Abstract
A significant proportion of the global diversity of flowering plants has evolved in recent geological time, probably through adaptive radiation into new niches. However, rapid evolution is at odds with recent research which has suggested that plant ecological traits, including the β- (or habitat) niche, evolve only slowly. We have quantified traits that determine within-habitat α diversity (α niches) in two communities in which species segregate on hydrological gradients. Molecular phylogenetic analysis of these data shows practically no evidence of a correlation between the ecological and evolutionary distances separating species, indicating that hydrological α niches are evolutionarily labile. We propose that contrasting patterns of evolutionary conservatism for α- and β-niches is a general phenomenon necessitated by the hierarchical filtering of species during community assembly. This determines that species must have similar β niches in order to occupy the same habitat, but different α niches in order to coexist.
Keywords: plant community structure, niche, evolution, rbcL, community assembly
1. Introduction
As the existence of community structure is increasingly tested against the foil of null models and neutral theory (Hubbell 2001), it becomes more and more apparent that species are not ecologically equivalent and that niche differences, mediated by interspecific competition, create significant structure in ecological communities (Silvertown et al. 1999; Gotelli & McCabe 2002; Clark & McLachlan 2003; Fargione et al. 2003; McGill 2003; Adler 2004). Hence the question arises, how do the traits evolve on which niche differences are based and upon which community structure is built? The answer is clearest in isolated archipelagos where communities are assembled gradually from species as they evolve in situ. In these cases the development of community structure is determined by the pace of adaptive radiation. In Darwin's finches in the Galapagos (Grant 1986), anolis lizards in the Carribean (Losos et al. 2003), spiders in Hawaii (Gillespie 2004) and plants in the Canary Islands (Francisco-Ortega et al. 1996), adaptive radiation has filled trophic or habitat niches with new species, replicating the process independently on different islands. Evolution has played a clear, historical role in creating community structure in such island communities, but what role does evolution play in the assembly of the more common situation of less isolated, continental communities (Losos 1996; Ackerly 2003)? We address ourselves to this question in the case of plants.
The majority of continental plant communities are assembled from species that have disparate evolutionary histories, as reflected in the taxonomy of their members which generally belong to a wide range of plant families. Temperate forests have well-documented post-glacial histories which show that species migrated at different rates and often from different sources into their present communities (Huntley & Birks 1983). Tropical tree species in Amazonia also responded individualistically to climate change in the Pleistocene (Bush et al. 2004). In SW Spain, Herrera (1992) found that Mediterranean shrub communities were assembled from lineages that arrived there at different times. North American desert plant communities are also post-glacial in formation (Van Devender 1986). In all these cases modern communities have been assembled very much more recently than the speciation events which generated their components. Perhaps only in the island continent of Australia and in the Cape floristic region are some present-day plant communities assembled mainly from species that evolved in situ. In other continental regions, the historical role of evolution in community assembly has been to stock the species pool from which community members have more recently been drawn. Therefore, the evolutionary question becomes, is the filtering process by which a community is assembled from the species pool biased towards certain lineages, or combinations of lineages, rather than others?
To date, the evidence has suggested that the answer to this question is ‘yes’. Prinzing et al. (2001) found that the distributions of European plants on various environmental axes, as estimated by Ellenberg indicator values which measure broad environmental tolerances to major environmental factors such as light, soil moisture and pH, have a strong phylogenetic component and thus appear to be evolutionarily conserved. Webb (2000) found that co-occurring tree species in a Bornean rainforest were more closely related than if they had been drawn randomly from the species pool. In 10 wet neotropical forest communities of woody plants, Chazdon et al. (2003) found that traits such as growth form, mating system and ecological distribution were evolutionarily conserved within lineages. Studying the ecology and distribution of birds, mammals and butterflies in southern Mexico, Peterson et al. (1999) concluded that recent speciation in these groups did not involve evolutionary changes in their niches which were similar between sister taxa. A recent review of the role of phylogeny in community ecology detected ‘a common pattern of phylogenetic conservatism in ecological character[s]’ (Webb et al. 2002).
