Abstract
Length-dependent helical propensities wAla(n,T) at T = 10, 25, and 60 °C are assigned from t/c values and NMR 13C chemical shifts for series 1 peptides TrpLysmInp2tLeu–AlantLeuInp2LysmNH2, n = 15, 19, and 25, m = 5, in water. Van’t Hoff analysis of wAla(n,T) show that α-helix formation is primarily enthalpy-driven. For series 2 peptides Ac–Trp Lys5Inp2tLeu–βAspHel–Alan–beta–tLeuInp2Lys5NH2, n = 12 and 22, which contain exceptionally helical Alan cores, protection factor-derived fractional helicities FH are assigned in the range 10–30 °C in water and used to calibrate temperature-dependent CD ellipticities [θ]λ,H,n,T. These are applied to CD data for series 1 peptides, 12 ≤ n ≤ 45, to confirm the wAla(n,T) assignments at T = 25 and 60 °C. The [θ]λ,H,n,T are temperature dependent within the wavelength region, 222 ± 12 nm, and yield a temperature correction for calculation of FH from experimental values of [θ]222,n,T,Exp.
Introduction
In this report, the last in a series1 that characterizes the helix-coil energetics for spaced, solubilized Alan peptides 1a,b in water, we assign temperature dependences to the alanine helical propensity and to the calibrating constants required to assign fractional helicities2 FH from circular dichroism CD ellipticities. As well as temperature, polypeptide helicity depends on length, on amino acid composition and sequence, and on the structures of N- and C-caps. Sets of energetic parameters that quantitate these dependences have been assigned by many workers from helicity changes caused by guest substitutions within helical host peptides or proteins.3 Unfortunately, for key sets, doubts and disagreements persist.4 Doubt resolution is a primary goal of new host–guest studies,5 which rely on Alan host peptides. Our first study pinpoints the need for new Alan hosts that, relative to 1a,b, retain helicity in water over a large temperature range, and herein, using CD temperature calibration data, we prove their feasibility.
Inp ≡ 4-carboxypiperidine tL ≡ tert-Leu beta ≡ β–aminoalanine
Long recognized as potentially ideal helical hosts, simple polyalanines are intractable in water. To solubilize our Alan constructs 1a,b and inhibit potential aggregation, we flank the Alan cores with charged Lysm regions,6 using, in effect, time-honored tri-block oligopeptide solubilization.7 Equally important, the cores are isolated from helix-perturbing charge by Inp2tL or tL3 spacing elements.6
Calibrations of experimental assignments of Alan fractional helicity FH and site helicites FHi are essential. Previously for constructs 1a,b, we calibrated8 the assignment of Alan core FH values9,10 from cap-corrected experimental ellipticities [θ]222. Along with three other quantitative helicity measures, we have used these [θ]222 to assign the alanine helical propensity wAla(n,2) at 2 °C, which proved to be length dependent.1
The following definitions and concepts are used in this report. A set of w values for a series of amino acids is easily transformed into a set of relative coil-helix free energies ΔΔG, defined with respect to one standard amino acid. Helicities from studies of guest substitutions within protein hosts are usually expressed in this form, but for peptide hosts, the Lifson–Roig11 helical propensity w is usually used. The w value is an equilibrium constant that reflects the tendency of a new amino acid residue to join a pre-existing helical conformation to which it is linked. As commonly modeled, w is temperature and residue dependent.
In the L–R model, a peptide helix of length n that lacks stabilization by tertiary packing consists of an ensemble of 2n equilibrated nonhelical, fully helical, and partially helical conformations, and the corresponding state sum SS assigns a state weight to each. It is conveniently calculated from a vector–matrix product and can be used to model any quantitative experimental helicity measurement. For L–R algorithms used in our studies, an Alan conformer containing k contiguous helical residues is weighted jυ2wkc, in which j and c are N- and C-capping parameters.12 The helix initiation parameter υ is usually taken as residue-independent, and υ, j, and c are assumed to be both length- and temperature-independent.
Temperature and Length Dependences for Alanine Helical Propensities wAla(n,T).
| (1) |
| (2) |
Four helicity data sets were used for our previous recursive wAla(n,2) assignment at 2 °C.1 For recursive assignment of 25 °C values, we use three of these: t/c data, 13C═O NMR chemical shifts, and [θ]222 values. We started with previously reported t/c data, assigned for peptides Ac–Hel–Alan–tLInp2Lys4Trp–NH2, 4 ≤ n ≤ 14.13 The value t/c is assigned as the ratio of integrated intensities of two types of 1H NMR resonances, attributable to slowly equilibrating t and c N-terminal amide rotamers for the acetamido group within the helix-inducing N-cap 4a,b, X = CH3. Only in its te state does the cap strongly initiate helices, and t/c, which is proportional to the ratio of respective helical state sums SSte/SScs, eq 1, thus provides a useful quantitative helicity measure. The t/c data of Figure 1, 4 ≤ n ≤ 14, were used to assign a length-dependent wAla(n,25) set, as previously described for 2 °C data.1
Figure 1.
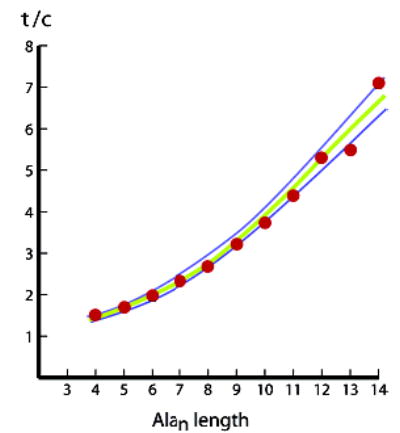
Red dots correspond to t/c data for the series Ac–Hel–Alan–tLInp2–Lys4Trp–NH2 n = 4–14, in water at 25 °C. Lines plot t/c values L–R-modeled from recursively assigned wAla(n,25) values. Previous parameter values13 at 2 °C were used to model the upper blue line: A = 0.80; B = 0.17, the initiation parameter for the t-state. The lower blue line and the best-fit green correspond to B = 0.15 and 0.16; j = 6.5, the N-capping Hel parameter for cs-state helices.
