Abstract
The extraordinary diversity of phytophagous insects may be attributable to their narrow specialization as parasites of plants, with selective tradeoffs associated with alternate host plants driving genetic divergence of host-associated forms via ecological speciation. Most phytophagous insects in turn are attacked by parasitoid insects, which are similarly specialized and may also undergo host-associated differentiation (HAD). A particularly interesting possibility is that HAD by phytophagous insects might lead to HAD in parasitoids, as parasitoids evolve divergent lineages on the new host plant-specific lineages of their phytophagous hosts. We call this process ‘cascading host-associated differentiation’ (cascading HAD). We tested for cascading HAD in parasitoids of two phytophagous insects, each of which consists of genetically distinct host-associated lineages on the same pair of goldenrods (Solidago). Each parasitoid exhibited significant host-associated genetic divergence, and the distribution and patterns of divergence are consistent with divergence in sympatry. Although evidence for cascading HAD is currently limited, our results suggest that it could play an important role in the diversification of parasitoids attacking phytophagous insects. The existence of cryptic host-associated lineages also suggests that the diversity of parasitoids may be vastly underestimated.
Keywords: insect diversification, host race formation, Copidosoma, sympatric speciation
1. Introduction
At least half of all animal species are parasites (Price 1980; Windsor 1998). Explanations for parasite diversity tend to revolve around three related factors: first, their often tight specialization, attributed to their intimate relationships with hosts (Price 1980); second, strong and disruptive selection pressures associated with specialization on different hosts that foster coevolutionary interactions and co-diversification (Maynard Smith 1966; Rice 1987; Futuyma & Moreno 1988; Thompson 1994); and third, reduced competition following specialization, which may allow coexistence of a diversity of ecologically similar parasite lineages (Simpson 1953; Mayr 1976; Price 1980). Genetic divergence among parasite populations driven by specialization on different hosts is a form of ecological speciation (sensu Schluter 1996, 2000), in which species divergence is driven by ecological selective pressures (with or without the additional influence of genetic drift or sexual selection). It has been posited that ecological speciation might occur in the absence of geographical barriers to gene flow (Schluter 2001). In parasites, sympatric divergence could be facilitated by host-associated shifts in phenology, mate selection, sensitivity to chemical compounds and other traits (Bush 1975; Poulin & Morand 2000; Berlocher & Feder 2002), encouraging speculation that ecological speciation (in sympatry) could have been a significant factor in promoting extensive diversification among parasitic organisms (de Meeûs et al. 1998; Via 2001; Stireman et al. 2005). Recent emphasis has been placed on the plant-feeding (phytophagous) insects as a hyperdiverse (Mitter et al. 1988) group of parasites in which ecological speciation may be important. In particular, researchers have stressed divergence of host-associated lineages of phytophagous insects (Mopper & Strauss 1998; Abrahamson et al. 2001; Via 2001; Funk et al. 2002; Stireman et al. 2005), including how such divergence may be driven by selection (e.g. phenology, natural enemies, plant chemistry).
The phytophagous insects are attacked by another hyperdiverse assemblage of specialized parasites: the insect parasitoids. Mechanisms of population and species divergence in parasitoids are largely unexplored, and a particularly interesting possibility is that host-associated differentiation (HAD) may cascade across trophic levels if HAD in a herbivore leads in turn to divergence of its parasitoids. In the major parasitoid clades Tachinidae (Diptera) and parasitic Hymenoptera, extensive adaptive radiation is evident: the Tachinidae are among the most speciose dipteran families (ca 10 000 described species, Irwin et al. 2003), and the hymenopteran parasitoids may account for up to 20% of all insect species (LaSalle & Gauld 1991). Parasitoids exhibit intimate associations with their hosts and most are highly specialized (Godfray 1994). In addition, many parasitoids are extremely sensitive to cues derived from particular herbivore–host plant interactions, relying on these cues to locate hosts (Vet & Dicke 1992; De Moraes et al. 1998; Dicke 2000; De Moraes & Mescher 2004). These characteristics of parasitoids suggest that host-related selection and genetic divergence could play an important role in their diversification. Cascading HAD, if widespread, could provide an important contribution to the extensive diversification of parasitoid lineages that attack phytophagous insects.
We tested for the existence of cascading HAD in two unrelated parasitoid species. Each parasitoid attacks a herbivore that itself consists of two host-associated lineages that have differentiated on closely related and sympatric plant hosts (Nason et al. 2002; Stireman et al. 2005). We predicted that, if cascading HAD is present, we should observe significant genetic differentiation associated with host plant use at sites of host plant sympatry and across the ranges of these parasitoid species. We find that each parasitoid exhibits morphologically cryptic genetic divergence consistent with cascading HAD: genetically distinct forms attacking the two host-specialist forms (races or cryptic species; Stireman et al. 2005) of their phytophagous insect hosts. To our knowledge, this is the first demonstration of cascading HAD in parasitoids. We argue that such cryptic divergence could be common in insect parasitoids, playing an important role in their diversification and, through a mechanism of positive feedback, diversification of their phytophagous insect hosts as well.
2. Study organisms
The goldenrod gallmakers Rhopalomyia solidaginis (Diptera: Cecidomyiidae) and Gnorimoschema gallaesolidaginis (Lepidoptera: Gelechiidae) are each composed of genetically differentiated lineages on the closely related and broadly sympatric host plants Solidago altissima and Solidago gigantea (Asteraceae) (McEvoy 1988; Miller 2000; Nason et al. 2002; Stireman et al. 2005). In each case, little, if any, morphological distinction exists between the host forms, which were previously thought to represent single species (Gagné 1989; Miller 2000; Nason et al. 2002; Stireman et al. 2005). Each gallmaker is attacked by a suite of wasp parasitoids. We tested for cascading HAD in one common parasitoid of each gallmaker: Platygaster variabilis Ashmead (Platygastridae, attacking Rhopalomyia) and Copidosoma gelechiae Howard (Encyrtidae, attacking Gnorimoschema). Both are polyembryonic (many clonal offspring develop from a single egg), attack their hosts in the egg stage (Leiby 1922), destroy the host larva in its final stadium, and emerge after pupating in the host remains in late summer or early fall. No other hosts are known for P. variabilis, but C. gelechiae is recorded from at least one other host (Gnorimoschema salinaris; Patterson 1915).