The apparent consensus that ecological traits are conservative in their evolution is difficult to reconcile with the equally common finding that plant communities have higher than expected species to genus ratios and are often structured by competition (Silvertown et al. 1999; Gotelli & McCabe 2002; Fargione et al. 2003). According to current, non-neutral models of coexistence, the maintenance of plant diversity requires the existence of trait or niche differences between species, usually involving trade-offs between traits (Silvertown 2004). Though models differ in the detailed mechanisms by which diversity is maintained within plant communities, none permits the stable coexistence of ecologically identical species. Thus, if the ecological traits that are important for coexistence were found to be conserved during evolution, this would challenge an important body of ecological theory. However, to date none of the traits that have been shown to display evolutionary conservatism have also been shown to be directly involved in coexistence within communities. For example, Ellenberg indicator values, which derive from the observed distributions of species on indirect gradients such as soil pH and light availability, pertain to differences between habitats. These traits therefore define what can be described as the ‘β niche’ because they refer to the scale at which β diversity is determined (Whittaker 1975; Pickett & Bazzaz 1978).
Do traits that determine within-habitat α diversity and which define the corresponding ‘α niche’ evolve as conservatively as β niches apparently do? To address this question we used a molecular phylogenetic analysis to test α niche traits for phylogenetic conservatism in two grassland plant communities. These α niche traits were chosen because previous analyses have demonstrated that they generate community structure (Silvertown et al. 1999). Interspecific competition causes niche shifts along the niche axes in question, strongly implicating segregation along these axes a role in coexistence (Silvertown 2004).
2. Material and methods
(a) Niche measurements
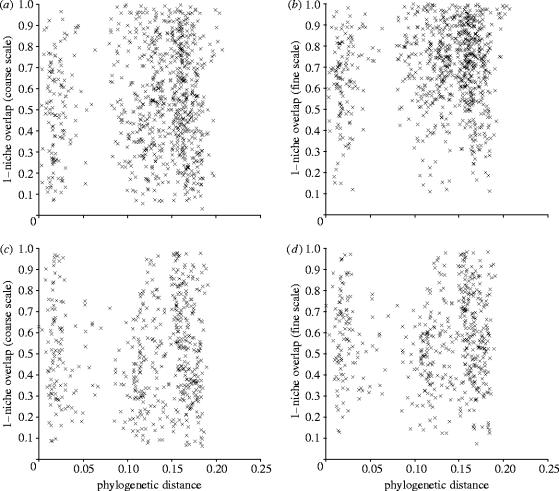
We sampled two mesotrophic grassland communities, at Tadham in Somerset and Cricklade in Wiltshire, England, classified as MG8 and MG4 types in the British National Vegetation Classification, respectively (Rodwell 1992). In a previous study, Silvertown et al. (1999) demonstrated that species in each of these communities segregated along hydrological gradients defined by two niche metrics. The niche metrics used in the study were sum exceedance values (SEV) which defined the drought (SEV drought) and aeration stress (SEV waterlogging) at each survey quadrat location at each of the two field sites. At the Cricklade site the two SEV axes are closely correlated, but at Tadham they are not. The Tadham site demonstrates that it is in fact possible, counter-intuitive though it may seem, for aeration stress and drought stress to be uncorrelated. This is because the water table fluctuates through the year and at a site like Tadham, which is very flat, an area far from a drainage ditch can be waterlogged in spring (causing aeration stress), but drought-stressed in summer. In the context of the present paper, the important thing is that our results at Tadham and Cricklade were the same (figure 1), and so do not depend upon the source of the correlation between SEV axes, even if this is different between sites. Further details are given by Silvertown et al. (2001).
Figure 1.
Pairwise α niche differences between species (1−Pianka's measure of niche overlap) plotted against the pairwise phylogenetic distance (branch lengths from the ML tree) between species at Tadham (a) coarse scale, (b) fine scale, Cricklade (c) coarse scale, and (d) fine scale.