Figure 4.
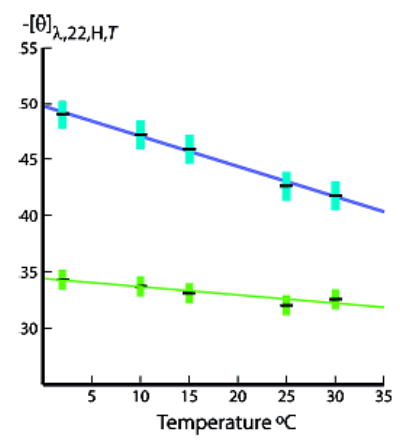
Temperature dependences of Figure 3b per-residue molar ellipticities [θ]222,H,22,T (blue) and [θ]208,H,22,T (green) for series 2 Ala22. Linear temperature regressions for Ala22: at 222 nm: intercept −49.7, slope +260; at 208 nm: intercept −34.6, slope +110. Linear regressions for Ala12 (data not shown): at 222 nm, intercept −44.5, slope +280; at 208 nm, intercept −34.7, slope +92. Units: intercept, 103 deg cm2 dmol−1; slope, deg cm2 dmol−1 K−1. Error limits are calculated from errors in FH values.
An extrapolation from this set generated preliminary values for a more extended series of wAla(n,25), 4 ≤ n ≤ 19, which was perturbed recursively to fit central-residue NMR 13C chemical shifts, measured at 25 °C for series 1b peptides, n = 15, 19, and 25. These shifts are proportional to site helicities FHi, eq 2, and their most accurate assignments result if the value of δ13CExp lies within 25–75% of the overall range of the calibrating chemical shifts δ13CHelix and δ13CCoil. At 25 °C, central residue shifts for series 1b peptides, n = 15 & 19, Table 1, meet this condition. We perturbed these using eq 2 starting at n = 14, to obtain L–R best-fits for experimental values of δ13CExp. The resulting wAla(n,25), 3 ≤ n ≤ 19, were again extrapolated into the range 15 ≤ n ≤ 25, and these were perturbed to fit δ13CExp for n = 25. Data of Table 1 compare δ13CExp and δ13CCalc pairs for n = 15, 19, and 25, which agree within measurement error. The green and blue lines of Figure 2 respectively plot the length dependences of wAla(n,25) and the previously assigned wAla(n,2).1 The cyan line plots wAla(n,10) values that were assigned by recursive perturbations of the wAla(n,2) set, using Table 1 chemical shift data.
Table 1.
Central Residue 13C NMR Chemical Shift Correlations for the Series TrpLysmtLeu3–AlantLeu3LysmNH2, n = 15, 19, and 25, T = 10, 25, and 60 °C
| temperature, °C | δ13CHelix, ppm | δ13CCoil, ppm | N = 15 δ13C | n = 19 δ13C | n = 25 δ13C |
|---|---|---|---|---|---|
| 1. 10 | 180.75a | 177.4 | exp 179.43 | exp 180.45 | exp 180.75 |
| calc 179.40 | calc 180.43 | calc 180.72 | |||
| 2. 25 | 180.70a | 177.3 | exp 178.87 | exp 179.87 | exp 180.53 |
| calc 178.83 | calc 179.90 | calc 180.53 | |||
| 3. 60 | 180.35a | 177.0 | exp 177.84 | exp 178.20 | exp 178.63 |
| calc 177.80 | calc 178.20 | calc 178.62 | |||
| w = 1.205b | w = 1.181b | w = 1.156b |
Limiting chemical shift values extrapolated from data measured at 2.3, 4.3, 6.3, and 8.3 °C. See Experimental Section for details.
Conventional length-independent w values assigned to 60 °C chemical shift values, n = 15, 19, 25; see text for a discussion.
Figure 2.
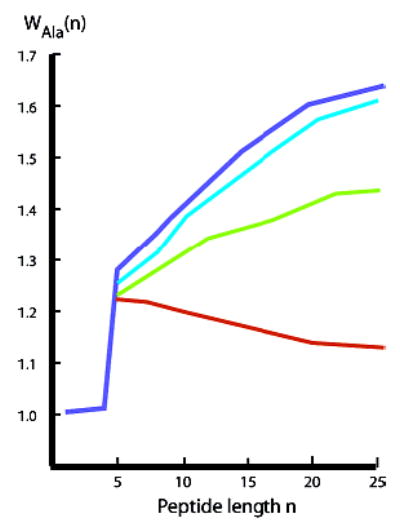
Temperature-dependent helical propensities wAla(n,T) for alanine residues in water within polyalanine contexts 1a,b; blue: T = 2 °C; cyan: T = 10 °C; green: T = 25 °C; red: T = 60 °C. Values assigned at or below 25 °C show a positive length dependence, but values assigned at 60 °C show a negative length dependence. CD data (shown below) show that for n > 25, all wAla(n,T) converge to an upper limit.
What loss of accuracy results if chemical shift data are modeled by fixed w values? The 25 °C, n =15 δ13CExp corresponds to a fixed w of 1.338. Applying this value to n = 19 and 25 yields, respectively, δ13Calc = 179.67 ppm (found: 179.87) and δ13CCalc = 180.25 ppm (found: 180.53). (Error limits: ± 0.02.)
Do t/c data, as used for early recursion stages, predetermine the subsequent 13C-based wAla(n,T) assignments for the entire length region? To address this question, we constructed a new wAla(n,T) series derived solely from shift data. A fixed w = 1.338 was used to model all peptide lengths n ≤ 15, and the 13C data for n = 15, 19, and 25 were then fitted recursively to assign wAla(n,T) within the length range 15 ≤ n ≤ 25. These assignments differed by less than 1% from those of Figure 2, implying that the t/c-derived wAla(n,25) for 3 ≤ n ≤ 14 have little effect on FH values assigned for the length range 15 ≤ n ≤ 25. This result is expected. The state sums of ensembles for these peptides, which have average w values in the range 1.3–1.4, are dominated by helical weights of the longer conformations.
