Abstract
Previously we identified a transcription factor, LPS-Induced TNF-α Factor (LITAF), mediating inflammatory cytokine expression in LPS-induced processes. To characterize the role of LITAF in vivo, we generated a macrophage-specific LITAF-deficient mouse (macLITAF−/−). Our data demonstrate that in macrophages (i) several cytokines (such as TNF-α, IL-6, sTNF-RII, and CXCL16) are induced at lower levels in macLITAF−/− compared with LITAF+/+ control macrophages; (ii) macLITAF−/− mice are more resistant to LPS-induced lethality. To further identify LITAF signaling pathways, we tested mouse TLR-2−/−, -4−/−, and -9−/− and WT peritoneal macrophages exposed to LPS. Using these cells, we now show that LITAF expression can be induced after challenge either with LPS from Porphyromonas gingivalis via agonism at TLR-2, or with LPS from Escherichia coli via agonism at TLR-4, both requiring functional MyD88. We also show that, in response to LPS, the MyD88-dependent LITAF pathway differs from the NF-κB pathway. Furthermore, using a kinase array, p38α was found to mediate LITAF phosphorylation and the inhibition of p38α with a p38-specific inhibitor (SB203580) blocked LITAF nuclear translocation and reduced LPS-induced TNF-α protein levels. Finally, macLITAF−/− macrophages rescued by LITAF cDNA transfection restored levels of TNF-α similar to those observed in WT cells. We conclude that LITAF is an important mediator of the LPS-induced inflammatory response that can be distinguished from NF-κB pathway and that p38α is the specific kinase involved in the pathway linking LPS/MyD88/LITAF to TNF.
Keywords: macrophage-specific, knockout mouse, Toll-like receptor, myeloid differentiation factor 88, p38α
LPS is a major integral structural component of the outer membrane of Gram-negative bacteria, and one of the most potent initiators of inflammation known. LPS activates monocytes and macrophages to produce cytokines such as TNF-α, IL-1, and IL-6 that, in turn, serve as endogenous inflammatory mediators (1, 2). Previously, we identified a transcription factor, LPS-induced TNF-α factor (LITAF), mediating the expression of inflammatory cytokines such as TNF-α in LPS-induced processes (3). LITAF was found to bind to STAT6B, a member of the STAT6 family forming a complex on the TNF-α promoter that modulates TNF activity (4, 5).
It is well known that the Toll-like receptors (TLRs) are integral components of the innate immune system, recognizing the presence of microbial invaders via molecules such as LPS (6–9). Recent studies indicate that TLRs share the capacity to bind the intracellular myeloid differentiation factor 88 (MyD88) (10, 11). This interaction of TLRs with MyD88 is involved in several well characterized pathways, including MyD88/IL-1R-associated kinase (IRAK)/TNF receptor-associated factor 6 (TRAF6) and the MAPK pathway (12, 13). More recently, studies have indicated that the 5-lipoxygenase-activating protein (FLAP) can act as a MyD88 partner and activator of NF-κB (14, 15), and that the flightless I homolog protein is a negative regulator of the TLR4-MyD88 pathway via its interaction with MyD88 (16). Therefore, identifying and characterizing the multiple proteins that function as MyD88 downstream partners should clarify the mechanisms through which specificity is conferred upon different TLR-mediated signaling pathways and further elucidate the LITAF signaling pathway.
The role of LITAF in vivo and its signal transduction pathway in LPS-induced inflammatory processes remain poorly defined. To characterize the role of LITAF in vivo, we generated a macLITAF−/− mouse in which LITAF is selectively ablated in macrophages. The signal transduction pathway involved in LPS-induced LITAF expression was further elucidated by using peritoneal macrophages extracted from these mice. Overall, our findings reveal a unique LITAF signaling pathway separate from NFkB and help to delineate its roles in the regulation of various inflammatory cytokines in response to LPS stimulation in mouse macrophages.
Results
Generation of macLITAF−/− Macrophages.
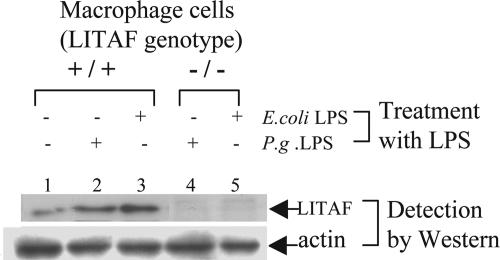
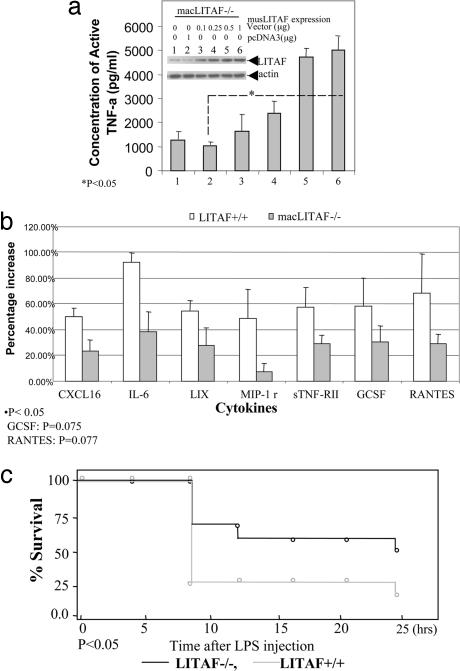
The innate immune function of LITAF in vivo was investigated after generating mice lacking LITAF in macrophages (macLITAF−/−) using the Cre-loxP system (Fig. 8, which is published as supporting information on the PNAS web site) (17). Western blot analysis showed that macLITAF−/− macrophages did not contain LITAF protein (Fig. 1, lanes 4 and 5) even after stimulation with Escherichia coli or Porphyromonas gingivalis LPS, in marked contrast to the response of cells from LITAF+/+ control mouse macrophages (lanes 2 and 3). Moreover, transient transfection of macLITAF−/− macrophages with pcDNA-musLITAF expression vector enhanced TNF-α protein levels (Fig. 2a). This finding is in agreement with our previous results showing that LITAF specifically activates TNF-α gene expression (18).