The abundance was recorded of each plant species in 844 1 m2 quadrats at Tadham and 644 1 m2 quadrats at Cricklade. Species occurring in less than 50 quadrats were removed from the analysis because they may bias niche overlap indices. The total niche space was divided into ‘boxes’ of size 0.5×0.5 SEV (fine scale) or 1×1 SEV units (coarse scale). Mean abundance for each species was calculated for these niche boxes simply by adding up the total abundance of that species from every quadrat in that box and dividing by the number of quadrats in the box. These mean abundances were then standardized so that they summed to 100% for each species across all niches before being analysed. Pairwise niche overlaps between species were calculated using Pianka's index (Pianka 1973).
A mean SEV for each species on each axis at each site was calculated as , where n was the number of quadrats in which a species was present, SEVi was the value at the location of quadrat i, and pi was the proportion of the species' total recorded abundance found in quadrat i.
(b) Phylogenetic analysis
Molecular phylogenetic trees for 55 species distributed among 42 genera (electronic supplementary material table 1 and fig. 1) found in the two meadow communities were produced from sequences of the plastid rbcL gene by maximum likelihood using Paup* software (Swofford 1996), with the fern Asplenium trichomanes specified as the outgroup (see electronic supplementary material for sequencing details). The evolutionary model used in estimating the phylogeny was the general time-reversible (6 ST) substitution model, with rate variation by codon position. Heterogeneous rates of rbcL evolution prevent use of substitution rates as a molecular clock.
(c) Statistical analysis
If the evolution of niches is slow and/or conservative, then we expect the similarity of their sequences (a measure of evolutionary distance) to be positively correlated with the similarity of their niches. Similarity between α niches was estimated in three ways, by the Euclidean distance between the centroids of species' niches and by pairwise niche overlaps at two scales of niche partitioning—coarse (niche space divided into SEV intervals of 1.0) and fine (niche space divided into SEV intervals of 0.5). An analysis using standard methods for assessing the significance of a correlation coefficient is inappropriate here, because the pairwise sequence similarities are not mutually independent, and nor are the pairwise niche overlaps or differences. We therefore used a permutational regression approach which allows for this dependence, as implemented in version 3.1 of the computer program Permute! (Lapointe & Legendre 1992; Legendre et al. 1994). One thousand ‘triple’ randomizations were run.
3. Results
Correlations between sequence differences, centroid differences and α niche overlaps at both scales were all positive but low: at Tadham, niche centroids: r=0.052 (p=0.136); niche overlaps: coarse r=0.063 (p=0.091), fine r=0.107 (p=0.015); at Cricklade, niche centroids: r=0.043 (p=0.208); coarse r=0.028 (p=0.310), fine r=0.023 (p=0.332) (figure 1). In one out of six tests (fine scale niches at Tadham) the correlation was significantly greater than zero but still very weak.
Mean values of the two niche metrics for the 55 species are given in electronic supplementary material table 1. Pairwise overlaps between all species used in the present study are given in electronic supplementary material 1.
Our new rbcL sequences may be found in GenBank under the accession numbers shown in electronic supplementary material table 1. The maximum likelihood phylogenetic tree (electronic supplementary material fig. 1) for the 55 species in our sample has a structure consistent with the accepted relationships among families (Bremer et al. 2003). Some congeneric species in Asteraceae and Poaceae are not grouped together, but these errors have only a very slight effect on the variable of interest in our analysis which is the phylogenetic distance between species. This depends mainly upon branch lengths deeper in the tree.
4. Discussion
There was practically no relationship between the evolutionary distance separating species, as measured by rbcL sequence divergence, and their ecological overlap in α niche space (figure 1), implying that plants' hydrological niches are evolutionarily labile. There is consequently a virtual absence of phylogenetic signal in the structure of these communities, even though the ecological structure is strong (Silvertown et al. 1999). The evolutionary lability of α niches is in accord with the ecological theory of adaptive radiation (Schluter 2000) and our results are almost identical to the conclusions reached in studies of anolis lizards (Losos et al. 2003). However, our findings for the α niche are at odds with the conclusions drawn by previous studies of the role of phylogeny in plant community structure in which β niche axes were analysed (e.g. Webb 2000; Prinzing et al. 2001; Chazdon et al. 2003; Ackerly 2003, 2004).