3. Material and methods
(a) Collections
To obtain Platygaster, we collected Rhopalomyia galls from S. altissima and S. gigantea across the upper Midwestern USA (Iowa, Minnesota, Nebraska, South Dakota), including four sites with sympatric collections from both host plants. For Copidosoma, pilot data suggested HAD was subtle, so we concentrated our efforts on three geographically distant sites where we could obtain large sympatric samples of Gnorimoschema galls from intermixed stands of S. altissima and S. gigantea: Fredericton (New Brunswick, Canada), Toronto (Ontario, Canada) and Milaca (Minnesota, USA). At each site, we collected galls in areas ranging from ca 1 to 5 ha, dispersing sampling across available plants to maximize genetic diversity and reduce the likelihood of collecting closely related parasitoid individuals. Parasitoids were flash-frozen in liquid nitrogen (for allozyme analysis) or stored in 95% ethanol (for mitochondrial DNA analysis). Platygaster specimens were identified by Matt McGown (Florida State Collection of Arthropods) and C. gelechiae by one of us (S. B. Heard). Voucher specimens of both hosts and parasitoids have been deposited in the Iowa State University Insect Collection. We refer to parasitoids attacking gallmakers on the two Solidago species as altissima and gigantea individuals or forms.
(b) Molecular methods
(i) MtDNA amplification
We extracted insect DNA using PUREGENE DNA extraction kits (Gentra Systems, Inc., Minneapolis, MN) and amplified 450–800 bp of cytochrome oxidase I (COI). DNA was amplified in 50 μl PCR reactions containing 5 μl genomic DNA, 5 μl (10×) PCR buffer (Invitrogen), 5 μl (10 mM) dNTP solution, 2.5–3.75 (50 mM) MgCl2, 2.5 μl of forward and reverse primers (5 pmol μl−1) and dH2O to 50 μl. The primers (C1j1751 and C1N2191 for Platygaster and ‘Pat’ and UEA7 for Copidosoma) were taken from Simon et al. (1994; first three) and Lunt et al. (1996). PCR conditions were: initial denaturing at 94 °C for 2 min, then 35 cycles of 94 °C for 30–45 s, 47–52 °C for 45–60 s, 72 °C for 1 min, and a final 72 °C extension period of 4 min. Sequencing of double-stranded PCR products was carried out on an automated ABI 377 Prism sequencer at the Iowa State University DNA sequencing facility, using ABI Prism Big Dye 3.1 and standard procedures. Sequences were examined with reference to chromatograms and initially aligned using AutoAssembler, with further manual alignment and sequence manipulation using MacClade (Maddison & Maddison 2000). Preliminary analyses revealed no mtDNA sequence variation among Copidosoma individuals from the sampled region (seven individuals collected from both hosts and from Minnesota to New Brunswick exhibited a single haplotype for 600 bp of COI; GenBank accession no. DQ267636), and so we do not report mtDNA sequence analysis for that species. We obtained mtDNA sequence data for 30 P. variabilis broods (16 from Rhopalomyia galls on S. altissima and 14 from S. gigantea; GenBank accession nos DQ267637–DQ267668).
(ii) Allozyme methods
Because C. gelechiae is polyembryonic, each allozyme genotype was assessed based on a (clonal) brood of individuals from a single G. solidaginis caterpillar. Broods were extracted in a plant extraction buffer (Nason et al. 2002). We resolved nine polymorphic enzyme loci: aconitate hydratase (ACOH, EC 4.2.1.3); glucose-6-phosphate isomerase (GPI, EC 5.3.1.9); glycerol-3-phosphate dehydrogenase (G3PDH, EC 1.1.1.8); d-2-hydroxy-acid dehydrogenase (HADH, EC 1.1.99.6); isocitrate dehydrogenase (IDH, EC 1.1.1.42); malate dehydrogenase (MDH, EC 1.1.1.37); phosphoglucomutase (PGM, EC 5.4.2.2); hexokinase (HK, E.C. 2.7.1.1); and lactate dehydrogenase (LDH, EC 1.1.1.27). Allozymes were run in 12% starch (Starch Art Corp.) gels and stained following Soltis et al. (1983), except G3PDH and HADH from Murphy et al. (1996). ACOH, HK and PGM were resolved in buffer system 11 of Soltis et al. (1983), while the remaining enzymes were resolved in a pH 6 morpholine–citrate buffer (Murphy et al. 1996). Banding patterns for each enzyme exhibited expected subunit structures and patterns of expression. These loci do not represent an exhaustive search for polymorphism in Copidosoma. Because COI sequence variation was informative for Platygaster, we did not pursue allozyme analysis for that species.