Fig. 1.
Confirmation of LITAF conditional knockout mice by Western blot. For Western blot analysis, macrophages (macLITAF−/− or LITAF+/+ as control) were stimulated with 0.1 μg/ml E. coli LPS for 16 h, and their extracts were detected by Western blot with antibody directed against murine LITAF or actin as control.
Fig. 2.
Phenotype of MacLITAF mouse. (a) ELISA of LITAF rescue. MacLITAF−/− macrophages were seeded in a six-well plate at 2 × 106 cells per well, and transiently transfected with varying concentrations of pcDNA-musLITAF expression vector DNAs or mock DNA (pcDNA3) as control, for 3 h, then washed with PBS and maintained overnight. The supernatants from each treated culture were used in triplicate ELISAs (Abraxis, Warminster, PA). ELISA immunoreactivity was quantified by using a VerSaDoc Imaging System (Bio-Rad) and graphed. The protein extracts (30 μg) from each treated group were analyzed by Western blot with antibody directed against LITAF or actin as control. (b) Murine cytokine antibody array. Membranes containing 62 cytokine antibodies (Array III and 3.1; RayBiotech, Norcross, GA) were blotted with equal amounts of conditioned medium from macrophages (macLITAF−/− or LITAF+/+ as control) after treatment with 0.1 μg/ml E. coli LPS, and were assessed according to the array manufacturer's protocol. Cytokine array experiments were performed three times, and the intensities of the relative expression levels of cytokines were quantified by densitometry (VerSaDoc Imaging System; Bio-Rad). All cytokine expression levels were normalized with positive signals obtained with biotin-conjugated IgG. The density value of each test sample was calculated and graphed. (c). MacLITAF−/− mice were more resistant to LPS-induce septic shock. Survival after LPS administration is shown. Age-matched male macLITAF−/− (n = 17) and LITAF+/+mice (n = 14) were injected i.p. with LPS (0.25 μg per mouse). Mortality was assessed every hour for 24 h. macLITAF−/− mice showed improved survival compared with LITAF+/+; P < 0.05. See Table 1.
Cytokine Responses in LITAF-Deficient Macrophages.
The levels of several cytokines were investigated by using an antibody array after macrophage stimulation with 0.1 μg/ml of E. coli LPS (Fig. 2b). After stimulation, the levels of seven cytokines were at least 40% lower (44% for CXCL16, 42% for IL-6, 53% for LIX, 86% for MIP-1r, 50% for sTNF-RII, 52% for G-CSF, and 40% for RANTES) in culture supernatants from macLITAF−/− macrophages, than from cultures of LITAF+/+ macrophages.
LITAF Deficiency and Endotoxic Shock.
The importance of LITAF in response to LPS in vivo was determined by comparing LPS-induced lethality in macLITAF−/− and LITAF+/+ control mice. After i.p. injection with d-galactosamine (d-GalN), followed by 0.25 μg of E. coli LPS per mouse, the animals were closely monitored. In murine models, it is well accepted that d-Ga1N dramatically sensitizes mice to the lethal effects of LPS via its toxic effects on hepatocytes (19). There is agreement that death in LPS/d-GalN-challenged animals is due to TNF toxicity (20) such that d-GalN-sensitized LITAF+/+ mice are sensitive to the lethal effect of LPS at a 100-fold lower dose than are unsensitized littermates (21). As shown in Fig. 2c and Table 1, which is published as supporting information on the PNAS web site, most deaths occurred between 4 and 8 h with proportions surviving in the two groups remaining quite parallel after that time. At 8 h, 11 of the initial 17 macLITAF−/− animals remained alive (64.7%), and only 4 of the 14 initial LITAF+/+ control mice (28.6%). No animals were censored or lost to follow-up. χ2 analysis gave a P value of 0.045. At 24 h, 9 of the initial 17 macLITAF−/− animals remained alive (52.9%), and only 3 of the initial 14 LITAF+/+ control mice (21.43%). χ2 analysis gave a P value of 0.073.
TLR Engagement in LITAF Signaling.
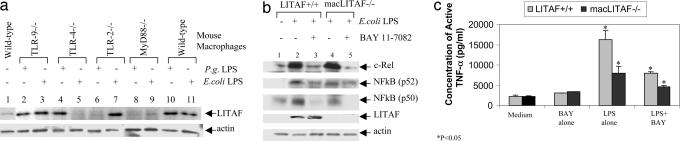
The LPS-dependent signaling pathway leading to LITAF activation was analyzed by investigating LITAF levels in response to LPS stimulation in mouse macrophages of various genotypes (TLR-2−/−, -4−/−, -9−/−, MyD88−/−, and WT controls) using Western blotting. No significant differences were observed in TLR-2 or -4 expression after LPS treatment, between macrophages either lacking LITAF or WT macrophages (Fig. 9, which is published as supporting information on the PNAS web site, lanes 7 and 8 or lanes 1 and 2). P. gingivalis LPS treatment induced LITAF levels in TLR-9−/− (lane 2), TLR-4−/− (lane 4), and WT (lane 10) macrophages, but not in TLR-2−/− (lane 6) or MyD88−/− (lane 8) macrophages (Fig. 3a). E. coli LPS treatment induced LITAF production in macrophages from TLR-9−/− (lane 3), TLR-2−/− (lane 7), and WT (lane 11) mice, but not in macrophages from TLR-4−/− (lane 5) or MyD88−/− (lane 9) animals.
Fig. 3.
