Abstract
Helicases are a ubiquitous class of enzymes involved in nearly all aspects of DNA and RNA metabolism. Despite recent progress in understanding their mechanism of action, limited resolution has left inaccessible the detailed mechanisms by which these enzymes couple the rearrangement of nucleic acid structures to the binding and hydrolysis of ATP1,2. Observing individual mechanistic cycles of these motor proteins is central to understanding their cellular functions. Here we follow in real time, at a resolution of two base pairs and 20 ms, the RNA translocation and unwinding cycles of a hepatitis C virus helicase (NS3) monomer. NS3 is a representative superfamily-2 helicase essential for viral replication3, and therefore a potentially important drug target4. We show that the cyclic movement of NS3 is coordinated by ATP in discrete steps of 11 ± 3 base pairs, and that actual unwinding occurs in rapid smaller substeps of 3.6 ± 1.3 base pairs, also triggered by ATP binding, indicating that NS3 might move like an inchworm5,6. This ATP-coupling mechanism is likely to be applicable to other non-hexameric helicases involved in many essential cellular functions. The assay developed here should be useful in investigating a broad range of nucleic acid translocation motors.
NS3 is a key component of the hepatitis C virus (HCV) RNA replication machinery and lies in a membrane-bound complex with other proteins7,8. NS3 is an NTPase with 3′ to 5′ helicase activity9,10, and it has been structurally characterized in various contexts11. We have developed a single-molecule12–16 assay for directly following the movement of full-length NS3 on its RNA substrate. Specifically, we use optical tweezers to apply a constant tension between two beads attached to the ends of a 60-base-pair (bp) RNA hairpin (Fig. 1a) and monitor the end-to-end distance change of the RNA as it is unwound by NS3. To establish the basis for interpretation of the enzymatic activity, we initially characterize the mechanical unfolding of the substrate in the absence of enzyme. The substrate unfolds at a force of 20.4 ± 0.2 pN (Fig. 1b). When the substrate is held at a constant force below 19 pN with the instrument’s force feedback mechanism, no unfolding takes place over periods of several minutes. Substrate unfolding at external forces below 19 pN must therefore be helicase-catalysed.
Figure 1. Assay with optical tweezers for assessing the mechanistic cycle of NS3.

a, Experimental design and attachment of the RNA substrate. Not to scale. b, Stages of an unwinding experiment: the substrate is first unfolded and refolded with mechanical force (green), next brought to a constant force chosen between 5 and 17 pN to monitor NS3-catalysed unwinding (red), then brought to 30 pN to probe its state (blue), and finally brought to 2 pN to allow refolding (yellow; 50% of traces, as this one, display incomplete substrate refolding because of NS3 binding). c, Representative trace of extension against time unwinding (15 pN, from b). d, Pairwise distance distribution for the unwinding trace in c (1-bp bins).
To follow NS3-catalysed unwinding, we flow NS3 (1–90 nM) and ATP (0.05–1 mM) together in buffer U (see Methods). We then hold the RNA substrate at a constant force of between 5 and 17 pN. NS3 loads on its substrate by means of a 3 ′ single-stranded RNA loading site. As NS3 unwinds the hairpin, the bead separation increases so as to hold the force on the molecule constant (Fig. 1b). The bead separation can be converted, at that force, into the number of RNA base pairs unwound as a function of time by using the worm-like-chain model of RNA elasticity17. The molecular geometry results in the release of two nucleotides (nt) for each base pair unwound, thereby amplifying the unwinding signal. The hairpin loop facilitates substrate reformation, allowing several unwinding traces to be collected with each RNA substrate. Unless otherwise noted, data are collected at 22 ±1 °C, 20 nM NS3, 1 mM ATP and 17 pN.
A rich variety of strictly ATP-dependent NS3 behaviours is observed throughout the unwinding traces obtained (N > 1000; Supplementary Fig. 1). The extension increases in sharp bursts of rapid strand separation (steps) followed by periods of constant extension (pauses) (Fig. 1c). A histogram of pairwise distances among all extensions of a given trace reveals that the distances between pauses are not randomly distributed but occur with well-defined periodicity (Fig. 1d). A Fourier analysis over each histogram yields an apparent unwinding step size of 11 ± 3 bp (Supplementary Discussion) over more than 100 traces under identical conditions. This step size appears to be intrinsic to NS3 because it is independent of substrate mechanical unfolding pattern and sequence, applied force, ATP concentration and NS3 concentration (Supplementary Figs 2 and 3)18. We observe rarer apparent backward15,16 steps by NS3, corresponding to stepwise refolding of the substrate after complete or partial unwinding (Supplementary Fig. 1c, d). It is possible that NS3 moves backwards (5′ to 3′) on the same strand19, or continues forwards (3′ to 5′) on the other strand, for example after turning around the tetraloop (Supplementary Fig. 1c). The backward step size, 12 ± 3 bp, is statistically indistinguishable from that of forward unwinding steps.
