–1011), the HCF-1C600 subunit (residues 1436–2035) or HCF-1ΔREP (with residues 1011–1435 deleted) were cloned between the SpeI and BamHI sites of pAPN. The HCF-1SR constructs were prepared by introducing nine silent mutations between HCF-1 cDNA nucleotides 124–144 (AAAGAATTAATTGTAGTTTTC; silent mutations underlined) into each pAPN-HCF-1 construct. For stable recombinant HCF-1 protein synthesis, HeLa cells were transfected by electroporation with 5 µg of the plasmid DNAs and transfected cells were selected in G418-containing medium (500 µg/ml) for 3 weeks. Drug-resistant colonies were tested for recombinant HCF-1 expression by HA tag immunoblot and immunofluorescence analyses. For each HCF-1 recombinant protein, three or more positive clones were selected and pooled.
siRNA preparation and transfection
A human HCF-1 siRNA duplex directed against the HCF-1 cDNA sequence AAGGAGCTCATCGTGGTGTTT (nucleotides 124–144) was checked against the human genome sequence to ensure that only the HCF-1 mRNA would be targeted. For the mouse (AAGGAG CTTATAGTGGTGTTT) and hamster (AAGGAGCTCATAGTGGTG TTT) HCF-1 siRNA duplexes, the respective 124–144 residues were used. Complementary RNA oligomers (Dharmacon) were annealed at 20 µM as described by the manufacturer. The firefly luciferase non-specific control siRNA duplex (Elbashir et al., 2001) was used. For siRNA transfection, 105 cells were seeded onto each well of 6-well plates in 2 ml of DMEM with 10% FBS, and transfected the following day with 10 µl of siRNA duplex and 5 µl of Oligofectamine in Opti-MEM medium as described (Invitrogen). At 12 h, cells were rinsed with PBS and then transfected again following the same protocol.
Semi-quantitative RT–PCR
To assay mRNA transcript levels, total RNAs were extracted with the Ultraspec-II RNA system (Biotecx). cDNA synthesis and PCR amplification were performed with Superscript one-step RT–PCR (Invitrogen), using 100 ng of total RNA. HCF-1 cDNA was amplified with sense (GTGGGAGTGGAAGAGACTCA) and antisense (GTGGGAGTGGA AGAGACTCA) primers for 30 cycles; exponential amplification was verified. β-actin cDNA was amplified for 24 cycles with the primers GGAGCACCCCGTGCTGCTGA and CCAGCCAGGTCCAGACGCA.
Immunofluorescence
For immunofluorescence, siRNA-transfected cells grown on glass coverslips were fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.2% Triton X-100 in PBS for 15 min, and then blocked for 60 min with 1% bovine serum albumin (BSA) at room temperature. Incubations with primary antibodies against HCF-1 (αN18, 1:800 dilution; αH12, 1:1000 dilution), HA tag [12CA5, 1:500 dilution; or polyconal (Babco) 1:250 dilution], Aurora-B (a kind gift of P.Sassone-Corsi, IGBMC, France; 1:200 dilution) and β-tubulin (Sigma; 1:500 dilution) were conducted at room temperature for 60 min. After washing, cells were incubated with Alexa 568- or Alexa 488-conjugated goat anti-mouse or goat anti-rabbit secondary antibodies (Molecular Probes) for 60 min. Cells were finally stained with DAPI and coverslips were mounted using Prolong Antifade (Molecular Probes). Samples were examined and pictures acquired on a ZeissAxiophot microscope equipped with a Photometrics Sensys (Oberckochen) cooled CDD camera using Image 2.0.5 software (ONCOR).
BrdU incorporation assays
For S phase analysis, siRNA-transfected cells grown on coverslips were incubated with BrdU (10 nM) and BrdU incorporation determined by immunofluorescence, using the BrdU labeling and detection kit I (Roche Molecular Biochemicals), which involves ethanol fixation. Subsequently, cells were co-stained with DAPI and αHCF-1 or αHA antibodies as described above.
Acknowledgments
Acknowledgements
We thank P.Sassone-Corsi for the Aurora-B antibody, E.De Stanchina and S.Lowe for the MEFs, D.Aufiero and R.Whitaker for technical support, the Herr laboratory members and P.Lassus and G.Pawlak for helpful discussions, and G.Hannon, N.Hernandez, S.Lowe and J.Wysocka for critical readings of the manuscript. E.J. was supported by a Leukemia & Lymphoma Society fellowship. This work was supported by PHS grants GM 54598 and CA13106.