Inspection of Figure 2 shows a significant decrease in the change in wAla(n,T) as the temperature is raised from 2 to 25 °C, suggesting that at higher temperatures, w values are length-independent. This hypothesis was tested for the 60 °C data of Table 1. For δ13CExp, n = 15, 19, and 25, we assigned fixed w values, shown in the last Table row, which correlate inversely with length. By perturbing the wAla(n,25) set, starting with low n, values for wAla(n,60), red line of Figure 2, were assigned recursively.
Positive hydrogen bonding cooperativity, a length-dependent increase in the strengths of α-helical amide–amide hydrogen bonds, provides the best explanation for an increase of w with length n, T ≤ 25 °C.14 How can one explain a negative length dependence for w, seen at 60 °C? Speculative hypotheses include a temperature-dependent change in the relative free energies of amide–water and amide–amide hydrogen bond formation or a temperature dependence of the helical initiation parameter t.15
A van’t Hoff analysis of the data of Figure 2 allows assignment of per-residue enthalpies of helix–coil propagation for alanine. Reported enthalpies lie in the range of −1.1 to −1.3 kcal/mol for experimental values,16a,d and −0.8 kcal/mol has been reported from gas-phase ab initio modeling.16b,c Our length-dependent enthalpies calculated for 10, 15, 20, and 25 residues are, respectively, −0.51, −0.91, −1.1, and −1.2 kcal/mol per residue, with an estimated uncertainty of ± 8–10%. Consistent with a recent literature review, α-helix formation by Alan peptides in water is primarily enthalpy-driven.16d
Temperature-Dependent FH Values for Series 2 Alan Peptides, 2–30 °C; Characterization of the Temperature Dependence of [θ]222,H,n,T; Extension of the Length Range of Temperature-Dependent wAla(n,T) Assignments; The Problem of Parametric Curvature; Feasibility of Polyalanine Second-Generation Helical Hosts for Assignments of Helical Propensities.
We previously proved that for alanine-rich peptides, experimental temperature-dependent CD ellipticities at 222 and 208 nm do not correlate linearly, and data of Figure 3a show that even at very low temperatures, the limiting ellipticity [θ]222,Hn,T remains temperature dependent.17 As seen in eqs 3 and 4, this parameter must be calibrated in water at normal temperatures before CD data can be used to extend the length range for wAla(n,T) assignments. Protection factors, PFi, resolve this problem.
Figure 3.
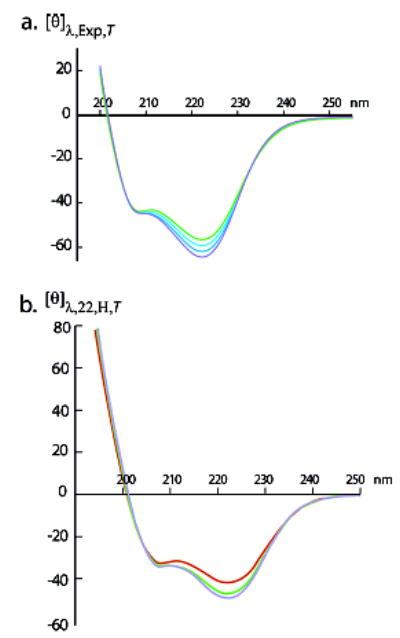
(a) Experimental per-residue molar ellipticities [θ]λ,22,T,Exp, units, for the peptide Ac–Hel(Ala4Lys)4Ala2NH2 in 32 mol % ethylene glycol–water at intervals of 10 °C from −20 °C (top green curve) to −50 °C (bottom blue curve).17 Convergence to temperature-independent values of [θ]λ is observed, except within the λ region: 222 ± 12 nm; the slope at 222 nm is +260 deg cm2 K−1 dmol−1. (b) Calculated values of [θ]λ,H,n,T for the completely helical conformations of the series 2 peptide βAspHel–Ala22–beta in water at 2 (blue curve), 10 (green curve), and 30 °C (red curve). Units: 103 deg cm2 dmol−1.
| (3) |
at 2° C,8
| (4) |
A PFi for the backbone NH at a peptide residue i equals the relative rate constant for NH → ND exchange in D2O, scaled by the corresponding rate constant for exchange of a solvent-exposed NH within an unstructured conformation.18 From the relationship FHNHi = 1 − 1/PFi, one can convert PFi into the site helicity measure FHNHi, the mole fraction of the site i NH that is helically hydrogen bonded.1b,8 We previously reported protection factors PFi measured in water at 2 °C for all alanine sites of representative series 1a and series 2 peptides;1,8 for the latter, the FH values approach 1.0.
The data of Table 2 show FH values calculated from PFi assignments for temperatures in the range of 10–30 °C for two series 2 peptides. In this section, we use these in four ways: first, a temperature calibration of [θ]222,H n,T and [θ]208,H n,T; second, an assignment of wAla(n,T) values within the length range 25 ≤ n ≤ 45 residues; third, a comparison of parametric curvature for series 1 and 2 peptides; and fourth, a feasibility demonstration for construction of new helical Alan hosts.
Table 2.
Temperature-Dependent Fractional Helicities Assigned to Series 2 Peptides Ala22 and Ala22 from Site-Protection Factors Assigned from Rates of Ala NH → ND Exchange in D2O at PD ≥ 4.5
| Temp (°C) | 2 | 10 | 15 | 25 | 30 |
|---|---|---|---|---|---|
| Ala22 | |||||
| 1.FHNH–corea | 0.992 | 0.990 | 0.985 | 0.97 | 0.93 |
| 2. FHCalcb | 0.96 | 0.96 | 0.95 | 0.94 | 0.90 |
| 3. FHSiteAvec | 0.96 | 0.95 | 0.93 | 0.94 | 0.91 |
| Ala12 | |||||
| 1. FHNH–corea | 0.99 | 0.99 | 0.97 | 0.94 | – |
| 2. FHCalcb | 0.92 | 0.92 | 0.92 | 0.89 | – |
| 3. FHSiteAvec | 0.93 | 0.92 | 0.91 | 0.88 | – |
The Alan core is defined as the (n − 8) central Alan residues.