The direct observation of helicase unwinding steps confirms the cyclical movement deduced from bulk measurements of several helicases (for example refs 1, 20, 21). More importantly, it gives direct access to individual mechanistic cycles of a helicase during translocation and unwinding and their coupling to ATP binding. To investigate the role of ATP in coordinating the mechanistic cycle of NS3, we measured the dependence of the cycle on [ATP]. We varied [ATP] below and above the Michaelis–Menten constant, K m (160 ± 6 μM for the helicase domain of NS3 (ref. 22)). We observe two effects of [ATP] on the stepping behaviour of NS3 (Fig. 2a). First, the mean pause duration of NS3 decreases with increasing [ATP], from 3.9 s at 0.05 mM ATP to 0.6 s at 1 mM ATP (Fig. 2b), indicating that exit from a pause requires ATP binding. The shapes of the pause duration distributions obtained between 0.05 and 1 mM ATP (Supplementary Fig. 4) indicate that ATP binding might not be the sole rate-limiting event required for pause exit. Rather, the data indicate a possible two-step kinetic mechanism to exit from the pause state, only one of which involves ATP binding. Globally fitting the data to such a mechanism reveals the rates of ATP binding (k b = (9.9 ± 1.0) × 103 M−1 s−1)23 and of an [ATP]-independent step2 (k o = 1.9 ± 0.1 s−1) during pause. Second, the stepping velocity of NS3 between two pauses (that is, the slope of the steps in Fig. 2a) increases with [ATP], from 19 ± 18 bp s−1 at 0.05 mM ATP to 51 ± 26 bp s−1 at 1 mM ATP (Fig. 2c). Dependence of the stepping velocity on [ATP] implies that each step observed here is not one elementary event with a constant velocity but is made up of substeps, each of which requires ATP binding. The dependence of the stepping velocity of NS3 on [ATP] (Fig. 2d) follows Michaelis–Menten kinetics with V max = 51 ± 3 bp s−1 and K m = 93 ± 21μM. Thus, ATP binding must also take place during each step, strongly indicating that each unwinding step may be composed of substeps, which is consistent with the free energy of hydrolysis of one ATP molecule not being sufficient to melt 11 bp.
Figure 2. [ATP] affects both NS3 pauses and steps.
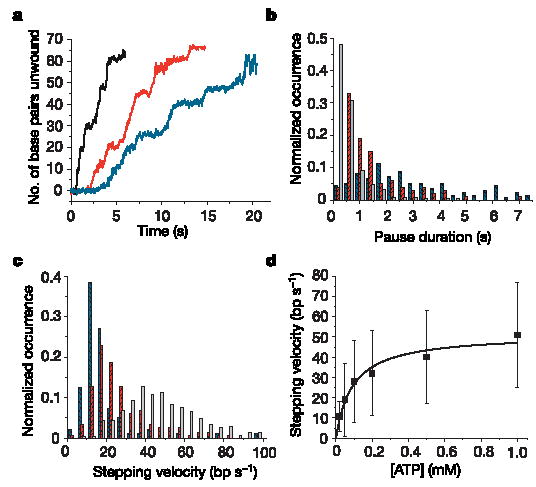
a, Representative traces of extension against time unwinding with 5 nM NS3 at 1 mM ATP (black), 0.1 mM ATP (red) and 0.05 mM ATP (blue). Traces are displaced along the time axis to avoid overlap. b, Histograms of pause durations (0.4-s bins) at 1 mM ATP (grey; 102 traces), 0.1 mM ATP (red; 52 traces) and 0.05 mM ATP (blue; 50 traces). c, Histograms of stepping velocities (5 bp s−1 bins) for the data used in b. d, Stepping velocity as a function of [ATP] with Michaelis–Menten fit. Error bars show s.d.
