Abstract
The rattlesnake fern (Botrychium virginianum (L.) Sw.) is obligately mycotrophic and widely distributed across the northern hemisphere. Three mitochondrial gene regions place this species with other ferns in Ophioglossaceae, while two regions place it as a member of the largely parasitic angiosperm order Santalales (sandalwoods and mistletoes). These discordant phylogenetic placements suggest that part of the genome in B. virginianum was acquired by horizontal gene transfer (HGT), perhaps from root-parasitic Loranthaceae. These transgenes are restricted to B. virginianum and occur across the range of the species. Molecular and life-history traits indicate that the transfer preceded the global expansion of B. virginianum, and that the latter may have happened very rapidly. This is the first report of HGT from an angiosperm to a fern, through either direct parasitism or the mediation of interconnecting fungal symbionts.
Keywords: biogeography, ferns, horizontal gene transfer, Ophioglossaceae, parasitic plants, phylogeography
1. Introduction
Horizontal gene transfer (HGT) appears to be common in plant mitochondrial (mt) genomes (Bergthorsson et al. 2003), but until very recently explanations of how such transfers occur have been speculative (Bergthorsson et al. 2003; Won & Renner 2003). Two studies show that parasitism of one plant by another is potentially an important mode of HGT in angiosperms. The first documented HGT from a host to its parasite (Davis & Wurdack 2004), and the second from a parasite to its host (Mower et al. 2004). This paper reports the first case of HGT from an angiosperm parasite to a putative fern host, and offers the first data on the geographic distribution of transgenes in a recipient species.
2. Material and methods
(a) Taxon sampling
Our analyses included four mt gene regions (atp1, atp6, matR and nad1B-C) spanning all major vascular plant clades (i.e. ferns, cycads, gymnosperms and angiosperms). The matR dataset included nearly all orders of flowering plants (APG 2003) and nad1B-C was similarly sampled for angiosperms and spanned most major eudicot clades. Each of these gene trees was rooted with Huperzia (atp1, matR and nad1B-C), Lycopodium (atp1), and Marchantia (atp6) following the large-scale phylogenetic studies by Karol et al. (2001) and Pryer et al. (2001). All primers and protocols used to generate these data, including those for RT-PCR and cDNA analyses described in §3 below, can be found in the supplementary online version accompanying this article.
Focused sampling for these four gene regions included 24 species (in 21 genera) representing all five families of Santalales, including representatives of the earliest diverging members of the order (Nickrent & Malécot 2001; APG 2003; Malécot et al. 2004). Our sampling of Loranthaceae spans the basal node of the family: the monospecific Australian root-parasitic Nuytsia is sister to the rest of the family and is included in our analyses (Vidal-Russell & Nickrent 2005). Along with the more derived aerial-parasitic loranths, root-parasitic Gaiadendron and Nuytsia are also included. We also sampled 12 fern species representing all three genera of Ophioglossaceae and species from all subgenera of Botrychium (Hauk et al. 2003), including the previously unplaced Afroasian subgenus Japanobotrychium (Hauk et al. 2003).
Population sampling of Botrychium virginianum (L.) Sw. for transgenic regions, matR and nad1B-C, included 34 accessions spanning its worldwide distribution, including North America (Canada, Mexico, United States), Central America (Costa Rica, Panama), South America (Bolivia, Brazil, Peru), the Greater Antilles (Dominican Republic), Europe (Austria, Germany, Switzerland) and Asia (China, Japan). For complete sampling information see table 1 in the Electronic Appendix.
We also included a fifth dataset, plastid rbcL, that included most major fern clades plus horsetails, and was rooted with Huperzia, Lycopodium, gymnosperms and cycads (Pryer et al. 2001). The purpose of including rbcL was to infer the biogeography of Botrychium, and to determine the approximate maximum age estimate for this HGT event (see §2d). The rbcL data are especially amenable to biogeographic analysis because: (i) the plastid genome does not appear to be susceptible to HGT across species boundaries and (ii) rbcL is sufficiently well sampled in ferns to provide a solid foundation for inferring the phylogeny and biogeography of Ophioglossaceae—the rbcL data set includes most major fern lineages, all genera of Ophioglossaceae, and nearly all species of Botrychium throughout their worldwide range (Hauk et al. 2003).