The value of FHcalc is assigned for a given n, under the assumption of a constant ratio of core FHi to overall FH.
The estimated errors in FH lie in the range of ± 0.02 (low temperature) to ± 0.03 (high temperature).
Because the Table 2 values of FH approach or exceed 0.90, the implicit two-state approximation of (5′) can be used to calculate [θ]λ,n,H,T from [θ]λ,U,T,19 FH(T), and [θ]λ,n,Exp,T. The resulting [θ]λ,22,H,T are plotted as functions of λ and T in Figure 3b. These spectra qualitatively resemble those previously reported, Figure 3a, for the alanine-rich peptide Ac–Hel(Ala4Lys)4Ala2NH2 in 32 mol % ethylene glycol–water for −20 ≥ T ≥ −50 °C.17
| (5) |
| (5′) |
Within error limits for [θ]λ,H,n,T, temperature-independence is seen except within the λ region, 222 ± 12 nm, implying that, as discussed previously,17 the n → π* CD transition is unusually responsive to an underlying temperature-dependent structural change. Figure 4 quantitates the temperature dependences of calculated values of [θ]λ,H,n,T at 208 and 222 nm, and eq 5 results if slope data are incorporated into eq 4.
| (6) |
Eq 6 allows use of CD data to confirm the wAla(n,25) assignments of Figure 2 and test the assumption that they converge to an upper limit at n = 25. The green circles of Figure 5 depict [θ]222,n,25,Exp data, measured in water at 25 °C for series 1a Alan peptides.6 To calculate [θ]222,T,Exp from eq 6 with Figure 2 wAla(n,25) values, we used the following 2 °C assignments: jtL = ctL = 0.5; and X = 6.5.1 The green curve shows the resulting [θ]222,25,Exp values, which have been extended into the length region 25 < n < 45. Although the data in this range are slightly overestimated, the overall fit lies within the [θ]222,T,Exp assignment error, estimated as ± 4%.
Figure 5.
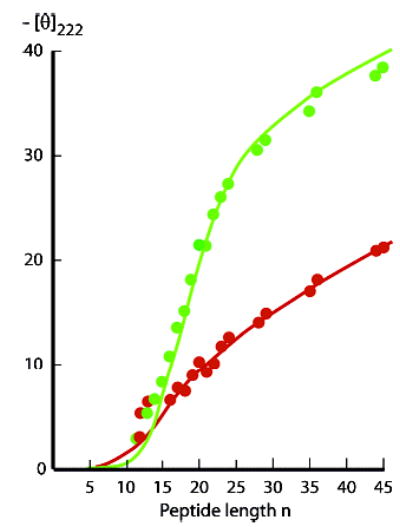
CD tests of wAla(n,T) data sets using [θ]222 data (green, 25 °C; red, 60 °C) measured in water for Alan 1a peptides.6 L–R modeling of [θ]222 at 25 °C (green line) used [θ]222,n,25 = (−60 500 + 260 (25−2))(1 − X/n), X = 6.5, as assigned at 2 °C.1 (fit: n = 18–45, SD: 1.2). At 60 °C (red line), [θ]222,n,60 = (−60 500 + 260 (60−2))(1 − X/n), X = 2.5. (fit: n = 16–45, SD: 0.5). Alternative (not shown): [θ]222,n,60 = (−60 500 + 260 (25−2))(1 − X/n), X = 3.7. (n = 16–45, SD: 0.9). Units: 103 deg cm2 dmol−1.
The red data points of Figure 5 depict [θ]222,n,60,Exp data. A rigorous value for [θ]222,H,n,60 has not been assigned, but we can model the data for two extremes: either the temperature correction of eq 6 can be extended to 60 °C or the correction converges to a high-temperature limit at 25 °C. For either choice, the 2 °C assignment of X = 6.5 generates models that underestimate all [θ]222,n,Exp values. However, if X is defined to minimize the standard deviations for each choice, then data can be fitted over the length range 12 ≤ n ≤ 45, as detailed in the Figure 5 legend.
| (7) |
| (8) |
The results of Figure 4 also provide insight into anomalous ellipticities observed at low temperatures for alanine-rich peptides. Normal CD spectra for highly helical peptides and proteins exhibit ellipticity minima of nearly equal intensity at 208 and 222 nm; the limiting value for R2 = [θ]222/[θ]208, introduced by Bruch et al.20 as a helicity measure, lies close to 1.0, with limiting [θ]222 intensities of (−37 to −44) × 103 deg cm2 dm−1.17,21 Our exemplar parameters for alanine-rich peptides are R2 = 1.3, and [θ]222,H,∞,2 = −60.5 × 103 deg cm2 dm−1, assigned for the extrapolated 2 °C spectrum for a completely helical alanine residue within an extended Alan sequence. Since publication of this assignment, it has been confirmed by oligopeptide studies22a and by low-temperature CD spectra of an alanine-rich antifreeze protein of the winter flounder.22b From Figure 4, these anomalies are predicted to be maximal at temperatures near 0 °C, and melting effects are expected to become dominant at higher temperatures.
As seen in Figure 6, plots of [θ]222,Exp,T vs [θ]208,Exp,T provide strong support for these predictions. Linear 222–208 nm temperature-derived parametric plots, with slopes within the range 1.1–1.3, are characteristic of helical protein fragments, but polyalanines exhibit parametric curvature, typified by Ala28 plot of Figure 6a.17 In high- and low-temperature regions, its local slopes are respectively 1.5 and 2.5; the first approaches the value for a normal helical heteropeptide, suggesting that melting dominates, but the high slope for the low-temperature region is best explained as largely reflecting the temperature dependence of the limiting helical ellipticity [θ]222,H,22,T.
Figure 6.