References
- Ajuh P.M., Browne,G.J., Hawkes,N.A., Cohen,P.T., Roberts,S.G. and Lamond,A.I. (2000) Association of a protein phosphatase 1 activity with the human factor C1 (HCF) complex. Nucleic Acids Res., 28, 678–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajuh P., Chusainow,J., Ryde,U. and Lamond,A.I. (2002) A novel function for human factor C1 (HCF-1), a host protein required for herpes simplex virus infection, in pre-mRNA splicing. EMBO J., 21, 6590–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A., Dean,N.M. and Honkanen,R.E. (2000) Serine/threonine protein phosphatase type 1γ1 is required for the completion of cytokinesis in human A549 lung carcinoma cells. J. Biol. Chem., 275, 1846–1854. [DOI] [PubMed] [Google Scholar]
- Cohen P.T. (2002) Protein phosphatase 1—targeted in many directions. J. Cell Sci., 115, 241–256. [DOI] [PubMed] [Google Scholar]
- Crosio C., Fimia,G.M., Loury,R., Kimura,M., Okano,Y., Zhou,H., Sen,S., Allis,C.D. and Sassone-Corsi,P. (2002) Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol., 22, 874–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Goto H., Motomura,S., Wilson,A.C., Freiman,R.N., Nakabeppu,Y., Fukushima,K., Fujishima,M., Herr,W. and Nishimoto,T. (1997) A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16f unction. Genes Dev., 11, 726–737. [DOI] [PubMed] [Google Scholar]
- Herr W. (1998) The herpes simplex virus VP16-induced complex: mechanisms of combinatorial transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol., 63, 599–607. [DOI] [PubMed] [Google Scholar]
- Hsieh J.J., Ernst,P., Erdjument-Bromage,H., Tempst,P. and Korsmeyer, S.J. (2003) Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol. Cell. Biol., 23, 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristie T.M. (1997) The mouse homologue of the human transcription factor C1 (host cell factor). Conservation of forms and function. J. Biol. Chem., 272, 26749–26755. [DOI] [PubMed] [Google Scholar]
- Kristie T.M., Pomerantz,J.L., Twomey,T.C., Parent,S.A. and Sharp,P.A. (1995) The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J. Biol. Chem., 270, 4387–4394. [DOI] [PubMed] [Google Scholar]
- LaBoissière S., Walker,S. and O’Hare,P. (1997) Concerted activity of host cell factor subregions in promoting stable VP16 complex assembly and preventing interference by the acidic activation domain. Mol. Cell. Biol., 17, 7108–7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassus P., Opitz-Araya,X. and Lazebnik,Y. (2002) Requirement for caspase-2 in stress-induced apoptosis before mitochondrial permeabilization. Science, 297, 1352–1354. [DOI] [PubMed] [Google Scholar]
- Luciano R.L. and Wilson,A.C. (2002) An activation domain in the C-terminal subunit of HCF-1 is important for transactivation by VP16 and LZIP. Proc. Natl Acad. Sci. USA, 99, 13403–13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S.S., Johnson,K.M. and Wilson,A.C. (2003) Molecular cloning of Drosophila HCF reveals proteolytic processing and self-association of the encoded protein. J. Cell. Physiol., 194, 117–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters J.R. (2002) HeLa cells 50 years on: the good, the bad and the ugly. Nat. Rev. Cancer, 2, 315–319. [DOI] [PubMed] [Google Scholar]
- Miller K. and Rizzino,A. (1996) Constitutive expression of fibroblast growth factor-4 does not alter the growth or the differentiation of embryonal carcinoma cells. Cell Growth Differ., 7, 203–211. [PubMed] [Google Scholar]
- Nakamura T. et al. (2002) ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell, 10, 1119–1128. [DOI] [PubMed] [Google Scholar]
- O’Keefe R.T., Henderson,S.C. and Spector,D.L. (1992) Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific α-satellite DNA sequences. J. Cell Biol., 116, 1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piluso D., Bilan,P. and Capone,J.P. (2002) Host cell factor-1 interacts with and antagonizes transactivation by the cell cycle regulatory factor Miz-1. J. Biol. Chem., 277, 46799–47808. [DOI] [PubMed] [Google Scholar]
- Reilly P.T. and Herr,W. (2002) Spontaneous reversion of tsBN67 cell proliferation and cytokinesis defects in the absence of HCF-1 function. Exp. Cell Res., 277, 119–130. [DOI] [PubMed] [Google Scholar]
- Reilly P.T., Wysocka,J. and Herr,W. (2002) Inactivation of the retinoblastoma protein family can bypass the HCF-1 defect in tsBN67 cell proliferation and cytokinesis. Mol. Cell. Biol., 22, 6767–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M., Lin,A.W., McCurrach,M.E., Beach,D. and Lowe,S.W. (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell, 88, 593–602. [DOI] [PubMed] [Google Scholar]
- Vogel J.L. and Kristie,T.M. (2002a) Autocatalytic proteolysis of the transcription factor-coactivator C1 (HCF): a potential role for proteolytic regulation of coactivator function. Proc. Natl Acad. Sci. USA, 97, 9425–9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J.L. and Kristie,T.M. (2002b) The novel coactivator C1 (HCF) coordinates multiprotein enhancer formation and mediates transcription activation by GABP. EMBO J., 9, 683–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A.C., LaMarco,K., Peterson,M.G. and Herr,W. (1993) The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell, 74, 115–125. [DOI] [PubMed] [Google Scholar]
- Wilson A.C., Peterson,M.G and Herr,W. (1995) The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev., 9, 2445–2458. [DOI] [PubMed] [Google Scholar]
- Wilson A.C., Freiman,R.N., Goto,H., Nishimoto,T. and Herr,W. (1997) VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol. Cell. Biol., 17, 6139–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J., Reilly,P.R. and Herr,W. (2001a) Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol. Cell. Biol., 21, 3820–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J., Liu,Y., Kobayashi,R. and Herr,W. (2001b) Developmental and cell-cycle regulation of Caenorhabditis elegans HCF phosphorylation. Biochemistry, 40, 5786–5794. [DOI] [PubMed] [Google Scholar]
- Wysocka J., Myers,P.M., Laherty,C.D., Eisenman,R.N. and Herr,W. (2003) Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev., 17, 896–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A., Kitabayashi,I., Ayton,P.M., Cleary,M.L. and Ohki,M. (2002) Leukemia proto oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood, 10, 3710–3718. [DOI] [PubMed] [Google Scholar]