Indeed, more than 50% of unwinding traces collected at 0.05 mM ATP show substeps that appear within the 11 ± 3-bp step (Fig. 3, arrows). Substeps are visible even under saturating [ATP] conditions, although with lower frequency. The fact that the motor is observed to take substeps in a wide range of [ATP] indicates that its fundamental mechanism of operation is conserved through all [ATP]. Increasing [ATP] simply decreases the time spent between substeps and thus the probability of observing them. Most substeps are between 2 and 5 bp (3.6 ± 1.3 bp; Supplementary Fig. 5), indicating the possible existence of three substeps per step on average. A Poisson analysis of stepping times at 0.05 and 0.1 mM ATP confirms that each step is indeed composed of three substeps (Supplementary Fig. 5c). The presence of substeps suggests that in a full mechanistic cycle, unwinding is achieved not in a single motion by the helicase but in discrete smaller motions coordinated and triggered by ATP binding.
Figure 3. NS3 steps are composed of substeps.

Number of base pairs unwound over time for representative steps observed with 5 nM NS3 at 1 mM ATP (a), 0.1 mM ATP (b) and 0.05 mM ATP (c and d, displaying different numbers of substeps). Arrows point to substeps. About 90% of the steps observed at 1 mM ATP show no visible substeps within the step.
How many translocation and unwinding cycles can NS3 perform before dissociating from its substrate? The processivity of NS3 was not found to be significantly dependent on either [ATP] or [NS3] (Supplementary Fig. 6). However, the processivity of NS3 is strongly dependent on force16 (Fig. 4a): whereas an average of 18 bp are unwound at 5 pN before the enzyme dissociates, an average of 53 bp are unwound at 17 pN. In contrast, force does not affect the pause duration or stepping velocity of NS3 (Fig. 4b), indicating that NS3 might be limited not by strand separation but rather by translocation. Alternatively, the rate of strand separation might not be affected by force either because of protection of the RNA by NS3 or because the strand separation reaction coordinate in the presence of NS3 is orthogonal to the external force. Taken together, these results imply that the applied force increases the average time that NS3 remains bound to its substrate (Fig. 4a, b). These results indicate a possible competition between strand reannealing and NS3 binding and that NS3 might be a processive single-strand translocase in the absence of competition from strand reannealing. Future experiments will probe to what extent NS3 uses a brownian ratchet mechanism and an active duplex-melting mechanism. The differential effects of force on NS3 velocity and processivity indicate that the forward movement and dissociation of NS3 are biochemically distinct events.
Figure 4. Effect of force on the behaviour of NS3, and proposed model of action.

a, Fraction of duplex RNA of different lengths that are unwound processively at different forces. The mean processivity of NS3 is 18, 20, 33, 46, 48 and 53 bp at 5, 9, 11, 13, 15 and 17 pN (21, 69, 64, 62, 69 and 67 traces, respectively; 1-bp bins). b, Stepping velocities (red squares; means ± s.d.) and pause duration (blue circles; means and 95% fit confidence intervals) in 5 nM NS3 at 9, 11, 13, 15 and 17 pN (18, 35, 19, 53 and 39 traces, respectively). Pause durations plotted here were determined from a single-exponential fit of the pause duration histograms (Supplementary Fig. 4). c, Proposed model of NS3 translocation and unwinding. The helix opener site (red ellipse) unwinds the substrate in substeps of 2–5 bp (x and y designate substep sizes) triggered by ATP (V max, K m). The translocator (blue circle) contacts dsRNA every 11 bp, and possibly elsewhere during the cycle as well, which requires ATP binding (k b) and an [ATP]-independent kinetic step (k o).
There are important similarities and differences between the NS3 activities reported here and those observed in previous bulk experiments. Kinetic features are similar: for example, the stepping velocity at saturating [ATP] is 35 ± 4 bp s−1 in bulk measurements and 51 ± 3 bp s−1 as measured here, and K m is 160 ± 6 μM in bulk measurements with the helicase domain of NS3 (ref. 22) whereas the apparent K m measured here is 163 ± 10 μM (Supplementary Discussion). However, previous measurements of step size (18 ± 2 bp)20 are larger than reported here (11 ± 3 bp). This difference probably stems from the fact that different NS3 oligomeric states are monitored in the two types of experiment (Supplementary Discussion). During single-cycle bulk assays of RNA unwinding, the functional species studied is an NS3 dimer. This dimer requires long 3′ single-stranded (ss) RNA overhangs (at least 14 nt) for initiation and displays a strictly concentration-dependent unwinding amplitude20; moreover, it has a second-order rate constant for the kinetics of functional complex formation9 (V.S. and A.M.P., unpublished observations). In the present assay, the active species is an NS3 monomer that initiates unwinding within seconds at short 3′ overhangs (4 nt or longer; Supplementary Fig. 7) and shows no concentration dependence of pause duration, stepping velocity or processivity (Supplementary Figs 6b and 8). In a different bulk assay we show that the NS3 monomer indeed has ATP-dependent helicase activity in the absence of force, and that it unwinds with processivity below that observed at 5 pN in the single-molecule assay (data not shown), consistent with force increasing the processivity of the NS3 monomer.