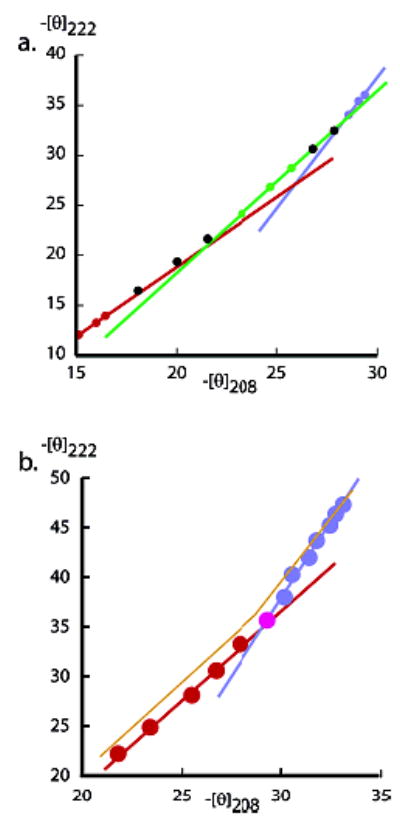
(a) A dual-wavelength parametric plot of cap-corrected [θ]222 vs [θ]208 measured in water for the series 1a peptide. Temperature is the implicit variable, and its range is 60–2 °C (respectively low and high −[θ] values), at intervals of 5 °C from 60 to 5 °C. Three local slopes are shown; 1.5 red; 1.8 green; 2.5, blue; each defined by three or four successive data points at high, medium, and low temperatures. The continuously increasing local slopes reflect the heterogeneity of helical lengths within the conformational ensemble. (The Ala28 spectra exhibit a 203 nm isoelliptic point.6) (b) Series 2 peptide βAspHel–Ala22–beta plot exhibits two regions of constant slope: 60–35 °C (red line and data points; C. C. = 0.997; slope 1.8) and 30–2 °C (blue line and data points; C. C. = 0.996; slope 3.2). Units: 103 deg cm2 dmol−1.
Figure 6b shows the characteristic parametric signature for a type 2 Alan peptide with n > 14.23 Data fall within two well-defined fixed-slope regions; from 2 to ca. 30 °C, [θ]222,Exp,T decreases by 20–25% with a slope of 3.2. The data of Table 2 and Figure 4 show that in this region, FH decreases by only 5–6%, but [θ]222,H,22,T decreases by ca. 16%. The slope within the 30–60 °C region of Figure 6b is 1.8, which is higher than that predicted for a normal linear parametric plot but suggests dominance by thermally induced FH changes. As seen in the next section, at low temperatures, the helical ensemble for this peptide is dominated by long helical conformers, which collectively can approximate a single helical state. At higher temperatures, the more normal “frayed” helical ensemble dominates, and the sharp transition between regions of fixed slopes in part reflects this transition.23
The take-home messages from these studies are clear. For any helical heteropeptide that lacks tertiary stabilization, a linear parametric 222 vs 208 nm ellipticity plot with a slope of 1.1–1.3 provides strong evidence that validates the helix–coil dichroic assumption that underlies eqs 4 and 5. Moreover, the helical ellipticity coefficients for these equations may be nearly temperature independent, and they can be assigned literature values calculated from the protein database.17,21 If, on the other hand, a helical alanine-rich peptide shows either linearity with an abnormally high slope or parametric curvature, then an accurate assignment of FH from CD data requires calibration. At least from zero to ambient temperatures, eq 6, derived from the 222 nm slope of Figure 4, should provide that calibration.
The data of Figure 6 also embody a proof-of-principle for construction of a needed new series of Alan helical hosts. Recently, we have measured helicity changes observed at 2, 25, and 60 °C for guest substitutions within a series 1a Ala19 host.5 Consistent with an early observation from the Scheraga group,24 our resulting sets of ΔΔG values are strongly temperature dependent. We believe this is an important finding, with the potential for resolving literature controversies that have resulted from comparisons of w or ΔΔG values, assigned at different temperatures.
For most hosts, intrinsic experimental factors allow helicity assignments only at fixed temperatures, and for the reported host–guest data sets, these vary widely. There is a need for host contexts that collectively allow accurate measurement of guest-induced helicity changes over the temperature range of 0–60 °C and that ensure a constant local structure at the site of guest substitution. Because many guest residues are helix breakers, to ensure accurate measurement of guest-induced helicity changes at high temperatures, the helicity of a candidate host must be relatively temperature insensitive. As seen in Figure 6a, by an ellipticity criterion, series 1 Alan peptides fail this test, because they lose ca. 70% of their helicity for a 2–60 °C, but Figure 6b suggests that over the same interval, a series 2 peptide retains more than 50% of its helicity. The following section provides independent support for this finding.
Discussion
The availability of the temperature- and length-dependent wAla(n,T) of Figure 2 allows L–R computational modeling of the melting properties of helically disposed short and medium Alan peptides that bear the helix-destabilizing caps of series 1a,b as well as the strongly helix stabilizing caps of series 2. Figure 7 plots L–R-modeled dependences of FH on w, T, and the capping parameters j = c, and the FH values for Alan peptides bearing helix weakly destabilizing or strongly stabilizing caps are seen to differ dramatically. For series 1a,b, at the broken lines in the Figure, at temperatures corresponding to w values close to 1.0 (higher temperatures), FH approaches zero for both Ala22 (broken magenta line) and Ala12 (broken orange line). Consistent with predictions from classical helix–coil theories,11 over the full w-range, these FH also depend strongly on length, and at high w, they do not converge to 1.0. The series 1 peptides display a sigmoid w dependence and a large slope change within the w-range of 1.1 to 1.5. The helical series 1 Ala28 peptide of Figure 3 is thus expected to melt progressively throughout the 2–60 °C temperature range, with a large overall change in FH that correlates with a gradual decrease in the mole fractions of longer helical conformers within its ensemble. The form of its curved parametric profile must then result from a combination of substantial melting throughout the temperature range with the significant temperature dependence of the limiting 222 nm helical ellipticity parameter [θ]222,H,22,T of Figure 4. Consistent with FH data of Table 2, the values for series 2 peptides in the high w region show insignificant length dependence and lie at or above 0.9. The classical dependence of FH on n and w is clearly overridden by the strong and selective cap stabilization of the longer series 2 helical peptide conformations that we have previously reported.1a,8
Figure 7.