LITAF signaling elements. (a) Detection of LITAF expression in mouse macrophages (TLR-2−/−, -4−/−, -9−/−, MyD88−/−, or WT as control) after LPS treatment. Proteins extracted from the LPS-stimulated mouse macrophages (from different genotypes) were analyzed by Western blot using antibody against LITAF or to actin as a control. (b and c) Analysis of the effect of BAY 11-7082 on the LPS-induced LITAF or NF-κB gene expression in macLITAF−/− cells. Proteins extracted from macrophages (macLITAF−/− or LITAF+/+ as control) that had undergone no treatment or treatment of 0.1 μg/ml E. coli LPS were measured by Western blot with antibody against LITAF, NF-κB p50, NF-κB p52, c-Rel, or actin (b). The supernatant from macLITAF−/− or LITAF+/+ cells treated with 0.1 μg/ml E. coli LPS alone or 5 μM BAY 11-7082 alone, or cotreated with 0.1 μg/ml E. coli LPS plus 5 μM BAY 11-7082 or untreated as control were used in triplicate ELISAs at the same conditions (Abraxis). (c) The immunoreactivity of each test sample was quantified by using a VerSaDoc Imaging System (Bio-Rad) and graphed. ∗, P < 0.05.
NF-κB in LITAF-Deficient Macrophages.
The amount of the NF-κB (p50, p52, or c-Rel) subunit was analyzed by Western blot in macrophages (macLITAF−/− or LITAF+/+ as controls). LPS-induced NF-κB (p50, p52, or c-Rel) proteins (Fig. 3b, lanes 2 and 4) were substantially inhibited by a highly specific pharmacological inhibitor of IkBα phosphorylation (BAY11-7082) (lane 3 and 5) (22) in either macLITAF−/− or LITAF+/+ macrophages. However, this inhibition did not decrease LITAF protein level in response to LPS stimulation (lane 3). Additionally, inhibition of NF-κB activation by BAY 11–7082 decreased LPS-induced TNF-α production by 50% in LITAF+/+ macrophages (Fig. 3c), suggesting that NF-κB mediates half of LPS-induced TNF-α production. On the other hand, LITAF alone or with another partner such as STAT6B (5) mediates the another half of LPS-induced TNF-α production. Furthermore, inhibition of NF-κB activation by BAY 11-7082 did not affect LPS-induced LITAF translocation into the nucleus (Fig. 4c, lane 2), suggesting that LITAF and NF-κB do not share the same signal pathway.
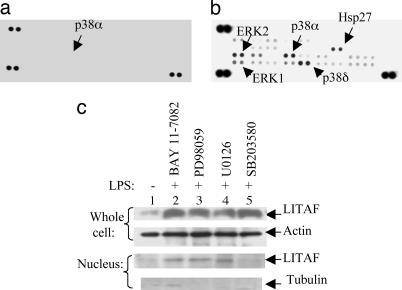
Fig. 4.
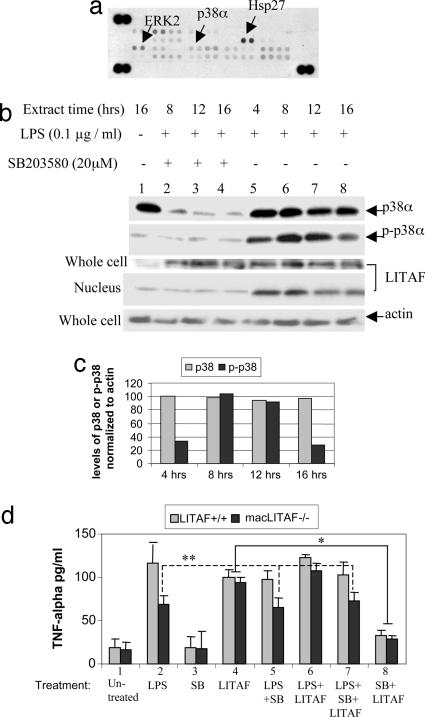
Kinase array. (a and b) The human phospho-MAPK array was used to detect multiple phosphorylated kinases in elutriated human monocytes, either untreated (a) or treated with 0.1 μg/ml E. coli LPS (b). (b) The strong signals of p38α/δ, ERK1/2 or Hsp27 in response to LPS treatment are indicated by arrows. (c) Analysis of the effects of kinase inhibitors on the LPS-induced LITAF nuclear translocation in WT mouse macrophages. Treatment with inhibitors, e.g., BAY 11-7082 (inhibits NF-κB, 5 μM), PD98059 (inhibits ERK1/2, 30 μM), U0126 (inhibits MEK, 10 μM) or did not show any effects on LITAF nuclear translocation, but SB203580 (inhibits p38 MAP kinase, 20 μM) completely blocked LITAF nuclear translocation, whereas the total level of LPS-induced LITAF expression was unchanged. Anti-β-tubulin antibody was used as a cytoplasmic marker to ensure the purity of the proteins extracted from nuclei because β-tubulin is expressed only in cytoplasm.
LPS-Induced LITAF Phosphorylation and Translocation.
LITAF translocation.
LITAF translocation in nucleus was tested in lysates from elutriated human monocytes and analyzed by both kinase array and Western blot. The phosphorylation levels of p38α/δ, Hsp27, or ERK1/2 were significantly increased in LPS-treated cells (Fig. 4 a and b) relative to untreated controls (Fig. 4a). Additionally, to determine the kinases involved in LITAF translocation, 20 available kinase inhibitors were tested, and the inhibition of LITAF nuclear translocation was analyzed by Western blot. Only SB203580, which specifically inhibits the p38 MAP kinase (23), completely blocked LITAF translocation, whereas in the whole cell, LITAF expression was unchanged (Fig. 4c, lane 5). No other inhibitor tested showed any effects. These results demonstrate that the p38 MAP kinase, p38α, participates in the phosphorylation of LITAF, and when inhibited by SB203580, LITAF does not translocate into the nucleus to activate cytokines including TNF-α and that LITAF is unable to be activated in the absence of p38α.
LITAF phosphorylation.