The similarities between the activities of an NS3 monomer and dimer provide a perspective on the role of helicase dimerization for NS3, and perhaps other helicases. Although multimeric helicase assemblies may benefit from an increased unwinding processivity24, oligomerization does not itself seem to introduce new features in the unwinding mechanism. The NS3 monomer may be the engine of the dimer, whose processivity can be increased by assistance either from another NS3 molecule (and perhaps even from a different protein species) or by external force. The basis for the observed difference in step size of an NS3 monomer and of a dimer is not currently well understood.
One possible model among others (Supplementary Discussion) to account for the observed 11-bp periodicity and its substep structure is shown in Fig. 4c, in which each NS3 monomer has two RNA-binding sites and in which ATP binding coordinates both translocation and unwinding in the fashion of an inchworm5,6 (Supplementary Discussion). We designate one site as the translocator and the other as the helix opener: for each mechanistic cycle, the translocator moves by an 11-bp step to contact double-stranded (ds) RNA ahead25,26 of the fork, and the helix opener moves by substeps of 2–5 bp. A lower rate of ATP binding during a pause ((9.9 ± 1.0) × 103 M−1 s−1; Supplementary Fig. 4) than before subsequent substeps (at least 5 × 104 M−1 s−1, Supplementary Fig. 5a) indicates that these two ATP-binding events might be distinct and that ATP binding might be required both for translocator movement (blue ATP in Fig. 4c) and for strand separation by the helix opener (red ATP in Fig. 4c; Supplementary Discussion).
With unprecedented spatial and temporal resolution, the present strand displacement assay provides a powerful new method of investigating individual mechanistic cycles of many other enzymes that separate nucleic acid strands in the course of their cellular functions, such as ribosomes and polymerases.
METHODS
RNA substrate
The hairpin substrate sequences were cloned into the EcoRI and HindIII sites of pBR322 (NEB). To generate the hairpin DNA fragment, four oligonucleotides (65–100 nt each) were sequentially annealed. After cloning, double digestion of the circular construct by NcoI (designed at the hairpin loop) and StyI yielded two DNA fragments, each containing half of the hairpin sequence. The two fragments were gel purified and sequenced separately, confirming the full hairpin sequence. A transcription template (pBR322 bases 3821–628 with hairpin insert) was obtained by polymerase chain reaction (PCR) with a T7 promoter-appended primer, and transcribed in vitro (Ambion). The 1,287-nt RNA product was annealed to pBR322-based DNA handles (Fig. 1a); the 599-bp handle had a 5′ biotin group (added through a PCR primer) and the 535-bp handle a 3′ digoxigenin group (added with exo-Klenow fragment (NEB) after 3′ recessed end-generation by EcoRI). The RNA sequence between the handles consisted of a 20-nt single-stranded spacer (shortened to 0 nt without effect), both arms of the 60-bp hairpin (underlined) separated by a tetraloop, and a 19-nt single-stranded spacer for NS3 loading (shortened to 4 nt without effect): 5 ′-UCUCAUGCAGGACAGUCGGAGGGAGCACUACGUUCGGACUAGUGUACUCUGACUUGAGACUACUGACAUCCAGAUCUCCCCCAUGGGAGAUCUGGAUGUCAGUAGUCUCAAGUCAGAGUACACUAGUCCGAAC GUAGUGCUCCCAGGAGCUCAGCUAUCAGAA-3′.
Unless otherwise mentioned, the above sequence (RNA1) was used. The pairing, geometry and attachment of the substrate were confirmed (Supplementary Discussion). Substrates of different sequences (RNA2, RNA3 and RNA4; Supplementary Fig. 2) were synthesized as above. Substrates had two helicase-loading sites, namely the ssRNA regions located 3′ to the hairpin and the 535-bp DNA/RNA handle. However, the extension change resulting from DNA/RNA unwinding was one-tenth of that for hairpin unwinding. Replacing the hairpin by a 29-nt ssRNA region resulted in no unwinding detection under standard conditions.