(c) Phylogenetic and population genetic analyses
Phylogenetic reconstruction of P. variabilis populations from mtDNA sequence data was conducted with maximum likelihood (ML) and Bayesian methods using PAUP 4.10 (Swofford 2001) and MrBayes 3.0b4 (Ronquist & Huelsenbeck 2003), respectively. An unidentified Platygaster species reared from Rhopalomyia lobata on Euthamia graminifolia (a close relative of Solidago) was used to root the tree. The model of sequence evolution for ML analyses was selected using Modeltest (Posada & Crandall 1998), with likelihoods of successively more complex models calculated on an initial neighbour-joining tree (Bio NJ, HKY distances, rate variation with gamma=0.5). We selected a K81uf model (Kimura 1981) with parameters (in PAUP format): Nst=6, rmat=(1.0000 9.3172 2.9684 2.9684 9.3172), pinvar=0, rates=gamma, shape=0.1986. ML analyses consisted of 50 replicate heuristic searches with TBR branch swapping. Bayesian analysis began with equiprobable priors and a character partition according to codon position. The analysis was run for 1 000 000 generations (sampled every 1000 generations) with eight heated chains and a burn-in of 10 000 generations. Likelihoods levelled off after ca 5000 generations. The proportion of trees from the posterior distribution containing a particular node was used to assess support for host-associated clades (i.e. posterior probability). Net divergence between host-associated clades was calculated using Mega 2.0 (Kumar et al. 2001). Within-clade average pairwise distances and confidence intervals were estimated using 1000 bootstrap replicates in Arlequin (Schneider et al. 2000).
We assessed relatedness among Copidosoma populations (across hosts and sites) using allozyme allele frequencies. For each site, we used GenePop (Raymond & Rousset 1995) to perform tests of genic (allelic) differentiation, relative to collection from S. altissima versus S. gigantea, for each allozyme locus, and a genotypic test for all loci pooled. We used our allozyme data to assess the likely number of HAD events. If HAD occurred once, then genetic variation among populations should be better explained by host plant than by geography. Alternatively, if HAD occurred repeatedly in different regions, then geography should explain genetic variation better. We tested these hypotheses by examining two hierarchical AMOVA models: one with host plant nested within site, and one with site nested within host plant. These analyses, along with estimates of pairwise Fst values between host forms (with significance estimates based on 1013 permutations) were conducted in Arlequin 2.000 (Schneider et al. 2000). We also constructed an unrooted neighbour-joining tree of Copidosoma populations, based on Cavalli-Sforza distances calculated from allele frequency data (Cavalli-Sforza & Edwards 1967), using Phylip (Felsenstein 2004). Neighbour-joining bootstrap support (1000 replicates) of nodes was assessed using the BootSeq module of Phylip.
4. Results
(a) Platygaster variabilis
Our ML analyses of P. variabilis resulted in a single tree of highest likelihood (−ln L=1121.78). Both ML and Bayesian trees support a strong division between P. variabilis reared from Rhopalomyia on S. gigantea and S. altissima regardless of geographical affinity (figure 1). Strong support for the gigantea clade is indicated by high percentages of neighbour-joining bootstraps and of trees sampled from Bayesian posterior distributions (100 and 99%, respectively). It is unclear whether the altissma and gigantea clades are sister, or whether the gigantea clade arose from within the altissima clade. The ML tree of highest likelihood and the summary of Bayesian trees suggest paraphyly of the altissima clade with respect to the gigantea clade (although with less than 50% support in Bayesian analysis), whereas bootstraps based on neighbour-joining recover a monophyletic altissima clade in 92% of replicates (figure 1). Divergence between the clades is quite strong (mean±s.e., 5.53±1.03% (uncorrected), 8.31±2.2% (corrected)) despite an apparent lack of distinguishing morphological characters (M. MacGown, personal communication). Within-race genetic diversity was higher for the altissima clade than the gigantea clade (average uncorrected pairwise divergence (±s.e.) 1.24±0.164 and 0.194±0.039%, respectively; p<0.05), paralleling patterns of genetic diversity in the Rhopalomyia host (Stireman et al. 2005).
Figure 1.
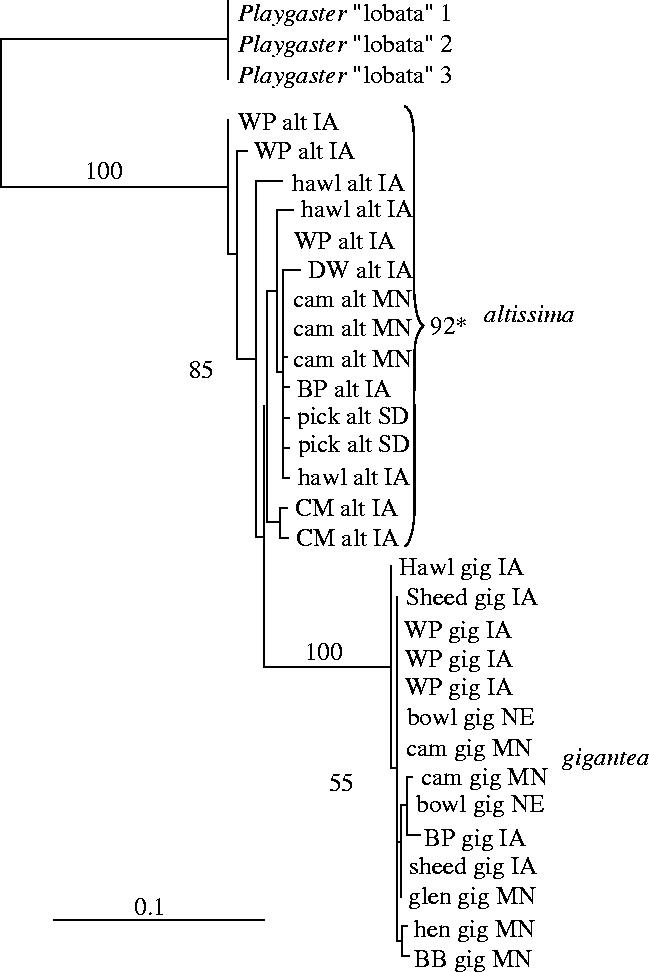
The ML tree of highest likelihood (−ln L=1121.78) for midwestern populations of Platygaster variabilis parasitoids of the gall midge Rhopalomyia solidaginis. Numbers above or to the side of clades indicate NJ bootstrap support. Numbers in parentheses indicate the percentage of trees containing the indicated clade that were sampled in the Bayesian phylogenetic analysis (see text). *Note that 92% of NJ bootstrap replicates indicated a monophyletic altissma clade despite the paraphyly inferred by ML and Bayesian methods.