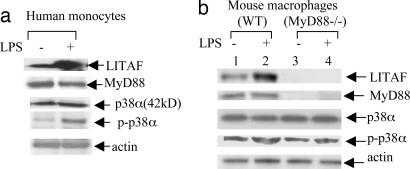
To investigate whether p38α activity was MyD88-dependent, proteins extracted from LPS-treated or untreated human monocytes or murine macrophages (WT or MyD88−/−) were analyzed by Western blot with antibodies against LITAF, MyD88, p38α, phospho-p38, or actin as control (Fig. 5 a and b). The results showed that LPS-induced p38α phosphorylation/activation is independent of MyD88 (Fig. 5b, lane 2 vs. 4). Furthermore, the influence of p38α phosphorylation on LITAF nuclear translocation was investigated by both kinase array and Western blot analysis. Treatment of human monocytes with 20 μM SB203580 for 8–16 h significantly reduced both p38α protein and phosphorylation levels (Fig. 6 a and b, lanes 2–4), thereby blocking LITAF translocation into the nucleus (Fig. 6b lanes 2–4 and d). There was no effect on LPS-induced LITAF protein levels in whole cells (Fig. 6b, lanes 2–4) in comparison to SB203580-untreated controls (lanes 6–8). To further examine whether p38α is involved in LITAF activation, we measured TNF-α by ELISA after treatment of mouse LITAF+/+ and macLITAF−/− macrophages with LPS and/or SB203580. In some cases, macLITAF−/− macrophages were first transiently transfected with 0.5 μg of pcDNA-musLITAF expression vector DNA to restore LITAF expression. Transient overexpression of LITAF DNA strongly induced production of TNF-α (Fig. 6d, condition no. 4, 78% for LITAF+/+ cells and 75% for macLITAF−/− cells) compared with the LPS-induced cells. However, TNF-α production was significantly reduced when transfectants were treated with SB203580 (25% reduction for LITAF+/+ cells, 23% for macLITAF−/− cells, Fig. 6d, condition no. 8). Additionally, no significant changes in TNF-α levels were observed in macLITAF−/− cells either treated with LPS alone (Fig. 6d, condition no. 2) or cotreated with SB203580 and LPS (condition no. 5) or with SB203580, LPS, and LITAF DNA (condition no. 7), demonstrating that p38α is involved in LPS-induced TNF-α production in LITAF+/+cells but not in LITAF-deficient cells.
Fig. 5.
Detection of p38α and phosphorylated p38α protein levels by Western blot in 0.1 μg/ml E. coli LPS-treated human monocytes (a) or mouse WT or MyD88−/− macrophages (b) with antibody against LITAF, MyD88, p38, phospho-p38, or actin as control.
Fig. 6.
LITAF phosphorylation and translocation. (a) The phosphorylated kinases were detected by human phospho-MAPK array after cotreatment of human monocytes with 0.1 μg/ml E. coli LPS plus 20 μM SB203580 for 4–16 h. No signal of p38α/δ was detected, in contrast to the strong signals of ERK2 or Hsp27, as indicated by arrows. (b) Western blot was used to detect translocation of p38α-mediated LITAF in mouse macrophages as follows. Protein extracts were collected at various times from whole cells or nuclei after cotreatment with LPS (lanes 2–8) and SB203580 (lanes 2–4). Proteins were detected with the following antibodies: LITAF, p38α, p-p38α and actin as control. (c) Levels of p38α protein and phosphorylated p38 detected by Western blot above (b) and normalized to actin and graphed. (d) Mouse macrophages were seeded in a 96-well plate at 2 × 104 cells per well, and transiently transfected with 0.5 μg of pcDNA-musLITAF expression vector DNAs or treated with 20 μM SB203580 alone or with 0.1 μg/ml E. coli LPS alone or cotreated with 0.5 μg of pcDNA-musLITAF expression vector DNAs and/or 0.1 μg/ml E. coli LPS and/or 20 μM SB203580, then maintained for 16 h. The supernatants from each treated culture were used in three separate ELISAs (Abraxis) to see effects of p38α inhibitor (SB203580) on TNF-α production in the treated macLITAF−/− or LITAF+/+ macrophages as control. ELISA immunoreactivity was quantified by using a VerSaDoc Imaging System (Bio-Rad) and graphed. ∗, P < 0.05; ∗∗, not significant.
Discussion
The present results have contributed to our understanding of the mechanism of LITAF expression leading to proinflammatory cytokine production. Namely, (i) inflammatory cytokines are induced at lower levels in macLITAF−/− macrophages than in LITAF+/+ control macrophages, whereas the restoration of the LITAF gene in macLITAF−/− macrophages rescues the deficiency; (ii) macrophage-specific LITAF-deficient mice are resistant to LPS-induced lethality. Although some LITAF-deficient mice did not survive LPS treatment advocating for the involvement of other transcription factors such as NFkB in this process, as a group they fared significantly better than WT mice. These data highlight the important participation of LITAF in an early response to endotoxin, and suggest the possibility of cytokine regulation through LITAF as a therapeutic intervention in LPS and TNF pathophysiological dysfunctions such as rheumatoid arthritis, Crohn's disease, innate immune dysregulation in CNS, or inflammatory changes in mesenteric fat. Indeed, recently we demonstrated a marked increase of LITAF expression in intestinal tissues obtained from patients with Crohn's disease compared with noninflamed matched control tissues (24).
Because TLRs are integral components of the innate immune system after LPS stimulation, and most TLRs share the capacity to bind to intracellular MyD88 (8, 9), we were interested in determining whether LITAF expression was induced by LPS through TLRs. Here we demonstrated that LITAF expression could be obtained after challenge with either the TLR2 agonist P. gingivalis LPS or the TLR4 agonist E. coli LPS; TLR9 is not involved. We also showed that both of these LPS-induced, LITAF-related signaling pathways converge at MyD88, as demonstrated by the absence of LITAF induction in cells lacking MyD88.