Taking all results together, the situation appears to be one in which evolution of the β niche is affected by phylogenetic conservatism, whereas the α niche is not. This hierarchical view of the evolution of ecological traits explains the frequent observation that species : genus ratios within communities are often higher than expected (Tofts & Silvertown 2000; Enquist et al. 2002). Our results indicate that the reason for this is that habitat-determining β niches evolve relatively slowly, while α niches on which coexistence depends evolve much more rapidly.
Why should the traits that determine α niches be less conservative in their evolution than those that determine β niches? We propose that the reason lies in the hierarchical nature of community assembly.
A plant's β niche defines the habitat or habitats in which it can survive. Habitats differ from one another along major, mainly physical, environmental gradients associated with hydrology, geology (and soils), fire and climate plus the overlaid effects of herbivores and succession. These gradients are large-scale, recur in the landscape and impose similar constraints upon all the species in them, leading to well-known convergences in traits such as succulence in arid environments or long-lived leaves in resource-poor habitats (Reich et al. 1999). These β niche traits are likely to be subject to phylogenetic niche conservatism (Harvey & Pagel 1991) which will constrain their evolution (though there may be exceptions such as in scrub jays; see Rice et al. 2003).
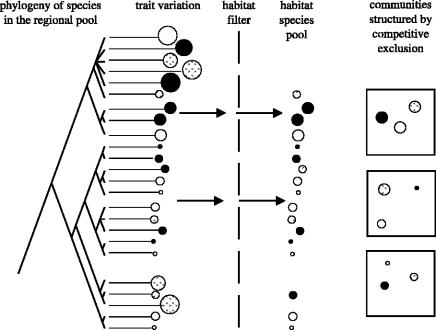
By contrast, α niches define the interactions within a community and according to most theories of coexistence (the unified neutral model being the main exception; Hubbell 2001) species must be different for coexistence to be possible (Chesson 2000; Silvertown 2004). Too great a degree of niche overlap leads to competitive exclusion. In essence, our argument is that for species to stably coexist within a habitat they must have the same β niche, but different α niches. Thus, if a trait is not evolutionarily labile, for whatever reason, it is very unlikely that it will play a role in defining a species α niche because the differences between species that are essential to coexistence will not evolve. In effect, we argue that two quite different filtering processes operate during community assembly (figure 2). A habitat filter excludes species that do not match certain habitat-specific physiological requirements and this filter operates as a conservative evolutionary force. The conservation of leaf traits found by Ackerly (2004) in his analysis of California chaparral communities provides a good example of this. At a smaller ecological scale a competitive exclusion filter operates in the assembly of communities, excluding combinations of species that are too alike in traits that influence coexistence, and this means that communities become structured by evolutionarily labile traits.
Figure 2.
A conceptual model of the hierarchical filtering process which occurs in community assembly, leading to conservatism in the traits defining the β niche and evolutionarily labile traits defining the α niche. Each ball represents a species whose diameter measures a trait such as its SEV for tolerance of soil waterlogging. Only species/balls below a critical diameter can pass through the habitat filter, but then the competitive exclusion filter prevents balls/species that are too similar (shown by their pattern) from coexisting within the same community or quadrat.
In an earlier analysis of the meadow communities at Tadham and Cricklade, Silvertown et al. (2001) used taxonomic ranks to calculate the degree of niche overlap between species within genera, genera within families, families within the eudicot and monocot clades, and between the clades themselves. Some degree of significant niche segregation was found to occur at each level of the taxonomic hierarchy, including between monocot and eudicot clades. It was concluded that the niche differences observed within a community can evolve at a range of phylogenetic levels including the deep past. This is not inconsistent with our present findings which indicate that some wide ecological divergences can be recent, while some evolutionarily distant taxa can be ecologically similar.