(b) Copidosoma gelechiae
For Copidosoma, each sympatric population pair exhibited significant host-associated genetic isolation at multiple allozyme loci (table 1). Estimates of Fst between host-associated population pairs were generally low, but varied more than an order of magnitude across sites (0.002–0.05). Despite significant host-associated structure at each site, in the neighbour-joining tree C. gelechiae populations grouped according to geography rather than host plant (figure 2; although bootstrap support for these clusters was only moderate (60–73%)). This pattern of host-associated genetic structure nested within geographical structure is supported by AMOVA analyses: the model with host nested within geography indicates significant genetic structure at both levels and explains more total variance, while the reverse model results in negative Fst values for between-host comparisons (table 2). Furthermore, the three population pairs differed in the allozyme loci that exhibited significant HAD (table 1). Together, these analyses suggest that HAD may have occurred independently in multiple geographical regions.
Table 1.
Individual genic and global genotypic tests of significant isolation relative to host plant (S. altissima or S. gigantea) for allozyme loci assayed for Copidosoma gelechiae in three sympatric populations. (Nalt and Ngig indicate the number of individuals sampled relative to host for each population. Values are p-values (bold indicates significance at p=0.05) and n.s. indicates p>0.1.)
| locus | Milaca (Minnesota, USA) | Toronto (Ontario, Can.) | Fredericton (New Bruns., Can.) |
|---|---|---|---|
| Nalt/Ngig | 71/66 | 88/82 | 44/58 |
| ACOH | 0.000 | n.s. | 0.027 |
| G3PDH | n.s. | 0.012 | n.s. |
| HADH3 | 0.014 | n.s. | n.s. |
| HK | — | n.s. | n.s. |
| IDH | 0.000 | n.s. | 0.075 |
| LDH | n.s. | 0.007 | n.s. |
| MDH | 0.001 | 0.001 | n.s. |
| PGI | n.s. | 0.083 | 0.001 |
| PGM | n.s. | n.s. | 0.076 |
| overall (genotypic) | 0.000 () | 0.001 () | 0.033 () |
| Fst | 0.0139 | 0.0510 | 0.0015 |
Figure 2.
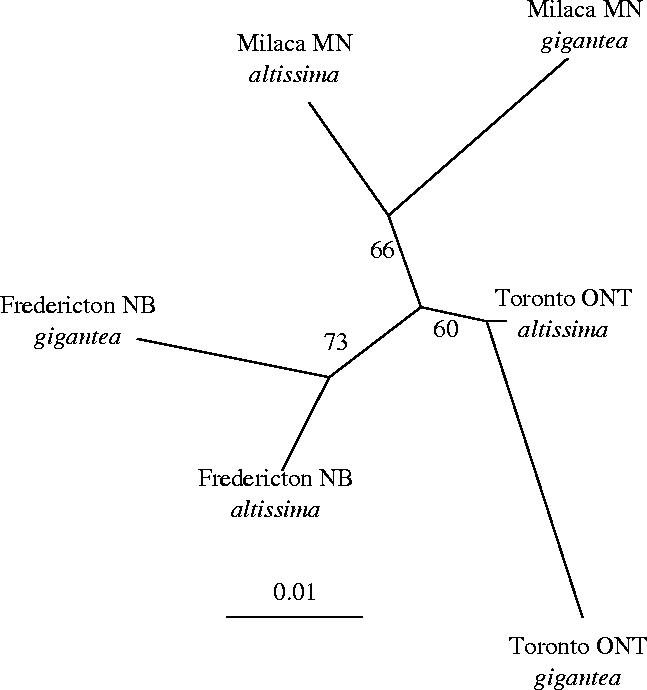
An unrooted neighbour-joining tree of Copidosoma gelechiae populations based on Cavalli-Sforza distances calculated from allozyme data (see text). Numbers at the nodes indicate bootstrap support. Note that despite significant evidence of host-associated genetic differentiation, all populations cluster according to geographical site.
Table 2.
Hierarchical AMOVA models for populations of Copidosoma gelechiae with either host plant (of host herbivores) nested within region (model A) or vice versa (model B).
| factor | d.f. | SS | variance component | percentage of variation | p |
|---|---|---|---|---|---|
| model A | |||||
| among regions | 2 | 16.89 | 0.0269 | 3.59 | 0.063 |
| between host plants (within regions) | 3 | 7.87 | 0.0174 | 2.32 | 0.000 |
| within host plants | 660 | 465.81 | 0.7058 | 94.09 | 0.000 |
| model B | |||||
| between host plants | 1 | 4.25 | −0.0037 | −0.50 | 0.717 |
| among regions (within host plants) | 4 | 20.51 | 0.0409 | 5.51 | 0.000 |
| within regions | 660 | 465.81 | 0.743 | 94.99 | 0.000 |
5. Discussion
Both P. variabilis and C. gelechiae exhibit morphologically cryptic genetic differentiation associated with use of the distinct altissima and gigantea races of their host gallmaking insects. In Platygaster, this divergence is relatively deep, indicating the existence of cryptic sibling species. In Copidosoma, divergence is subtle, perhaps reflecting initial stages of population divergence, in which significant gene flow still occurs between populations. While other studies have documented HAD in parasitoids (for instance, Morehead et al. 2001 for ant-attacking phorids and Aldrich & Zhang 2002 for Hemiptera-attacking tachinids), we believe our data provide the first evidence presented for cascading HAD from herbivores to their parasitoids. That cascading HAD may occur in other insect guilds as well is suggested by recent evidence of host-associated genetic and phenotypic divergence in a facultative predator of Eurosta gall flies (Mordellistena convicta) in this same Solidago system (Eubanks et al. 2003; Blair et al. 2005).