Several studies indicated that NF-κB is an important factor linking the MyD88-mediated signaling pathways (11). Because our studies showed that MyD88 is also involved in the LITAF signaling pathway, it was of particular interest to investigate whether LITAF production depends on NF-κB activity in macrophages. The data presented here permit the conclusion that LITAF and NFkB have separate induction pathways that are not affected by each other. Several lines of evidence support this conclusion. No significant changes of NF-κB gene expression level were observed in response to the transient overexpression of LITAF in either macLITAF−/− or LITAF+/+ macrophages (data not shown). Also, we found that BAY 11-7082, which specifically inhibits NF-κB (c-Rel, p52, or p50) activation, did not alter LPS-induced LITAF production (Fig. 3b) and translocation (Fig. 4c) even as it reduced TNF-α levels in either macLITAF−/− or LITAF+/+ macrophages (Fig. 3c).
It is well known that macrophage cells are pivotal in innate immune response, particularly where TLRs are sentinel receptors for microbial pathogens. Upon engagement with different microbial ligands, different TLRs initiate a canonical signal transduction where MyD88 serves as an adaptor protein at the downstream of TLRs. TLRs recruits MyD88 and subsequently IRAKs. Then the activated IRAK1 is released from receptor complex and forms complex with TRAF6, which mediates activation of MAPK and NF-κB forming the following signaling pathway, MyD88-IRAK-TRAF6-NIK-IκB/NFκB as well as MAPK (12, 13). Recently, a newly identified protein, FLAP-1, was found to be a NF-κB linking MyD88-FLAP-1-NF-κB (14, 15). However, although LITAF expression was found to be MyD88-dependent, neither LITAF production nor LITAF-induced TNF production was found to be affected by NFκB inhibitor BAY 11-7082. Therefore, we propose that LITAF does not directly link to MyD88-dependent NF-κB activation and conclude that the MyD88-dependent LITAF pathway differs from the MyD88-dependent NF-κB pathway in response to LPS. Taken together, these findings indicate that TLR-2/4-mediated signaling pathway involves MyD88 is an adaptor upstream of LITAF and, in turn, LITAF nuclear translocation regulates TNF-α gene expression. This pathway seems different from the characterized pathways such as MyD88-IRAK-TRAF6 and MyD88-FLAP-1-NF-κB
As for other transcription factors, phosphorylation is an important event for translocation from cytoplasm to nucleus. Indeed, recently, we reported that, upon LPS stimulation, LITAF binds to STAT6B as a heterodimer and subsequently the complex LITAF-STAT6(B) translocates into the nucleus where it significantly increases transcription of several inflammatory cytokines (5). Additionally, it is well known that the STAT family of transcription factors translocate from cytoplasm to nucleus, where they are phosphorylated and become homo- or heterodimers (25). We wanted to establish whether a specific kinase induced by LPS is required for LITAF phosphorylation and translocation. Thus, a kinase array was used in this study to identify this kinase. It was clearly observed that signals from phosphorylated forms of p38α/δ, Hsp27, or ERK1/2 were significantly increased in LPS-treated cells compared with controls (Fig. 4b). To confirm this, a panel of kinase-specific inhibitors was tested. Only inhibition of p38 MAP kinase by SB203580 completely blocked LPS-induced LITAF translocation into the nucleus in comparison to controls, whereas LITAF protein levels were unaffected. Because p38δ was not affected in response to LITAF translocation (data not shown), it suggests that p38α, and not p38δ or other kinases, participates in the phosphorylation of LITAF. Additionally, the changes of p38α protein and phosphorylation levels detected from the LPS-treated or untreated macrophages (WT or MyD88−/−) indicate that both p38α production and LPS-induced p38α phosphorylation are MyD88-independent. Interestingly, the p38α phosphorylation level reached a peak at 8 h after LPS treatment and then gradually declined, whereas the p38α protein level was not changed during this time, a time-course that is similar to the protein level changes for LITAF translocation reported by Tang et al. (5). This finding shows that p38α activation upon LPS stimulation results in LITAF phosphorylation/activation in human monocytic cells as well as mouse macrophages.
Analysis of the effects of p38α on LITAF-dependent TNF-α secretion in LITAF+/+ or macLITAF−/− macrophages showed that LPS-induced TNF-α levels were reduced by 30% in macLITAF−/− cells compared with LITAF+/+ cells. Furthermore, no significant changes of TNF-α protein levels were observed in macLITAF−/− cells either treated with LPS alone or cotreated with SB203580 plus LPS or SB203580 plus LPS plus LITAF DNA). In addition, the TNF-α secretion was strongly induced after LITAF DNA transfection in LITAF+/+ and macLITAF−/− cell, but significantly reduced (by >75%) after SB203580 treatment. Because SB203580 is a specific inhibitor of p38α MAP kinase, we conclude that p38α is the kinase specifically involved in the LITAF/TNF-α pathway in response to LPS stimulation.
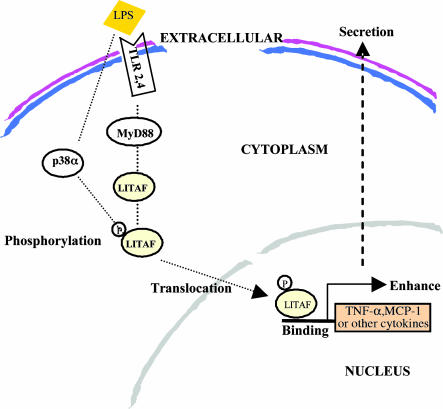
Together, the present data provide evidence for the activation of the pathway connecting TLR-2/4, MyD88, p38α, LITAF, and TNF-α upon LPS stimulation (Fig. 7). This pathway highlights the multiple sites of potential therapeutic interventions, including regulation of p38α, aimed at reducing the deleterious events associated with inflammatory conditions and should be instrumental in the design and development of target agents affecting LITAF activity.
Fig. 7.
Diagram of the proposed LITAF signaling pathway. LITAF and STAT6B (5) are induced by P. gingivalis LPS via TLR-2 or by E. coli LPS via TLR-4. Their production is MyD88-dependent. Subsequently, they are phosphorylated by p38α before protein–protein interactions aimed at forming a complex. This phosphorylation leads to the sequestration of the complex in the cytoplasm before translocation of the molecules to the nucleus. In the nucleus, the complex most likely separates to allow for LITAF alone to bind to the specific sequence (CTCCC) (4) of various cytokine genes and thus to activate their transcription.