(b) Phylogenetic analysis
Maximum likelihood (ML) optimization was implemented for all independent (atp1, atp6, matR and nad1B-C) and combined (matR plus nad1B-C) datasets in PHYML ver. 2.4.4 (Guindon & Gascuel 2003) under the general time reversible model (GTR), or a submodel of the GTR model, as determined by ModelTest 3.6 (Posada & Crandall 1998). ML support values were estimated from 100 bootstrap replicates in PHYML, and parsimony support was estimated from 1000 bootstrap replicates in PAUP*4b10 (Swofford 2003). Parsimony bootstrap replicates were implemented with heuristic searches using 10 random taxon additions per replicate, tree-bisection–reconnection, MULPARS, and holding 10 trees at each taxon addition. To assess the alternate topological placement of B. virginianum in single data set analyses we employed the Kishino–Hasegawa (Kishino & Hasegawa 1989), Shimodaira–Hasegawa (Kadowaki et al. 1996) and parametric bootstrap tests (Huelsenbeck et al. 1996) using ML.
(c) Statistical phylogeography
We calculated Tajima's D and mismatch distribution with ARLEQUIN (Schneider et al. 2000) for all samples of B. virginianum. The demographic expansion model of Rogers and Harpending (Harpending 1994; Rogers 1995) was used to analyse the pairwise mismatch distributions. Twenty-five polymorphic sites were obtained for combined matR and nad1B-C. There were 19 and 6 polymorphic sites representing 11 and 7 haplotypes across individuals for transgenic matR and nad1B-C, respectively.
(d) Biogeographic Analyses
To infer the location of disjunctions of Ophioglossaceae clades, ancestral areas were reconstructed on the ML rbcL topology with dispersal–vicariance analysis (DIVA) as implemented in DIVA ver. 1.1 (Ronquist 1997). Our data matrix used to assess ancestral areas was constructed by scoring each species for its presence in seven major continental areas of endemism: Africa, Asia, Australia, Central America, Europe, North America and South America. Distributional data were obtained from Clausen (1938) and Wagner & Wagner (1993) (see supplementary online materials for trees and scorings). To corroborate results obtained using DIVA we similarly analysed these data using parsimony as implemented with the default ‘trace’ optimization of MacClade.
To estimate divergence times, branch lengths and an associated likelihood score were calculated for the respective models of sequence evolution in the absence of a molecular clock for plastid rbcL data following Davis et al. (2002). Finding that a clock was rejected (p < 0.05) we used penalized likelihood (Sanderson 2002) to estimate divergence times, focusing specifically on the split between B. virginianum and its closest Botrychium relatives. Since this transfer event is restricted to B. virginianum (see §3), the divergence time for this node provides a maximum age estimate for this event. The transfer can be younger than this node, but not older. To estimate standard errors associated with divergence times, we used the parametric bootstrapping strategy outlined in Davis et al. (2002).
We applied 12 minimum fossil age constraints recently reviewed by Schneider et al. (2004) to the rbcL data to estimate absolute divergence times (see Electronic Appendix). In addition, we also included the minimum age constraint of 57.8 Myr (time-scale sensu Berggren et al. 1995) for stem group Botrychium based on reliable Paleocene Botrychium fossils from western North America (Rothwell & Stockey 1989). We also separately enforced two maximum age constraints for the root of our tree, which corresponds to the euphyllophyte clade (Pryer et al. 2001)—380 and 450 Myr. The first age constraint corresponds to the earliest known euphyllophyte fossils (Schneider et al. 2004), and the second corresponds to the approximate maximum age for all land plants based on fossil and molecular divergence time estimates (Sanderson 2003). The cross validation procedure for these data yielded an optimal smoothing value of 1000.
3. Results and discussion
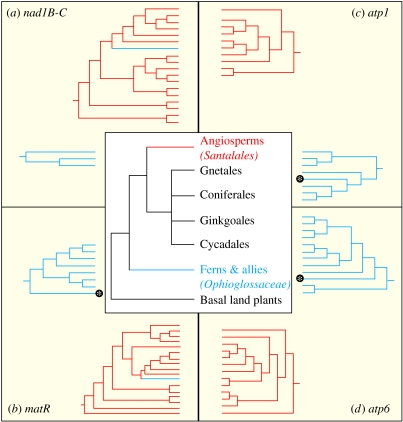
While reconstructing the phylogenies of all four mt gene regions we uncovered two strongly discordant placements for the rattlesnake fern, B. virginianum. The genes atp1, atp6, and one copy of matR placed B. virginianum with its closest relatives as a member of the fern family Ophioglossaceae (figure 1b–d). In contrast, nad1B-C and a second copy of matR placed it as a member of the parasitic angiosperm order Santalales (figure 1a,b), which includes the sandalwoods and mistletoes. We sought angiosperm matR and nad1B-C in other Ophioglossaceae (i.e., representatives of all three genera of Ophioglossaceae plus all subgenera of Botrychium sensu Hauk et al. 2003, including the closest relatives of B. virginianum) using a battery of angiosperm specific primers for these gene regions. In all sampled Ophioglossaceae, angiosperm-like copies were detected only in B. virginianum. In addition, universal matR primers designed to amplify both native and transgene copies of matR uncovered only the native copy in other Ophioglossaceae.