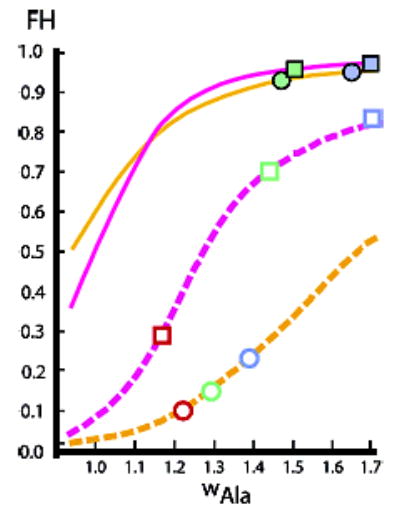
Parameter dependences of Lifson–Roig-modeled fractional helicities FH for Ala22 and Ala12 peptides. The temperature dependence of FH values is modeled by varying the length-independent values for w. Magenta lines are calculated for Ala22, orange lines for Ala12. Continuous lines correspond to FH for strong cap stabilization: j = 200, c = 7; broken lines correspond to weak cap destabilization: j = c = 0.5. Symbols drawn as squares (Ala22) and circles (Ala12) report FH calculated from the wAla(n,T) of Figure 2: red: T = 60 °C, green: T = 25 °C, blue: T = 2 °C. Filled symbols correspond to j = 200, c = 7; open symbols correspond to j = c = 0.5. Symbols have been positioned on appropriate line sites at which FH values are equal; thus, an average w (and temperature) is assigned to the wAla(n,T)-derived FH. These sites thus define approximate fixed-w assignments for the four peptides.
Experimental Section
Synthesis, Purification, and Characterization of Peptides
Peptides were synthesized on a 0.02–0.03 mole scale by automated continuous-flow solid-phase synthesis using a PE Biosystems Pioneer Peptide Synthesizer with standard 9-fluorenylmethyleneoxy-carbonyl (Fmoc/HATU) chemistry. All peptides were cleaved from the resin, purified, and characterized by MS, NMR, and AUC as previously reported.1,8
NMR and CD Measurements; Lifson–Roig Modeling
As reported previously,1,8 temperature dependences of 13C chemical shifts at 2.3, 4.3, 6.3, and 8.3 °C were measured at Ala site 5 for the 13C, 15N-labeled series 2 Ala12 peptide on a Bruker Avance 600 instrument equipped with four channels and a pulsed field gradient triple probe with z gradients. For assignment of chemical shifts for selectively 13C-labeled members of peptide series 1, samples were dissolved in water at concentrations in the range of 2–10 mM, and 13C spectra were taken on a 500 MHz Oxford Magnet with a Unity INOVA console using a Varian 500 SW/PFG broadband probe. Previously reported parameters and temperature assignment methods were used.1,8 CD data were collected on a calibrated Aviv 62DS spectrometer, and peptide concentrations were assigned from the Trp chromophore.1,6,8 The All L–R modeling was carried out using previously reported, tailored algorithms.1a,12
Assignment of Temperature-Dependent wAla(n,T)
The values for j and c, the N- and C-capping parameters for the tL residue, are set as 0.5 for all wAla(n,T) assignments.1 As described previously, wAla(n) values were tailored to increase monotonically with convergence to a limiting value for sufficiently large n, under the constraint of small local changes in slope. Initial values for wAla(n,T) were perturbed and applied to the variable-w Lifson–Roig algorithm to achieve best-fit approximations to the t/c and 13CExp chemical shift data.1 Values for wAla(n,25) were assigned from 25 °C t/c data7 for the length series Ac–Hel–Alan–tLInp2–Lys4Trp–NH2, n = 4–14, and from 25 °C values for central residue 13C NMR chemical shifts δ13CExp, measured for the series W–Lys5–tL3–Alan–tL3–Lys5NH2, n = 15, 19, 25. Temperature-dependent values for the limiting chemical shift parameter δ13CCoil of eq 6 were assigned experimentally as the central Ala5 residue 13C chemical shifts for W–Lysm–tL3–Ala5–tL3–LysmNH2. Values for δ13CHelix, Table 1, were calculated from the equation, δ13CHelix(ppm) = 180.825–0.0080 T. The parameters for this equation were extrapolated linearly from measurements at 2.3, 4.3, 6.3, and 8.3 °C as noted above. This extrapolation is least reliable for the assignment of δ13CHelix at 60 °C, and we addressed this problem by carrying out assignments of FHi and corresponding eq 1-derived δ13CCalc for a series of plausible values for δ13CHelix. The final value was chosen that minimizes the temperature dependence for wAla(n,60) for n = 15–25; coincidentally, this is also the value calculated in the above equation. Values previously assigned for wAla(n,2) are in numerical order from 1–25: 1, 1, 1, 1, 1.28, 1.31, 1.33, 1.34, 1.39, 1.41, 1.425, 1.455, 1.48, 1.505, 1.525, 1.54, 1.565, 1.58, 1.59, 1.60, 1.61, 1.62, 1.625, 1.63, 1.635;2,3 for wAla(n,10): 1, 1, 1, 1, 1.25, 1.28, 1.30, 1.32, 1.35, 1.38, 1.40, 1.42, 1.435, 1.47, 1.49, 1.50, 1.51, 1.53, 1.55, 1.575, 1.575, 1.605, 1.615, 1.62, 1.63; for wAla(n,25): 1, 1, 1, 1, 1.23, 1.25, 1.27, 1.28, 1.295, 1.31, 1.325, 1.35, 1.36, 1.365, 1.37, 1.375, 1.38, 1.385, 1.395, 1.41, 1.42, 1.43, 1.43, 1.435, 1.435; for wAla(n,60): 1, 1, 1, 1, 1.22, 1.22, 1.22, 1.21, 1.20, 1.20, 1.19, 1.19, 1.18, 1.17, 1.17, 1.16, 1.15, 1.15, 1.15, 1.14, 1.14, 1.14, 1.13, 1.13, 1.13.