The rate at which plant niches evolve is fundamental to our understanding of the origin of plant diversity and how this is maintained in species-rich communities. Both the present-day global diversity of flowering plants, perhaps exceeding 422 000 named species (Govaerts 2001), and the high species-richness of many plant communities suggest that the traits which determine a plant's niche must be evolutionarily labile and evolve rapidly. Much of the species diversity in the largest angiosperm clade, the eudicots (Magallon et al. 1999), and in the grasses (Kellogg 2000) which comprise a significant proportion of the monocots, is relatively recently evolved (Davies et al. 2004). If the ecological traits that define a plant's niche were not evolutionarily labile, adaptive radiation could not easily occur, and it would be necessary to account for the observed global diversity of flowering plants by some other mechanism.
Phylogeny reconstruction involved two limitations. Mainly for logistical reasons, we used only one gene in the reconstruction of the ML tree, but this did not compromise our results because rbcL is known to yield structures that are highly concordant with multi-gene trees (Soltis et al. 2000). We have been unable to measure actual rates of α niche evolution because the rate of sequence evolution in rbcL is too slow and variable between clades. This limitation does not prevent us testing the specific null hypothesis that the distance separating species in niche space is independent of their evolutionary distance. The finding that we cannot reject this null hypothesis in five out of six tests indicates that α niches are evolutionarily labile. This discovery lifts any obstacle to rapid evolution and, if found to be general, explains how the diversification of the flowering plants was possible.
Our finding that α niches are more evolutionarily labile than β niches ought to be a very general one if our explanation of this pattern is correct. It can be tested directly by using phylogenetic reconstruction of adaptive radiations to compare the number of evolutionary transitions that involve species entering a novel habitat, and therefore evolving a new β niche, versus the number of new species which evolve without a change of β niche but which are observed to coexist with other members of the clade. If, as we predict, β niche evolution is conservative while α niche evolution is not, then new β niches should evolve in only a minority of all speciation events. The ideal type of community in which to test these predictions would be one that contains a more complete sampling of the clades from which the community is assembled than do the English meadows that we have analysed here.
Acknowledgments
This study was supported in part by a Defra contract BD1310 to David Gowing and by NERC grant NER/A/S/1999/00054 to Michael Fay, Mark Chase, Andy Purvis and Mick Crawley. We thank David Ackerly, Mark Chase, Rob Freckleton, Jonathan Losos, Mark Pagel, Andy Purvis and Campbell Webb for comments on drafts of the manuscript.
Supplementary Material
References
- Ackerly D.D. Community assembly, niche conservatism, and adaptive evolution in changing environments. Int. J. Plant Sci. 2003;164:S165–S184. 10.1086/368401 [Google Scholar]
- Ackerly D.D. Adaptation, niche conservatism, and convergence: comparative studies of leaf evolution in the California chaparral. Am. Nat. 2004;163:654–671. doi: 10.1086/383062. 10.1086/383062 [DOI] [PubMed] [Google Scholar]
- Adler P.B. Neutral models fail to reproduce observed species-area and species-time relationships in Kansas grassland. Ecology. 2004;85:1265–1272. [Google Scholar]
- Bremer B, et al. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot. J. Linn. Soc. 2003;141:399–436. 10.1046/j.1095-8339.2003.t01-1-00158.x [Google Scholar]