Experimental Procedures
Bacteria and Cell Lines.
All bacterial cloning constructs used E. coli strain DH5α (Invitrogen, Carlsbad, CA). Strain 381 of P. gingivalis was grown in brain heart infusion broth with hemin (5 μg/ml) and menadione (1 μg/ml) in an anaerobic atmosphere (85% N2/10% H2/5% CO2) for 24–48 h at 37°C before preparation of LPS as we have described (26). The human monocytes, purchased from AB (Columbia, MD) were grown in RPMI medium 1640 supplemented with 10% FBS and were maintained in a humidified atmosphere of 5% CO2 at 37°C.
Kinase Inhibitors.
SB203580, PD98059, U0126, and BAY 11-7082 were purchased from EMD Biosciences (San Diego, CA). Human monocytes or mouse macrophages (WT or MyD88−/− or macLITAF−/−) were treated with 20 μM SB203580 (a p38 MAPK inhibitor), 30 μM PD98059 (MEK inhibitor) (27), 10 μM U0126 (ERK inhibitor) (28), or 5 μM BAY 11-7082 (inhibitor of IkBα phosphorylation).
Mice.
Founder mice of the TLR-2−/−, -4−/−, -9−/−, or MyD88−/− strains, and their corresponding WT controls were obtained from S. Akira (Osaka University, Osaka, Japan). The LITAF conditional knockout mouse strain (macLITAF−/−) was generated as described below. LITAF+/+ animal were used as WT animals. All animals were maintained at the Boston University transgenic facility. Mice used in experiments were 8–12 weeks of age, and were kept under strict specific pathogen-free (SPF) conditions. All procedures involving animals were approved by the Institutional Animal Care and Use Committee at Boston University Medical Center.
Macrophages.
Details regarding macrophages are given in Supporting Text, which is published as supporting information on the PNAS web site.
LPS Purification and Stimulation in Macrophages.
E. coli LPS serotype O55:B5 LPS (catalog no. L2880; Sigma, St. Louis, MO) was dissolved in PBS (5 mg/ml) by sonication for 2 min, aliquoted, and stored at −80°C until use. P. gingivalis LPS (P. gingivalis 381) was purified as described (26). All LPS preparations were free of protein or lipoprotein contaminants. The precultured macrophages were washed with PBS once and resuspended in 1 ml of RPMI medium 1640 with 10% FCS. The cells were stimulated with 100 ng/ml LPS (E. coli or P. gingivalis) for 3 h and washed with PBS once, then maintained in a humidified atmosphere of 5% CO2 at 37°C overnight.
Plasmid Constructs.
The mouse LITAF DNA (GenBank accession no. AF230522) in-frame DNA fragments were generated from a mouse spleen cDNA library (Stratagene, La Jolla, CA) by PCR with the primers 5′-AAGATGTCTAATGAGCCACC-3′ and 5′-TTAGCACAAGCGCTTGTATG-3′, and subcloned into the pCDNA3 vector (Invitrogen) to generate pcDNA-musLITAF expression vector.
Generation of LITAF Conditional Knockout Mice.
A mouse ES-129 P1 genomic library (Genome Systems, St. Louis, MO) was screened with the mouse LITAF cDNA. A 4.2-kb HindIII–NaeI fragment containing exons 2–4 of the mouse LITAF gene was subcloned into a modified pGEM-3Zf vector (Promega, Madison, WI). The targeting vector was made by inserting the first loxP site 1 kb upstream of exon 2, and inserting the second loxP site 1 kb downstream of exon 4. The loxP sites were used for the homologous recombination. A 2-kb neomycin resistance cassette (neo) was inserted between the first loxP site and exon 2. J1 embryonic stem (ES) cells were electroporated with the linearized targeting construct. The ES cells were scored for homologous recombination by Southern blotting. EcoRI-digested genomic DNAs were hybridized with a 3-kb SacI–DNA 5′ probe that contained a partial LITAF gene. Positive clones containing both loxP sites plus the neomycin gene (Neo) were screened as described (29). Clone 156 (of 264 ES cells screened) harboring a homologous recombination was identified. This clone was injected into C57BL/6 blastocysts, and two chimeric mice successfully transmitted the floxed (fl) LITAF allele through the germ line. F1 LITAFfl/+ mice were intercrossed to generate LITAFfl/fl mice. LysM-cre mice expressing Cre recombinase under the control of the mouse lysozyme M gene regulatory region (30, 31) were intercrossed to LITAFfl/fl mice to generate mice carrying both the Cre transgene and two floxed LITAF alleles: LysM-cre LITAFfl/fl mice, referred below as macLITAF−/−. The LITAFfl/fl mice and their peritoneal macrophages were used as controls and are referred as LITAF+/+ (Fig. 1a). The detection of neomycin or LITAF DNA segments in macrophages of mice was performed by PCR with the following primer pairs: 5′-AGGATCTCCTGTCATCTCACCT-3′ and 5′-ATGGGTCACGACGAGATCCT-3′ for generation of a neomycin DNA segment (266 bp) or 5′-CTTTAAGGCTGAGATAGA-3′ and 5′-CTAAGGGCAGAAGACAGC-3′ for generation of a LITAF DNA segment (205 bp).
Injection of LPS into Mice and LPS Lethality Test.
At the age of 8–12 weeks, macLITAF−/− mice along with control animals (LITAF+/+) weighing 20–25 g were injected i.p. with a single dose of d-galactosamine (25 mg; Sigma) followed by an i.p. injection of P. gingivalis LPS (0.1 and 10 μg/kg) in a total volume of 0.1 ml of PBS containing 1% BSA. All animals were continuously monitored for LPS-induced d-galactosamine-dependent lethality for 24 h after LPS challenge (n = 14 per treatment group).
Preparation of Extracts and Western Blot Analysis.