(a) Mode of differentiation
Genetic and ecological evidence suggests the possibility that the phytophagous hosts of Platygaster and Copidosoma have evolved host-associated lineages in sympatry (Stireman et al. 2005). Could divergence of the parasitoids have occurred in sympatry as well? Parasitoids in general are highly specialized and intimately associated with their hosts—life-history traits thought to facilitate sympatric divergence (Berlocher & Feder 2002). Current geographical barriers to gene flow in Platygaster and Copidosoma are probably negligible given the broadly sympatric and often syntopic distribution of their hosts (and host's food plants), and given that such small insects often have excellent dispersal capability (e.g. Antolin & Strong 1987).
Unfortunately, inferring modes of divergence from current geographical and genetic data is extremely difficult and often controversial (Losos & Glor 2003). However, two major lines of evidence suggest that sympatric divergence is possible, at least for Copidosoma. First, divergence between altissima and gigantea lineages of Copidosoma is much shallower than the corresponding divergence in its host, G. gallaesolidaginis. Gnorimoschema gallaesolidaginis has strong HAD in both mtDNA (Φst =0.544) and allozymes (Fst =0.159), while C. gelechiae exhibits little mtDNA variation (J. O. Stireman, unpublished data) and local host-associated allozyme Fst values of 0.002–0.05. (For Platygaster, evidence for non-concordance is equivocal, with 5.5% net divergence between host forms, versus 6.7% between the Rhopalomyia hosts; large bootstrap variances and the likelihood of different rates of molecular evolution in host and parasitoid (Arbogast et al. 2002) prevent an inference of relative timing.) Second, allozyme data from our widely spaced (more than 1000 km) study populations suggest that Copidosoma has experienced multiple independent occurrences of HAD, whereas Gnorimoschema shows evidence of a single event (Stireman et al. 2005). These results are inconsistent with the concurrent divergence expected under the simplest vicariant allopatric model, although of course more complicated allopatric models could be invoked in which multiple vicariant events and changing geographical distributions could produce similar genetic patterns. Further behavioural, ecological and genetic work will be needed to establish the mechanisms responsible divergence in Copidosoma and Platygaster (and such work is underway).
If ecological selection is involved in divergence of host-associated parasitoid races, what might be the source of this selection? Selection for divergence may be associated with physiological incompatibilities and reduced hybrid fitness in the parasitoid–host insect interaction, analogous to those argued for host-race formation in phytophagous insects (Drés & Mallet 2002). Alternatively, selection may be associated with the two different plant species on which insect hosts are found, such as tradeoffs associated with detecting and locating hosts (and perhaps mates) via plant cues (e.g. Bernays 2001). In the latter case, HAD by parasitoids could be sparked simply by diet expansion of the host insect, without actually requiring genetic or phenotypic differentiation of the host insect relative to plant use. Our current data do not support this scenario, since divergence of the parasitoids examined here is less than (Copidosoma) or at least no greater than (Platygaster) that of their hosts. However, they also do not reject it, as we have not yet tested for HAD in parasitoids of generalist goldenrod herbivores (e.g. the tortricid moth gallmaker Epiblema). Finally, parasitoid divergence might be driven by neither host nor host's host plant alone, but by their interaction. For example, parasitoid differentiation might be encouraged by allochronic isolation due to phenological differences in the hosts' development resulting from traits of their host plants.
(b) Cascading host-associated differentiation and parasitoid diversification
It is unclear how frequently HAD occurs in phytophagous insects, but our previous analysis of the herbivore community associated with S. altissima and S. gigantea suggests it may be relatively widespread (Stireman et al. 2005). If herbivore HAD frequently cascades to parasitoids, this process could be integral to the extraordinary radiations of parasitoid lineages that attack phytophagous insects (e.g. most Chalcidoidea, Ichneumonoidea, Tachinidae). The results of several studies hint at strong ecological pressures, favouring population divergence in parasitoids. In Agathis braconid parasitoids of Greya moths, Althoff & Thompson (2001) found localized morphological differentiation (ovipositor length) and patterns of searching behaviour specific to alternate host plants (Heuchera spp.) of the Greya hosts, although there was no evidence of neutral genetic structure among geographically separated parasitoid populations. Similarly, in a study of Diaeretiella braconid wasps attacking cabbage aphids and Russian wheat aphids, Baer et al. (2004) detected significant local population divergence and fitness tradeoffs among wasps using different hosts (though they failed to find evidence of host races at a larger scale).
Well-documented examples of host–parasite cospeciation are more widespread in interactions involving parasites of vertebrates (Hafner & Nadler 1988; McCoy et al. 2001) and bacterial endosymbionts (Clark et al. 2000, Lo et al. 2003). The high host specificity that makes this possible is a trait shared by many parasitoids. However, the cascading HAD that we have identified differs from these interactions in several important respects. First, unlike traditional parasites, parasitoids almost always kill their hosts, preventing vertical transmission; the free-living adult stage that must then locate a new host provides opportunities for transmission among taxa. Second, cascading HAD takes place in a tritrophic context, where the interaction between plant and insect may contribute to diversification in the parasitoids. When herbivores expand their host range, parasitoids experience an entirely new adaptive environment that may include novel plant volatile and surface chemicals that might be used in host location, novel plant toxins ingested by hosts, novel plant morphologies and altered insect and plant phenologies. All of these factors could create divergent selection pressures and contribute to HAD.