Assignment of Series 2 Ala12 and Ala22 Protection Factors PFi and FH Values at Temperatures in the Range 10–30 °C
The NH→ND exchange kinetics and calculation of PFi closely followed published protocols used for the 2 °C study, in which PFi was measured for Ala NH sites of eleven series 2 Alan peptides, 8 ≤ n ≤ 24.8 Measurements were made in D2O at a pD ≥ 4.5, at which the βAsp function of the N-cap is fully ionized and maximizes helix initiation. As previously reported, exchange kinetics were followed by monitoring both peak height and curve-fitted resonance intensities. These methods gave excellent data agreement. The following sequence regularities facilitated site assignments in this study: for the 2 °C study, an average PFi of ca. 70 was observed for NH sites 1–4; at sites (n − 3) to n, the PF decreased monotonically from ca. 80 to 20; and at site (n + 1), the NH of β-alanine, exchange was rapid, and the FHNH(n+1) ≡ 1 − 1/PF(n+1) was estimated to lie in the range 0.1–0.3. The remaining (n − 8) PFi corresponds to the nearly coincident NH resonances of the Alan core, and averages lie within the range 100–125. Two methods were used to calculate FH for each peptide from these data. In the first, PFi in the core region, at site 3, and at the last five NH sites were converted successively to FHNHi, to FHi, and to FHCalc values using the previously reported inequalities.8 For the 2 °C length series, the ratio of FHNHi for the core region to the overall FH was found to be 0.965 ± 0.01, and in the second method we used this ratio to transform the following Ala22 PFi-derived core FHNHi from this study into FHSiteAve estimates: 10 °C, 0.990, 0.96; 15 °C, 0.985, 0.95; 25 °C, 0.97, 0.94; 30 °C, 0.93, 0.90. The errors in PFi for these data translate into an error of ca. ± 0.02 in FH. The first method was also used at these temperatures, relying on our previous 2 °C NH chemical shift assignments and extrapolating temperature-dependent chemical shifts where necessary. Values for FHCalc and FHSiteAve, reported in Table 2, are in reasonable agreement. The FH for the Ala12 peptide were analyzed similarly, as seen in Table 2. A larger temperature gradient was seen for core FHNHi and the terminal NH residues, which restricted the temperature range in which exchange could be monitored accurately.
Modeling of Figure 3 Limiting Slopes
L–R modeling for the series 1 Ala28 peptide, using the Figure 2 wAla(n,T), provided respective temperature-dependent FH at 2, 20, 25, and 60 °C: 0.86, 0.82, 0.80, 0.36. Equation 6 was used to calculate values at these temperatures for [θ]222,H,28,T: −46.5, −43.8, −40.0, −31. Replacement of the slope parameter +260 by +92 in eq 6 and setting [θ]208,H,∞,T = −46 000 deg cm2 dmol−1 allowed calculation of the corresponding series for [θ]208,H,28,T: −33.8, −32.9, −32.6, −39.4. Units: 103 deg cm2 dmol−1. In both cases, the temperature correction was applied to the full temperature series. From these values, the FH and [θ]U19 introduced into eqs 4 and 5 provided the following estimates for [θ]222,28,T,Exp at 2, 10, 25, and 60 °C: −39.7, −35.7, −32.0, −12.4; and for [θ]208,28,T,Exp: −29.9, −28.1, −26.5, −14.7. The 2–10 °C parametric slope of Figure 6a is then modeled as (−39.7 + 35.7)/(−29.9 + 28.1) = 2.2, and the 25–60 °C slope is (−32.0 + 12.4)/(−26.5 +14.7) = 1.6, in reasonable agreement with the experimental limiting slopes.
Supplementary Material
Acknowledgments
Financial support was provided primarily by NIH Grant GM13453.
Footnotes
Supporting Information Available: Two Figures with legends: temperature-dependent CD spectra for an unstructured Ala residue and Parametric 222 nm vs 208 nm ellipticity comparison, 2–60 °C, for the series 2 Ala12 peptide. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kennedy RJ, Walker SM, Kemp DS. J Am Chem Soc. 2005;127:16961–16968. doi: 10.1021/ja054645g. (a) [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kennedy. R. J.; Miller, J. S.; Kemp. D. S. Submitted for publication
- 2.The fractional helicity FH of a potentially helical peptide is the atom fraction of its backbone α-carbons that belong to helical conformations under particular experimental conditions. The site helicity FHi is the corresponding atom fraction of the α-carbons at a particular residue site i. The average of all FHi over a particular helical region equals the overall FH for that region.
- 3.Kallenbach NR, Spek EJ. Methods Enzymol. 1998;285:26–41. doi: 10.1016/s0076-6879(98)95033-9. For a review through 1997 see: [DOI] [PubMed] [Google Scholar]
- 4.For example: (a) Scheraga, H. A. The Intrinsic Tendency Toward α-Helix Formation. In Perspectives in Structural Biology; Vijayan, M., Yathindra, N., Kolaskar, A. S., Eds.; Indian Academy of Sciences: Bangalore, 1999; pp 275–282.; Creamer TP, Rose GD. Proteins: Struct, Funct, Genet. 1994;19:85–97. doi: 10.1002/prot.340190202. (b) [DOI] [PubMed] [Google Scholar]; Myers JK, Pace CN, Scholtz JM. Biochemistry. 1997;36:10923–10929. doi: 10.1021/bi9707180. (c) [DOI] [PubMed] [Google Scholar]
- 5.Moreau, R. J.; Nasr, K. A.; Török, M.; Miller, J. S.; Schubert, C.; Kennedy, R. J.; Kemp, D. S. In preparation
- 6.Miller JS, Kennedy RJ, Kemp DS. Biochemistry. 2001;40:305–309. doi: 10.1021/bi0019500. (a) [DOI] [PubMed] [Google Scholar]; Miller JS, Kennedy RJ, Kemp DS. J Am Chem Soc. 2002;124:945–962. doi: 10.1021/ja011726d. (b) [DOI] [PubMed] [Google Scholar]
- 7.Ingwall RT, Scheraga HA, Lotan N, Berger A, Katchalski E. Biopolymers. 1968;6:331–368. doi: 10.1002/bip.1968.360060308. (a) [DOI] [PubMed] [Google Scholar]; Gratzer W, Doty P. J Am Chem Soc. 1963;95:1193–1197. (b) [Google Scholar]; Rothwarf DM, Davenport VG, Shi PT, Peng JL, Scheraga HA. Biopolymers. 1996;39:531–536. (c) [Google Scholar]
- 8.Heitmann B, Job GE, Kennedy RJ, Walker SM, Kemp DS. J Am Chem Soc. 2005;127:1690–1704. doi: 10.1021/ja0457462. (a) [DOI] [PMC free article] [PubMed] [Google Scholar]; Job GE, Heitmann B, Kennedy RJ, Walker SM, Kemp DS. Angew Chem, Int Ed. 2004;43:5649–5651. doi: 10.1002/anie.200460536. (b) [DOI] [PubMed] [Google Scholar]
- 9.A length series of Alan peptides 2 was used for CD calibration, and peptide 2, n = 12, provided calibration at 2 °C for 13C═O NMR chemical shifts. As defined by protection factor measurements,8 the FH values for these peptides at 2 °C approach 1.0, owing to the presence of the charged helix-enhancing caps, βAsp-Hel (4a,b, X–CO– ≡ NH–CH(CO2–)CH2CO–) and beta ≡ β-amino-alanine.