- Bush M.B, Silman M.R, Urrego D.H. 48,000 years of climate and forest change in a biodiversity hot spot. Science. 2004;303:827–829. doi: 10.1126/science.1090795. 10.1126/science.1090795 [DOI] [PubMed] [Google Scholar]
- Chazdon R.L, Careaga S, Webb C.O, Vargas O. Community and phylogentic structure of reproductive traits of woody species in wet tropical forests. Ecol. Monogr. 2003;73:331–348. [Google Scholar]
- Chesson P. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 2000;31:343–366. 10.1146/annurev.ecolsys.31.1.343 [Google Scholar]
- Clark J.S, McLachlan J.S. Stability of forest biodiversity. Nature. 2003;423:635–638. doi: 10.1038/nature01632. 10.1038/nature01632 [DOI] [PubMed] [Google Scholar]
- Davies T.J, Barraclough T.G, Chase M.W, Soltis P.S, Soltis D.E, Savolainen V. Darwin's abominable mystery: insights from a supertree of the angiosperms. Proc. Natl Acad. Sci. USA. 2004;101:1904–1909. doi: 10.1073/pnas.0308127100. 10.1073/pnas.0308127100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist B.J, Haskell J.P, Tiffney B.H. General patterns of taxonomic and biomass partitioning in extant and fossil plant communities. Nature. 2002;419:610–613. doi: 10.1038/nature01069. 10.1038/nature01069 [DOI] [PubMed] [Google Scholar]
- Fargione J, Brown C.S, Tilman D. Community assembly and invasion: an experimental test of neutral versus niche processes. Proc. Natl Acad. Sci. USA. 2003;100:8916–8920. doi: 10.1073/pnas.1033107100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco-Ortega J, Jansen R.K, Santos-Guerra A. Chloroplast DNA evidence of colonization, adaptive radiation, and hybridization in the evolution of the Macaronesian flora. Proc. Natl Acad. Sci. USA. 1996;93:4085–4090. doi: 10.1073/pnas.93.9.4085. 10.1073/pnas.93.9.4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie R. Community assembly through adaptive radiation in Hawaiian spiders. Science. 2004;303:356–359. doi: 10.1126/science.1091875. 10.1126/science.1091875 [DOI] [PubMed] [Google Scholar]
- Gotelli N.J, McCabe D.J. Species co-occurrence: a meta-analysis of J.M. Diamond's assembly rules model. Ecology. 2002;83:2091–2096. [Google Scholar]
- Govaerts R. How many species of seed plants are there? Taxon. 2001;50:1085–1090. [Google Scholar]
- Grant P.R. Princeton University Press; Princeton, NJ: 1986. Ecology and evolution of Darwin's finches. [Google Scholar]
- Harvey P.H, Pagel M.D. Oxford University Press; Oxford, UK: 1991. The comparative method in evolutionary biology. [Google Scholar]
- Herrera C.M. Historical effects and sorting processes as explanations for contemporary ecological patterns—character syndromes in Mediterranean woody plants. Am. Nat. 1992;140:421–446. 10.1086/285420 [Google Scholar]
- Hubbell S.P. Princeton University Press; Princeton, NJ: 2001. The unified neutral theory of biodiversity and biogeography. [DOI] [PubMed] [Google Scholar]
- Huntley B, Birks H.J.B. Cambridge University Press; Cambridge, UK: 1983. Past and present pollen maps for Europe 0–13,000 years ago. [Google Scholar]
- Kellogg E.A. The grasses: a case study in macroevolution. Annu. Rev. Ecol. Syst. 2000;31:217–238. 10.1146/annurev.ecolsys.31.1.217 [Google Scholar]
- Lapointe F.-J, Legendre P. A statistical framework to test the consensus among additive trees (cladograms) Syst. Biol. 1992;41:158–171. [Google Scholar]
- Legendre P, Lapointe F.-J, Casgrain P. Modeling brain evolution from behavior: a permutational regression approach. Evolution. 1994;48:1487–1499. doi: 10.1111/j.1558-5646.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- Losos J.B. Phylogenetic perspectives on community ecology. Ecology. 1996;77:1344–1354. [Google Scholar]