Figure 1.
Phylogenetic evidence for the discordant placements of Botrychium virginianum. Central tree depicts our current understanding of major land plant relationships. Each pair of trees in the corners is extracted from the larger ML analyses of (a) nad1B-C (from 100 total taxa), (b) matR (from 208 total taxa), (c) atp1 (from 145 total taxa), and (d) atp6 (from 74 total taxa). Santalales are depicted in red, Ophioglossaceae in blue. Transgenic copies of matR and nad1B-C are shown in Santalales in blue. Asterisks indicate the placement of B. virginianum in Ophioglossaceae based on vertically inherited copies of these genes; we did not recover a native copy of nad1B-C. See Electronic Appendix for detailed tree topologies and statistics.
This anomalous phylogenetic placement is robust, according to the Kishino–Hasegawa, Shimodaira–Hasegawa and parametric bootstrap tests: transgenic matR and nad1B-C favoured (p<0.05 for all tests) the placement of B. virginianum with Santalales rather than with Ophioglossaceae; similarly, native matR, atp1 and atp6 favoured (p<0.05 for all tests) the placement of B. virginianum with Ophioglossaceae rather than with Santalales. These results cannot be attributed to contamination, which can be ruled out for these reasons: (i) DNA extractions of B. virginianum were done independently in the laboratories of Davis and Wurdack, prior to any extraction of Santalales; (ii) all samples of B. virginianum possessed the same transgenes and (iii) these transgenes are different from any Santalales we sampled—if contamination had occurred we would expect sequences from B. virginianum to match those of Santalales extracted in our labs, but they do not. The most reasonable explanation for our results is that part of the genome in B. virginianum was acquired from Santalales via HGT. While other studies have reported HGT between angiosperms and gymnosperms (Won & Renner 2003) and between angiosperms and mosses (Bergthorsson et al. 2004), this is the first report of gene transfer between angiosperms and ferns.
The two transgenic regions found in B. virginianum, nad1B-C and matR, both reside within the nad1 gene (Dombrovska & Qiu 2004). The similar phylogenetic placement of B. virginianum using each gene region indicates that transgenes in this species were most likely transferred together in a single event. The native matR gene that places B. virginianum in Ophioglossaceae appears to be functional, while the copy of matR that nests within Santalales is not. RT-PCR products and cDNA sequences were only recovered for native matR, and were absent for the Santalalean copy. The pseudogenic nature of the latter is further confirmed by the presence of several internal termination codons and by the loss of reading frame in this sequence. The exonic region of nad1B-C was similarly undetected in RT-PCR/cDNA analysis suggesting that the coding portion of this transgene region is also non-functional.
Botrychium virginianum is a terrestrial fern common in temperate forests throughout the northern hemisphere and extends south through America in moist upland habitats to Bolivia and Brazil (Clausen 1938; Hauk et al. 2003). The nad1B-C and matR transgenic regions are restricted to B. virginianum within Ophioglossaceae and were present in all 34 individuals we sampled across the range of B. virginianum. Therefore, HGT probably occurred after B. virginianum diverged from its closest relatives and before it expanded into its present global distribution. Ancestral area reconstructions (using Diva and MacClade) and molecular divergence time estimates based on plastid rbcL data indicate that this divergence most likely occurred in Asia, and that the transfer event is no older than the Eocene (34.9±3.5 Myr, or 44.2±6 Myr; older maximum age constraints (e.g. 500 Myr) for the root node in our analysis push this age estimate slightly older, but the age of interest is still within the Eocene). Alternatively, the transfer event may have occurred in a single population of an already widespread B. virginianum, followed by spread of the transgenic regions throughout the species via selective gene sweep. Since neither of these transgene regions appears to be functional in B. virginianum, they are unlikely to confer a selective advantage, which favours the hypothesis that the transfer preceded the expansion of B. virginianum.