NS3 protein
Full-length NS3 from HCV genotype 1a was expressed (pQE40 plasmid, N-terminal His6 tag) and purified with a protocol described elsewhere27, with small modifications. [NS3] was measured with the Bradford assay. Although some earlier bulk measurements were done with NS3 genotype 1b20, all bulk measurements stated here refer to NS3 genotype 1a unless otherwise mentioned.
Experiments with optical tweezers
We used a dual-beam optical tweezers instrument28 to manipulate individual RNA molecules. Molecule characterization was performed as described elsewhere29. The RNA was unfolded and refolded by moving (200 nm s−1) the 2.2-μm streptavidin-coated bead (Spherotech), connected to a piezoelectric stage through a micropipette, relative to the 2.9-μm anti-digoxigenin-coated bead30, held in the laser trap. The distance between the folded state and the transition state29 of the 60-bp hairpin was large (more than 10 nm), resulting in a narrow force range over which unfolding took place. Force-catalysed unfolding at 1.4 pN or more below the mean unfolding force was not observed. Buffer U consisted of 20 mM MOPS, 30 mM NaCl, 0.9% v/v glycerol, 0.75 mM MgCl2, 0.1% Tween 20, 2 mM dithiothreitol, pH 6.5 at 22 °C. Data were collected at 60 Hz and the force detector signal was smoothed with an RC filter with a time constant of 14 ms. In constant-force feedback mode, force was constant to 0.1 pN. In the absence of NS3, two standard deviations in extension covered 1.4 nm at 60 Hz, which translates to a 2-bp resolution under our molecular geometry at 17 pN tension (Supplementary Discussion). Data were collected in the 10 min after injection of NS3 and ATP (Supplementary Discussion). Putative unwinding was confirmed by pulling the substrate after the constant-force period (Fig. 1b): a shortened mechanical unfolding transition or its absence indicated partial or full unwinding, respectively. For each RNA molecule attached between beads, we collected an average of ten unwinding traces.
Data analysis
The worm-like chain model17 was used with a ssRNA contour length of 0.59 nm per nucleotide and a persistence length of 1 nm (ref. 29). NS3 binding to ssRNA did not significantly change its contour and persistence lengths (Supplementary Discussion). The step size was the spatial frequency with the highest power (Fourier analysis) of a pairwise distance distribution from a single trace. An identical step size was obtained if the power spectra of individual pairwise distance distributions were summed (Supplementary Fig. 2f) and the highest power was identified. Steps were detected by scanning for maxima in the slope of the unwinding curve within a running-window (MATLAB custom-written programs). The window size was larger than step durations, and data points within maximal-slope windows were cropped on the basis of the mean and standard deviation of constant-extension regions. Pause durations were taken as the intervals between two successive steps and the velocity of a step was its best linear-fit slope (of the raw data). As [ATP] decreased, the number of data points per step increased and the best linear-fit R 2 decreased. The maximum velocity that the constant-force feedback could follow was 172 ± 17 nm s−1, well above the maximum observed NS3 velocity. Substeps were required to belong to a previously detected step and were detected with a smaller running window than steps, whereas subpauses (periods between substeps within a step) were required to be longer than 80 ms.