One particularly interesting implication of this tritrophic perspective is that it may create positive feedback between parasitoid and herbivore diversification. If enemy-free space (sensu Jeffries & Lawton 1984) is an important factor in facilitating herbivore HAD (e.g. Brown et al. 1995; Berdegue et al. 1996), but parasitoids often ‘catch up’ by forming host races on these plant-specific herbivore lineages, then repeated cycles of shifting and HAD may result in ever increasing diversity of both groups. This provides a potentially powerful and testable explanation for the exceptional diversification of insect taxa possessing phytophagous and parasitoid life histories.
One line of work that challenges our argument for the importance of cascading HAD is Wiegmann et al.'s (1993) comparative analysis, which found no overall evidence that the parasitoid habit was associated with increased diversity. However, Wiegmann's result may be due to the inclusion of many parasitoid groups that attack non-phytophagous or host-generalist phytophagous insects. In such cases, the critical interaction between herbivore and host plant is absent or mitigated. The patterns of parasitoid diversity and the potential for HAD we demonstrate here suggest that the impressive radiations of parasitic Hymenoptera and Diptera might arise in part from their parasitism of specialist phytophagous insects. Furthermore, the two cases of morphologically cryptic parasitoid HAD we document represent relatively ancient, well-differentiated and reproductively isolated lineages (Platygaster) as well as recent differentiation (Copidosoma). That deeply divergent parasitoid lineages can remain morphologically cryptic suggests that parasitoid clades may harbour substantial unappreciated diversity. If so, then insect parasitoids might even surpass the phytophagous insects in the number of genetically and ecologically distinct lineages.
Acknowledgments
For permission to collect galls, we thank Gerald and Bonnie Kragt, the City of Fredericton, the Iowa Department of Natural Resources, and the Toronto and Region Conservation Authority. Graham Cox, Kevin Day, William Godsoe, Kristie Heard, Michelle LeBlanc, Katie Richardson, Kristie Richardson, David Smith, Paige Smith, Susan Timmons, Dennis Wong, and three generations of Terpstras and Hanenburgs assisted in field and lab. This research was funded by the Natural Sciences and Engineering Research Council (Canada; Discovery Grant to S.B.H.) and by the National Science Foundation (USA; grants DEB 0707752 to S.B.H. and DEB 0107938 to J.D.N.).
References
- Abrahamson W.G, Eubanks M.D, Blair C.P, Whipple A.V. Gall flies, inquilines, and goldenrods: a model for host-race formation and sympatric speciation. Am. Zool. 2001;41:928–938. [Google Scholar]
- Aldrich J.R, Zhang A. Kairomone strains of Euclytia flava (Townsend), a parasitoid of stink bugs. J. Chem. Ecol. 2002;28:1565–1582. doi: 10.1023/a:1019972312132. doi:10.1023/A:1019972312132 [DOI] [PubMed] [Google Scholar]
- Althoff D.M, Thompson J.N. Geographic structure in the searching behaviour of a specialist parasitoid: combining molecular and behavioural approaches. J. Evol. Biol. 2001;14:406–417. doi:10.1046/j.1420-9101.2001.00286.x [Google Scholar]
- Antolin M.F, Strong D.R. Long-distance dispersal by a parasitoid Anagrus delicates Mymaridae and its host. Oecologia. 1987;73:288–292. doi: 10.1007/BF00377520. doi:10.1007/BF00377520 [DOI] [PubMed] [Google Scholar]
- Arbogast B.S, Edwards S.V, Wakeley J, Beerli P, Slowinski J.B. Estimating divergence times from molecular data on phylogenetic and population genetic timescales. Annu. Rev. Ecol. Syst. 2002;33:707–740. doi:10.1146/annurev.ecolsys.33.010802.150500 [Google Scholar]
- Baer C.F, Tripp D.W, Bjorksten T.A, Antolin M.F. Phylogeography of a parasitoid wasp (Diaeretiella rapae): no evidence of host-associated lineages. Mol. Ecol. 2004;13:1859–1869. doi: 10.1111/j.1365-294X.2004.02196.x. doi:10.1111/j.1365-294X.2004.02196.x [DOI] [PubMed] [Google Scholar]
- Berdegue M, Trumble J.T, Hare J.D, Redak R.A. Is it enemy-free space? The evidence for terrestrial insects and freshwater arthropods. Ecol. Entomol. 1996;21:203–217. [Google Scholar]
- Berlocher S.H, Feder J.L. Sympatric speciation in phytophagous insects: moving beyond controversy? Annu. Rev. Entomol. 2002;47:773–815. doi: 10.1146/annurev.ento.47.091201.145312. doi:10.1146/annurev.ento.47.091201.145312 [DOI] [PubMed] [Google Scholar]
- Bernays E.A. Neural limitations in phytophagous insects: implications for diet breadth and evolution of host affiliation. Annu. Rev. Entomol. 2001;46:703–727. doi: 10.1146/annurev.ento.46.1.703. doi:10.1146/annurev.ento.46.1.703 [DOI] [PubMed] [Google Scholar]
- Blair C.P, Abrahamson W.G, Jackman J.A, Tyrrell L. Cryptic speciation and host-race formation in a purportedly generalist tumbling flower beetle. Evolution. 2005;59:304–316. [PubMed] [Google Scholar]
- Brown J.M, Abrahamson W.G, Packer R.A, Way P.A. The role of natural enemy escape in a gallmaker host-plant shift. Oecologia. 1995;104:52–60. doi: 10.1007/BF00365562. doi:10.1007/BF00365562 [DOI] [PubMed] [Google Scholar]
- Bush G.L. Sympatric speciation in phytophagous parasitic insects. In: Price P.W, editor. Evolutionary strategies of parasitic insects and mites. Plenum Press; New York: 1975. pp. 187–207. [Google Scholar]