- 10.Chen YH, Yang JT, Martinez HM. Biochemistry. 1972;11:4120–4131. doi: 10.1021/bi00772a015. (a) [DOI] [PubMed] [Google Scholar]; Woody RW, Tinoco I., Jr J Chem Phys. 1967;46:4927–4945. (b) [Google Scholar]; Johnson WC, Jr, Tinoco I., Jr J Am Chem Soc. 1972;94:4389–4390. doi: 10.1021/ja00767a084. (c) [DOI] [PubMed] [Google Scholar]
- 11.Lifson S, Roig A. J Chem Phys. 1961;34:1963–1874. (a) [Google Scholar]; (b) Poland, D.; Scheraga, H. A. Statistical Mechanical Theory of Order–Disorder Transitions in Biological Macromolecules; Academic Press: New York, 1970; Doig AJ, Baldwin RL. Protein Sci. 1995;4:1325–1336. doi: 10.1002/pro.5560040708. (c) [DOI] [PMC free article] [PubMed] [Google Scholar]; Qian H, Schellman JA. J Phys Chem. 1992;96:3987–2994. (d) [Google Scholar]
- 12.Kemp DS. Helv Chim Acta. 2002;85:4392–4423. We replace the conventional –R weight jυ2w(n−2)c by jυ2wnc, which has significant advantages: [Google Scholar]
- 13.Kennedy RJ, Tsang KY, Kemp DS. J Am Chem Soc. 2002;124:934–944. doi: 10.1021/ja016285c. [DOI] [PubMed] [Google Scholar]
- 14.Wieczorek R, Dannenberg JJ. J Am Chem Soc. 2003;125:8124–8129. doi: 10.1021/ja035302q. (a) [DOI] [PubMed] [Google Scholar]; Guo H, Karplus M. J Phys Chem. 1994;98:7104–7105. (b) [Google Scholar]; Mehler EL. J Am Chem Soc. 1980;102:4051–4056. (c) [Google Scholar]
- 15.We employ a fixed υ = 0.048 to assign stabilities of single or paired residues with helical φ, ψangles, but to model helical end effects, we have replaced υ by t. A fixed w = 1.108 and a temperature-dependent t of 0.07 (a 45% increase from t = υ = 0.048 at 2 °C) successfully models Table 1 60 °C chemical shifts.
- 16.Scholtz JM, Marqusee S, Baldwin RL, York EJ, Stewart JM, Santoro M, Bolen DW. Proc Natl Acad Sci USA. 1991;88:2854–2858. doi: 10.1073/pnas.88.7.2854. (a) [DOI] [PMC free article] [PubMed] [Google Scholar]; Ooi T, Oobatake M. Proc Natl Acad Sci USA. 1991;88:2859–2863. doi: 10.1073/pnas.88.7.2859. (b) [DOI] [PMC free article] [PubMed] [Google Scholar]; Wieczorek R, Dannenberg JJ. J Am Chem Soc. 2005;127:14534–14535. doi: 10.1021/ja053839t. (c) [DOI] [PubMed] [Google Scholar]; Richardson JM, Lopez MM, Makhatadze GL. Proc Natl Acad Sci USA. 2005;102:1413–1418. doi: 10.1073/pnas.0408004102. (d) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallimann P, Kennedy RJ, Miller JS, Shalongo W, Kemp DS. J Am Chem Soc. 2003;125:1203–1220. doi: 10.1021/ja0275360. [DOI] [PubMed] [Google Scholar]
- 18.Bai Y, Milne JS, Mayne L, Englander SW. Proteins: Struct, Funct, Genet. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.For CD spectra for [θ]λ,U,T, see the Supporting Information.
- 20.Bruch MD, Dhingra MM, Gierasch LM. Proteins: Struct, Funct, Genet. 1991;10:130–139. doi: 10.1002/prot.340100206. [DOI] [PubMed] [Google Scholar]
- 21.Besley NA, Hirst JD. J Am Chem Soc. 1999;121:9636–9644. [Google Scholar]
- 22.Farmer RS, Kiick KL. Biomacromolecules. 2005;6:1432–1539. doi: 10.1021/bm049216+. (a) [DOI] [PMC free article] [PubMed] [Google Scholar]; Marshall CB, Chakrabartty A, Davies PL. J Biol Chem. 2005;280:17920–17929. doi: 10.1074/jbc.M500622200. (b) [DOI] [PubMed] [Google Scholar]
- 23.Four series 2, Alan peptides with n > 13 (temperature range 2–60 °C) showed double-slope 222 vs 208 nm parametric plots similar to that of Figure 6b. Three cases with n < 13 showed single-slope plots with slope ranges 2.2–2.3. A brief discussion of these, with an example, appears in the Supporting Information.
- 24.Gō M, Hesselink FT, Gō N, Scheraga H. Macromolecules. 1974;7:459–467. doi: 10.1021/ma60040a013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