Supplementary Material
Acknowledgments
We thank H. V. Le from the Schering-Plough Research Institute for the NS3 plasmid; S. B. Smith, P. T. X. Li, Y. R. Chemla and J.-C. Liao for discussions and technical help; T. M. Lohman for critical reading of the manuscript, and members of our laboratories for discussions and critical reading of the manuscript. This research was supported by CIHR and FQRNT doctoral fellowships (S.D.), an NIH postdoctoral fellowship (R.K.B.), NIH (I.T., A.M.P., C.B.), DOE (C.B.), and HHMI grants to investigators A.M.P. and C.B.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 2.Lucius AL, Lohman TM. Effects of temperature and ATP on the kinetic mechanism and kinetic step-size for E. coli RecBCD helicase-catalyzed DNA unwinding. J Mol Biol. 2004;339:751–771. doi: 10.1016/j.jmb.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Kolykhalov AA, Mihalik K, Feinstone SM, Rice CM. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J Virol. 2000;74:2046–2051. doi: 10.1128/jvi.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frick DN. Helicases as antiviral drug targets. Drug News Perspect. 2003;16:355–362. doi: 10.1358/dnp.2003.16.6.829307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 6.Bianco PR, Kowalczykowski SC. Translocation step size and mechanism of the RecBC DNA helicase. Nature. 2000;405:368–372. doi: 10.1038/35012652. [DOI] [PubMed] [Google Scholar]
- 7.Delagoutte E, von Hippel PH. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: Integration of helicases into cellular processes. Q Rev Biophys. 2003;36:1–69. doi: 10.1017/s0033583502003864. [DOI] [PubMed] [Google Scholar]
- 8.Dimitrova M, Imbert I, Kieny MP, Schuster C. Protein-protein interactions between hepatitis C virus nonstructural proteins. J Virol. 2003;77:5401–5414. doi: 10.1128/JVI.77.9.5401-5414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pang PS, Jankowsky E, Planet PJ, Pyle AM. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 2002;21:1168–1176. doi: 10.1093/emboj/21.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levin MK, Gurjar M, Patel SS. A Brownian motor mechanism of translocation and strand separation by hepatitis C virus helicase. Nature Struct Mol Biol. 2005;12:429–435. doi: 10.1038/nsmb920. [DOI] [PubMed] [Google Scholar]
- 11.Kim JL, et al. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 12.Dohoney KM, Gelles J. Chi-sequence recognition and DNA translocation by single RecBCD helicase/nuclease molecules. Nature. 2001;409:370–374. doi: 10.1038/35053124. [DOI] [PubMed] [Google Scholar]
- 13.Bianco PR. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 14.Ha T, et al. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 15.Perkins TT, Li HW, Dalal RV, Gelles J, Block SM. Forward and reverse motion of single RecBCD molecules on DNA. Biophys J. 2004;86:1640–1648. doi: 10.1016/S0006-3495(04)74232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dessinges MN, Lionnet T, Xi XG, Bensimon D, Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc Natl Acad Sci USA. 2004;101:6439–6444. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 18.Levin MK, Yang YH, Patel SS. The functional interaction of the hepatitis C virus helicase molecules is responsible for unwinding processivity. J Biol Chem. 2004;279:26005–26012. doi: 10.1074/jbc.M403257200. [DOI] [PubMed] [Google Scholar]
- 19.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 20.Serebrov VS, Pyle AM. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature. 2004;430:476–480. doi: 10.1038/nature02704. [DOI] [PubMed] [Google Scholar]
- 21.Ali JA, Lohman TM. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science. 1997;276:377–380. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- 22.Levin MK, Patel SS. Helicase from hepatitis C virus, energetics of DNA binding. J Biol Chem. 2002;277:29377–29385. doi: 10.1074/jbc.M112315200. [DOI] [PubMed] [Google Scholar]
- 23.Levin MK, Gurjar MM, Patel SS. ATP binding modulates the nucleic acid affinity of hepatitis C virus helicase. J Biol Chem. 2003;278:23311–23316. doi: 10.1074/jbc.M301283200. [DOI] [PubMed] [Google Scholar]
- 24.Tackett AJ, Chen Y, Cameron CE, Raney KD. Multiple full-length NS3 molecules are required for optimal unwinding of oligonucleotide DNA in vitro. J Biol Chem. 2005;280:10797–10806. doi: 10.1074/jbc.M407971200. [DOI] [PubMed] [Google Scholar]
- 25.Tackett AJ, Wei L, Cameron CE, Raney KD. Unwinding of nucleic acids by HCV NS3 helicase is sensitive to the structure of the duplex. Nucleic Acids Res. 2001;29:565–572. doi: 10.1093/nar/29.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bianco PR. Hepatitis C NS3 helicase unwinds RNA in leaps and bounds. Lancet. 2004;364:1385–1387. doi: 10.1016/S0140-6736(04)17238-3. [DOI] [PubMed] [Google Scholar]
- 27.Yao N, et al. Structure of the hepatitis C virus RNA helicase domain. Nature Struct Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 28.Smith SB, Cui Y, Bustamante C. Optical-trap force transducer that operates by direct measurement of light momentum. Methods Enzymol. 2003;361:134–162. doi: 10.1016/s0076-6879(03)61009-8. [DOI] [PubMed] [Google Scholar]
- 29.Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 30.Bryant Z, et al. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–341. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