- Cavalli-Sforza L.L, Edwards A.W.F. Phylogenetic analysis: models and estimation procedures. Evolution. 1967;21:550–570. doi: 10.1111/j.1558-5646.1967.tb03411.x. [DOI] [PubMed] [Google Scholar]
- Clark M.A, Moran N.A, Baumann P, Wernegreen J.J. Cospeciation between bacterial endosymbionts (Buchnera) and a recent radiation of aphids (Uroleucon) and pitfalls of testing for phylogenetic congruence. Evolution. 2000;54:517–525. doi: 10.1111/j.0014-3820.2000.tb00054.x. [DOI] [PubMed] [Google Scholar]
- de Meeûs T, Michalakis Y, Renaud F. Santa Rosalia revisited: or why are there so many kinds of parasites in ‘the garden of earthly delights’? Parasitol. Today. 1998;14:10–13. doi: 10.1016/s0169-4758(97)01163-0. doi:10.1016/S0169-4758(97)01163-0 [DOI] [PubMed] [Google Scholar]
- De Moraes C.M, Mescher M.C. Biochemical crypsis in the avoidance of natural enemies by an insect herbivore. Proc. Natl Acad. Sci. USA. 2004;101:8993–8997. doi: 10.1073/pnas.0403248101. doi:10.1073/pnas.0403248101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Moraes C.M, Lewis W.J, Pare P.W, Alborn H.T, Tumlinson J.H. Herbivore-infested plants selectively attract parasitoids. Nature. 1998;393:570–573. doi:10.1038/31219 [Google Scholar]
- Dicke M. Chemical ecology of host-plant selection by herbivorous arthropods: a multitrophic perspective. Biochem. Syst. Ecol. 2000;28:601–617. doi: 10.1016/s0305-1978(99)00106-4. doi:10.1016/S0305-1978(99)00106-4 [DOI] [PubMed] [Google Scholar]
- Drés M, Mallet J. Host races in plant feeding insects and their importance in sympatric speciation. Phil. Trans. R. Soc. B. 2002;357:471–492. doi: 10.1098/rstb.2002.1059. doi:10.1098/rstb.2002.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubanks M.D, Blair C.P, Abrahamson W.G. One host shift leads to another? Evidence of host-race formation in a predaceous gall-boring beetle. Evolution. 2003;57:168–172. doi: 10.1111/j.0014-3820.2003.tb00226.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Department of Genome Sciences; University of Washington, Seattle: 2004. PHYLIP (phylogeny inference package) version 3.6. [Google Scholar]
- Funk D.J, Filchak K.E, Feder J.L. Herbivorous insects: model systems for the comparative study of speciation ecology. Genetica. 2002;116:251–267. doi:10.1023/A:1021236510453 [PubMed] [Google Scholar]
- Futuyma D.J, Moreno B. The evolution of ecological specialization. Annu. Rev. Ecol. Syst. 1988;19:207–233. doi:10.1146/annurev.es.19.110188.001231 [Google Scholar]
- Gagné R.D. Cornell University Press; Ithaca, NY: 1989. The plant-feeding gall midges of North America. [Google Scholar]
- Godfray H.C.J. Princeton University Press; Princeton, NJ: 1994. Parasitoids: behavioral and evolutionary ecology. [Google Scholar]
- Hafner M.S, Nadler S.A. Phylogenetic trees support the coevolution of parasites and their hosts. Nature. 1988;332:258–259. doi: 10.1038/332258a0. doi:10.1038/332258a0 [DOI] [PubMed] [Google Scholar]
- Irwin M.E, Schlinger E.I, Thompson F.C. Diptera, true flies. In: Goodman S.M, Benstead J.P, editors. The natural history of Madagascar. University of Chicago Press; Chicago, IL: 2003. pp. 692–702. [Google Scholar]
- Jeffries M.J, Lawton J.H. Enemy free space and the structure of ecological communities. Biol. J. Linn. Soc. 1984;23:269–286. [Google Scholar]
- Kimura M. Estimation of evolutionary distances between homologous nucleotide sequences. Proc. Natl Acad. Sci. USA. 1981;78:454–458. doi: 10.1073/pnas.78.1.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen I.B, Nei M. Arizona State University; Tempe, AZ: 2001. MEGA2: molecular evolutionary genetics analysis software. [DOI] [PubMed] [Google Scholar]
- LaSalle J, Gauld I.D. Parasitic Hymenoptera and the biodiversity crisis. Insect Parasitoids, 4th European Workshop. Redia. 1991;74:315–334. [Google Scholar]
- Leiby R.W. The polyembryonic development of Copidosoma gelechiae with notes on its biology. J. Morphol. 1922;37:195–249. doi:10.1002/jmor.1050370104 [Google Scholar]
- Lo N, Bandi C, Watanabe H, Nalepa C, Beninati T. Evidence for cocladogenesis between diverse dictyopteran lineages and their intracellular endosymbionts. Mol. Biol. Evol. 2003;20:907–913. doi: 10.1093/molbev/msg097. doi:10.1093/molbev/msg097 [DOI] [PubMed] [Google Scholar]
- Losos J.B, Glor R.E. Phylogenetic comparative methods and the geography of speciation. Trends Ecol. Evol. 2003;18:220–227. doi:10.1016/S0169-5347(03)00037-5 [Google Scholar]
- Lunt D.H, Zhang D.H, Szymura J.M, Hewitt G.M. The insect cytochrome oxidase I gene: evolutionary patterns and conserved primers for phylogenetic studies. Insect Mol. Biol. 1996;5:153–165. doi: 10.1111/j.1365-2583.1996.tb00049.x. [DOI] [PubMed] [Google Scholar]
- Maddison W.P, Maddison D.R. Sinauer Associates; Sunderland, MA: 2000. MacClade 4.01b. Analysis of phylogeny and character evolution. [Google Scholar]
- Maynard Smith J. Sympatric speciation. Am. Nat. 1966;100:637–650. doi:10.1086/282457 [Google Scholar]
- Mayr E. Harvard University Press; Cambridge, UK: 1976. Evolution and the diversity of life. [Google Scholar]
- McCoy K.D, Boulinier T, Tirard C, Michalakis Y. Host specificity of a generalist parasite: genetic evidence of sympatric host races in the seabird tick Ixodes uriae. J. Evol. Biol. 2001;14:395–405. doi:10.1046/j.1420-9101.2001.00290.x [Google Scholar]
- McEvoy, M. V. 1988 The gall insects of goldenrods (Compositae: Solidago) with a revision of the species of Rhopalomyia (Diptera: Cecidomyiidae). M.S. thesis, Cornell University, New York.