Mouse macrophages (TLR-2−/−, TLR-4−/−, TLR-9−/−, MyD88−/−, WT, macLITAF−/− or LITAF+/+) were stimulated for 16 h at 37°C with 0.1 μg/ml of LPS (from E. coli or P. gingivalis) or transiently transfected with DNAs using Lipofactamine (Invitrogen) for 3 h before LPS treatment. Both treated cells and untreated control cells were cultured in RPMI medium 1640 with 10% FCS. Elutriated human monocytes (2 × 106) were either left untreated or were treated with 0.1 μg/ml of LPS alone or cotreated with 0.1 μg/ml of LPS and 20 μM SB203580, then incubated at 37°C (5% CO2). The cells were harvested at various times, and the proteins from whole cell or nucleus were fractionally purified as described (32). The purification of nuclear proteins is briefly described as follows. The treated or untreated cells were scraped and pellets were resuspended in 400 μl of cold buffer A (10 mM Hepes, pH 7.9/10 mM KCl/0.1 mM EDTA/0.1 mM EGTA/1 mM DTT/0.5 mM phenylmethylsulfonyl fluoride/1 μg/ml pepstatin A/10 μg/ml leupeptin/10 μg/ml aprotinin) on ice for 15 min in the presence of 25 μl 1% Nonidet P-40. Then, samples were vortexed and centrifuged for 1 min at 10,000 × g, and the pellet was resuspended with 100 μl of buffer B (20 mM Hepes, pH 7.9/400 mM NaCl/1 mM EDTA/1 mM EGTA/1 mM DTT/0.5 mM phenylmethylsulfonyl fluoride/1 μg/ml pepstatin A/10 μg/ml leupeptin/10 μg/ml aprotinin). After shaking on a rocker platform for 15 min at 4°C, samples were centrifuged for 15 min at 10,000 × g at 4°C and readied for Western blot analysis. Cell lysates either from whole cell or nucleus (30 μg total protein per lane) were applied to SDS polyacrylamide gels, and proteins were detected by Western blotting with the following antibody directed against LITAF (611615; BD Biosciences, Franklin Lakes, NJ), MyD88 (sc-8197; Santa Cruz Biotechnology, Santa Cruz, CA), NF-κB p50 (sc-1190; Santa Cruz Biotechnology), NF-κB p52 (sc-298), c-ReL (sc-71), TLR-2 (sc-10739), TLR-4 (sc-16240), or actin (C-11; Santa Cruz Biotechnology) as control.
ELISA.
Primary mouse macrophages (macLITAF−/− or LITAF+/+) or human monocytes were seeded (2 × 104 cells in 96-well plate or 2 × 106 cells in six-well plate) and were either stimulated with 0.1 μg/ml of E. coli LPS (Sigma) or cotreated with 0.1 μg/ml of LPS and inhibitor (20 μM SB203580, 30 μM PD98059, 5 μM BAY 11-7082, or 10 μM U0126) or were transiently transfected with 1 μg of DNA using Lipofactamine (Invitrogen) for 3 h before LPS treatment, then incubated at 37°C, 5% CO2 for 16 h. Culture supernatants were harvested and centrifuged at 1,500 × g to remove cell debris. Concentrations of mouse or human TNF-α in the supernatant of each well of treated and untreated control cells were measured by ELISA (Abraxis, Warminster, PA). The ELISA immunoreactivity was quantified by using a VerSaDoc Imaging System (Bio-Rad, Hercules, CA) and graphed.
Mouse Protein Cytokine Array.
Macrophages (macLITAF−/− or LITAF+/+), seeded in a 96-well plate at 2 × 104 cells per well, were stimulated with 0.1 μg/ml of E. coli LPS (Sigma). The treated cells were cultured in RPMI medium 1640 with 10% FCS. After incubation for 16 h at 37°C, 5% CO2, the conditioned medium was harvested and centrifuged at 1,500 × g to remove cell debris before being applied to a mouse protein cytokine array (RayBiotech, Norcross, GA). The array membranes were processed according to the manufacturer's instructions. Briefly, membranes were blocked with a blocking buffer, and then 1 ml of medium from each culture of treated cells was individually added and incubated at room temperature for 2 h. Finally, the results with immunoreactivity were assessed and quantified by using a VerSaDoc Imaging System, (Bio-Rad) and graphed.
Human Phospho-MAPK Array.
Elutriated human monocytes (2 × 106 in a six-well plate) were either left as untreated controls or were treated with 0.1 μg/ml E. coli LPS alone or cotreated with 0.1 μg/ml LPS and 20 μM SB203580 (EMD Biosciences), then incubated at 37°C, 5% CO2 for 16 h. The total protein from whole cells was purified. Cell lysates (200 μg total proteins per array) were applied following the manufacturer's instructions (R & D Systems, Minneapolis, MN). The array with immunoreactivity was quantified using a VerSaDoc Imaging System (Bio-Rad) and graphed.
Supplementary Material
Acknowledgments
We thank Dr. S. Akira for providing TLR-2−/−, TLR-4−/−, TLR-9−/−, and MyD88−/−mice. This work was supported by National Institute of Dental and Craniofacial Research Grant R01 DE14079.