- Losos J.B, Leal M, Glor R.E, de Queiroz K, Hertz P.E, Rodriguez Schettino L, Chamizo Lara A, Jackman R.R, Larson A. Niche lability in the evolution of a Caribbean lizard community. Nature. 2003;424:542–545. doi: 10.1038/nature01814. 10.1038/nature01814 [DOI] [PubMed] [Google Scholar]
- Magallon S, Crane P.R, Herendeen P.S. Phylogenetic pattern, diversity, and diversification of eudicots. Ann. Mo. Bot. Gard. 1999;86:297–372. [Google Scholar]
- McGill B.J. A test of the unified neutral theory of biodiversity. Nature. 2003;422:881–885. doi: 10.1038/nature01583. 10.1038/nature01583 [DOI] [PubMed] [Google Scholar]
- Peterson A.T, Soberon J, Sanchez-Cordero V. Conservatism of ecological niches in evolutionary time. Science. 1999;285:1265–1267. doi: 10.1126/science.285.5431.1265. 10.1126/science.285.5431.1265 [DOI] [PubMed] [Google Scholar]
- Pianka E.R. The structure of lizard communities. Annu. Rev. Ecol. Syst. 1973;4:53–74. 10.1146/annurev.es.04.110173.000413 [Google Scholar]
- Pickett S.T.A, Bazzaz F.A. Organization of an assemblage of early successional species on a soil moisture gradient. Ecology. 1978;59:1248–1255. [Google Scholar]
- Prinzing A, Durka W, Klotz S, Brandl R. The niche of higher plants: evidence for phylogenetic conservatism. Proc. R. Soc. B. 2001;268:2383–2389. doi: 10.1098/rspb.2001.1801. 10.1098/rspb.2001.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich P.B, Ellsworth D.S, Walters M.B, Vose J.M, Gresham C, Volin J.C, Bowman W.D. Generality of leaf trait relationships: a test across six biomes. Ecology. 1999;80:1955–1969. [Google Scholar]
- Rice N.H, Martinez-Meyer E, Peterson A.T. Ecological niche differentiation in the Aphelocoma jays: a phylogenetic perspective. Biol. J. Linn. Soc. 2003;80:369–383. 10.1046/j.1095-8312.2003.00242.x [Google Scholar]
- Rodwell J.S. Grasslands and montane communities. vol. 3. Cambridge University Press; Cambridge, UK: 1992. British plant communities. [Google Scholar]
- Schluter D. Oxford University Press; Oxford, UK: 2000. The ecology of adaptive radiation. [Google Scholar]
- Silvertown J. Plant coexistence and the niche. Trends Ecol. Evol. 2004;19:605–611. 10.1016/j.tree.2004.09.003 [Google Scholar]
- Silvertown J, Dodd M.E, Gowing D, Mountford O. Hydrologically-defined niches reveal a basis for species-richness in plant communities. Nature. 1999;400:61–63. 10.1038/21877 [Google Scholar]
- Silvertown J, Dodd M, Gowing D. Phylogeny and the niche structure of meadow plant communities. J. Ecol. 2001;89:428–435. doi: 10.1098/rspb.2005.3288. 10.1046/j.1365-2745.2001.00553.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D.E, et al. Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Bot. J. Linn. Soc. 2000;133:381–461. 10.1006/bojl.2000.0380 [Google Scholar]
- Swofford D.L. Sinauer; Sunderland, MA: 1996. PAUP*: phylogenetic analysis using parsimony (and other methods), version 4.0. [Google Scholar]
- Tofts R, Silvertown J. A phylogenetic approach to community assembly from a local species pool. Proc. R. Soc. B. 2000;267:363–370. doi: 10.1098/rspb.2000.1010. 10.1098/rspb.2000.1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Devender T.R. Climatic cadences and the composition of Chihuahuan desert communities: the Late Pleistocene packrat midden record. In: Diamond J, Case T.J, editors. Community ecology. Harper & Row; New York: 1986. pp. 285–299. [Google Scholar]
- Webb C.O. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am. Nat. 2000;156:145–155. doi: 10.1086/303378. 10.1086/303378 [DOI] [PubMed] [Google Scholar]
- Webb C.O, Ackerly D.D, McPeek M.A, Donoghue M.J. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 2002;33:475–505. 10.1146/annurev.ecolsys.33.010802.150448 [Google Scholar]
- Whittaker R.H. Macmillan; NY: 1975. Communities and ecosystems. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.