- Miller W.E. Thomas Say Publications in Entomology; Lanham, MD: 2000. A comparative taxonomic–natural history study of eight Neartic species of Gnorimoschema that induce stem galls on Asteraceae, including descriptions of three new species (Lepidoptera: Gelechiidae) [Google Scholar]
- Mitter C, Farrell B, Weigmann B. The phylogenetic study of adaptive zones: has phytophagy promoted insect diversification? Am. Nat. 1988;132:107–128. doi:10.1086/284840 [Google Scholar]
- Mopper S, Strauss S.Y. Chapman & Hall; New York: 1998. Genetic structure and local adaptation in natural insect populations: effects of ecology, life history and behavior. [Google Scholar]
- Morehead S.A, Seger J, Feener D.H, Jr, Brown B.V. Evidence for a cryptic species complex in the ant parasitoid Apocephalus paraponerae (Diptera: Phoridae) Evol. Ecol. Res. 2001;3:273–284. [Google Scholar]
- Murphy R.W, Sites J.W, Buth D.G, Haufler C.H. Isozyme electrophoresis. In: Moritz C, Hillis D.M, Marble B.K, editors. Molecular systematics. Sinauer Associates; Sunderland, MA: 1996. pp. 51–120. [Google Scholar]
- Nason J.D, Heard S.B, Williams F.R. Host-associated genetic differentiation in the goldenrod elliptical-gall moth, Gnorimoschema gallaesolidaginis (Lepidoptera: Gelechiidae) Evolution. 2002;56:1475–1488. doi: 10.1111/j.0014-3820.2002.tb01459.x. [DOI] [PubMed] [Google Scholar]
- Patterson J.T. Observations on the development of Copidosoma gelechiae. Biol. Bull. 1915;29:333–360. [Google Scholar]
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Poulin R, Morand S. The diversity of parasites. Q. Rev. Biol. 2000;75:277–293. doi: 10.1086/393500. doi:10.1086/393500 [DOI] [PubMed] [Google Scholar]
- Price P.W. Princeton University Press; Princeton, NJ: 1980. Evolutionary biology of parasites. [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Heredity. 1995;86:248–249. [Google Scholar]
- Rice W. Speciation via habitat specialization: the evolution of reproductive isolation as a correlated character. Evol. Ecol. 1987;1:301–315. doi:10.1007/BF02071555 [Google Scholar]
- Ronquist F.R, Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. doi:10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Schluter D. Ecological causes of adaptive radiation. Am. Nat. 1996;148:S40–S64. doi:10.1086/285901 [Google Scholar]
- Schluter D. Oxford University Press; Oxford, UK: 2000. The ecology of adaptive radiation. [Google Scholar]
- Schluter D. Ecology and the origin of species. Trends Ecol. Evol. 2001;16:372–381. doi: 10.1016/s0169-5347(01)02198-x. doi:10.1016/S0169-5347(01)02198-X [DOI] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Genetics and Biometry Laboratory; University of Geneva, Switzerland: 2000. Arlequin 2.000, a software for population genetics data analysis. [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994;87:651–701. [Google Scholar]
- Simpson G.G. Columbia University Press; New York: 1953. The major features of evolution. [Google Scholar]
- Soltis D.E, Haufler C.H, Darrow D.C, Gastony G.J. Starch gel electrophoresis of ferns, a compilation of grinding buffers, gel and electrode buffers, and staining schedules. Am. Fern J. 1983;73:9–15. [Google Scholar]
- Stireman J.O, III, Nason J.D, Heard S.B. Host-associated genetic differentiation in phytophagous insects: general phenomenon or isolated exceptions? Evidence from a goldenrod insect community. Evolution. 2001;59:2573–2587. [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, MA: 2001. PAUP*. Phylogenetic analysis using parsimony *and other methods. Version 4b10. [Google Scholar]
- Thompson J.N. University of Chicago Press; Chicago, IL: 1994. The coevolutionary process. [Google Scholar]
- Vet L.E.M, Dicke M. The ecology of infochemical use by natural enemies in a tritrophic context. Annu. Rev. Entomol. 1992;37:141–172. doi:10.1146/annurev.en.37.010192.001041 [Google Scholar]
- Via S. Sympatric speciation in animals: the ugly duckling grows up. Trends Ecol. Evol. 2001;16:381–390. doi: 10.1016/s0169-5347(01)02188-7. doi:10.1016/S0169-5347(01)02188-7 [DOI] [PubMed] [Google Scholar]
- Wiegmann B.M, Mitter C, Farrell B. Diversification of carnivorous parasitic insects: extraordinary radiation, or specialized dead end? Am. Nat. 1993;142:737–754. doi:10.1086/285570 [Google Scholar]
- Windsor D.A. Most of the species on earth are parasites. Int. J. Parasitol. 1998;28:1939–1941. doi: 10.1016/s0020-7519(98)00153-2. doi:10.1016/S0020-7519(98)00153-2 [DOI] [PubMed] [Google Scholar]