Glossary
Abbreviations
- LITAF
LPS-induced TNF-α factor
- TLR
Toll-like receptor
- MyD88
myeloid differentiation factor 88
- IRAK
IL-1R-associated kinase
- TRAF
TNF receptor-associated factor
- FLAP
5-lipoxygenase-activating protein.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Beutler B, Hoebe K, Du X, Ulevitch RJ. J Leukoc Biol. 2003;74:479–485. doi: 10.1189/jlb.0203082. [DOI] [PubMed] [Google Scholar]
- 2.Morrison SG, Morrison RP. Infect Immun. 2005;175:7536–7542. doi: 10.4049/jimmunol.175.11.7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Myokai F, Takashiba S, Lebo R, Amar S. Proc Natl Acad Sci USA. 1999;96:4518–4523. doi: 10.1073/pnas.96.8.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang X, Fenton MJ, Amar S. Proc Natl Acad Sci USA. 2003;100:4096–4101. doi: 10.1073/pnas.0630562100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang X, Marciano DL, Leeman SE, Amar S. Proc Natl Acad Sci USA. 2005;102:5132–5137. doi: 10.1073/pnas.0501159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 7.Fan J, Malik AB. Nat Med. 2003;9:315–321. doi: 10.1038/nm832. [DOI] [PubMed] [Google Scholar]
- 8.Dobrovolskaia MA, Medvedev AE, Thomas KE, Cuesta N, Toshchakov V, Ren T, Cody MJ, Michalek SM, Rice NR, Vogel SN. J Immunol. 2003;170:508–519. doi: 10.4049/jimmunol.170.1.508. [DOI] [PubMed] [Google Scholar]
- 9.Suri SS, Janardhan KS, Parbhakar O, Caldwell S, Appleyard G, Singh B. Vet Res. 2006;37:541–551. doi: 10.1051/vetres:2006017. [DOI] [PubMed] [Google Scholar]
- 10.Kawai T, Sato S, Ishii KJ, Coban C, Hemmi H, Yamamoto M, Terai K, Matsuda M, Inoue JI, Uematsu S, Takeuchi O, Akira S. Nat Immunol. 2004;5:1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- 11.Peng G, Guo Z, Kiniwa Y, Voo KS, Peng W, Fu T, Wang DY, Li Y, Wang HY, Wang RF. Science. 2005;309:1380–1384. doi: 10.1126/science.1113401. [DOI] [PubMed] [Google Scholar]
- 12.Covert MW, Leung TH, Gaston JE, Baltimore D. Science. 2005;309:1854–1857. doi: 10.1126/science.1112304. [DOI] [PubMed] [Google Scholar]
- 13.Chen BC, Wu WT, Ho FM, Lin WW. J Biol Chem. 2002;277:24169–24179. doi: 10.1074/jbc.M106014200. [DOI] [PubMed] [Google Scholar]
- 14.Serio KJ, Reddy KV, Bigby TD. Am J Physiol. 2005;288:C1125–C1133. doi: 10.1152/ajpcell.00296.2004. [DOI] [PubMed] [Google Scholar]
- 15.Wang T, Gu S, Ronni T, Du YC, Chen X. J Proteome Res. 2005;4:941–949. doi: 10.1021/pr050031z. [DOI] [PubMed] [Google Scholar]
- 16.Wang T, Chuang TH, Ronni T, Gu S, Du YC, Cai H, Sun HQ, Yin HL, Chen X. J Immunol. 2006;176:1355–1362. doi: 10.4049/jimmunol.176.3.1355. [DOI] [PubMed] [Google Scholar]
- 17.Eriksson B, Bergqvist I, Eriksson M, Holmberg D. FEBS Lett. 2000;479:106–110. doi: 10.1016/s0014-5793(00)01893-7. [DOI] [PubMed] [Google Scholar]
- 18.Bolcato-Bellemin AL, Mattei MG, Fenton M, Amar S. J Endo Res. 2004;10:15–23. doi: 10.1179/096805104225003780. [DOI] [PubMed] [Google Scholar]
- 19.Silverstein R. J Endotoxin Res. 2004;10:147–162. doi: 10.1179/096805104225004879. [DOI] [PubMed] [Google Scholar]
- 20.Maruyama H, Kikuchi S, Kawaguchi K, Hasunuma R, Ono M, Ohbu M, Kumazawa Y. Shock. 2000;13:160–165. doi: 10.1097/00024382-200013020-00011. [DOI] [PubMed] [Google Scholar]
- 21.Osakabe N, Yasuda A, Natsume M, Sanbongi C, Kato Y, Osawa T, Yoshikawa T. Free Rad Biol Med. 2002;33:798–806. doi: 10.1016/s0891-5849(02)00970-x. [DOI] [PubMed] [Google Scholar]
- 22.Kim E, Kim SH, Kim S, Kim TS. J Immunol. 2006;176:256–264. doi: 10.4049/jimmunol.176.1.256. [DOI] [PubMed] [Google Scholar]
- 23.Mazharian A, Roger S, Maurice P, Berrou E, Popoff MR, Hoylaerts MF, Fauvel-Lafeve F, Bonnefoy A, Bryckaert M. J Biol Chem. 2005;280:26002–26010. doi: 10.1074/jbc.M414083200. [DOI] [PubMed] [Google Scholar]
- 24.Stucchi A, Reed K, O'Brien M, Cerda S, Andrews C, Gower A, Bushell K, Amar S, Leeman S, Becker J. Inflamm Bowel Dis. 2006;12:581–587. doi: 10.1097/01.MIB.0000225338.14356.d5. [DOI] [PubMed] [Google Scholar]
- 25.Rane SG, Reddy EP. Oncogene. 2002;21:3334–3358. doi: 10.1038/sj.onc.1205398. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Q, Desta T, Fenton M, Graves DT, Amar S. Infect Immun. 2005;73:935–943. doi: 10.1128/IAI.73.2.935-943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao Y, Xu Q, Kwon MJ, Matta R, Liu Y, Hong SC, Chang CH. J Immunol. 2006;177:70–76. doi: 10.4049/jimmunol.177.1.70. [DOI] [PubMed] [Google Scholar]
- 28.Cao J, He J, Ding H, Zeng Y. Pain. 2005;118:336–349. doi: 10.1016/j.pain.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 29.Chatterjee PK, Coren J.S. Nucleic Acids Res. 1997;25:2205–2212. doi: 10.1093/nar/25.11.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Transgenic Res. 1999;4:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 31.Lauth M, Spreafico F, Dethleffsen K, Meyer M. Nucleic Acids Res. 2002;30:115–118. doi: 10.1093/nar/gnf114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarkar S, Lyer G, Wu J, Glass N.L. EMBO J. 2002;21:4841–4850. doi: 10.1093/emboj/cdf479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.