Two lines of evidence suggest that B. virginianum may have achieved its global distribution very rapidly, perhaps in thousands of years rather than millions. A rapid expansion would fit with the life history of this species. Like many ferns, including Ophioglossaceae, B. virginianum is almost certainly easily dispersed over long distances by its small spores (Peck et al. 1990; Barrington 1993). Additionally, the ability of single spores to establish new colonies through its inbreeding bisexual gametophytes (Soltis & Soltis 1986) should also speed the spread of this species. Rapid expansion is similarly supported at the molecular level by pairwise mismatch distributions (Rogers & Harpending 1992) and Tajima's D (Tajima 1989) for all sampled individuals of these transgenes (analysed independently and in combination). When we pooled all samples and used mismatch distributions to infer population expansion within B. virginianum none of the datasets were able to reject a unimodal distribution (i.e. expanding population model), providing evidence for a rapid population expansion in B. virginianum (matR (p=0.10), nad1B-C (p=0.59) and combined matR plus nad1B-C (p=0.94)). Similarly, significantly negative values of Tajima's D provide evidence of rapidly expanding populations, and were suggested for matR (D=−1.62, p=0.03), and combined matR plus nad1B-C (D=−1.82, p=0.02), and were marginally insignificant for nad1B-C (D=−1.35, p=0.07).
Phylogenetic analyses of the nad1B-C and matR transgenes (analysed independently and in combination) place B. virginianum as sister to the hemiparasitic family Loranthaceae within the order Santalales (figure 2). While most Loranthaceae are aerial stem parasites, three monospecific genera (Atkinsonia, Gaiadendron and Nuytsia) are root parasites (Kuijt 1969), and both morphological (Feuer & Kuijt 1980) and molecular (Nickrent 2001; Nickrent 2002) evidence indicates that root parasitism is ancestral in the family. B. virginianum is a terrestrial fern that spends part of its life cycle as a subterranean gametophyte and juvenile sporophyte (Johnson-Groh et al. 2002), and the rhizome is hardly, if at all, emergent at maturity (Gifford & Foster 1989). While it follows that a root parasite is the most likely donor of the transgenes found in B. virginianum, none of the three root-parasitic loranth species are presently found in Asia. Given the evidence cited above that B. virginianum originated in Asia, and the fact that our analyses (figure 2) show that the transgenes are not sister to any extant genus of Loranthaceae, it seems most likely that the transgenic donor was a root-parasitic Asian loranth that is now extinct. Nevertheless, the Neotropical Gaiadendron punctatum (Ruiz & Pavón) G. Don may serve as a model for how this gene transfer could have occurred. Gaiadendron represents one of the earliest-diverging extant members of Loranthaceae (Kuijt 1963; Feuer & Kuijt 1980; Nickrent 2002), and is the only root-parasitic loranth sympatric with B. virginianum (TROPICOS 2005). While we do not know any report of a parasitic relationship between Loranthaceae and B. virginianum, root parasitism is very cryptic and would probably go unnoticed if such a relationship existed, and Gaiadendron is known to parasitize ferns in the mountains of Costa Rica, where B. virginianum occurs (Kuijt 1963; TROPICOS 2005).
Figure 2.
Phylogeny of Santalales based on combined nad1B-C and matR. Likelihood and parsimony bootstrap values, respectively, are given for those clades supported at greater than 50%. Asterisks indicate less than 50% bootstrap support. See Electronic Appendix for detailed tree topologies and statistics.
Another possibility is that there was never a direct parasitic connection between B. virginianum and terrestrial Loranthaceae. Instead, HGT may have been indirect via a shared fungal symbiont. The subterranean gametophyte of B. virginianum lacks chlorophyll and must be infected by an endophytic fungus in order to grow (Gifford & Foster 1989). The intracellular fungus forms a mycorrhiza-like association with the fern gametophyte and transfers to it carbohydrates from nearby mycorrhizal photosynthetic plants, a relationship dubbed epiparasitism or mycoheterotrophy (Schmid & Oberwinkler 1994; Smith & Read 1997). At maturity, the roots of the sporophyte of B. virginianum lack root hairs and depend on their fungal symbiont for water and minerals (Kovács et al. 2003). Mycorrhizae have been reported in Santalales (Landis et al. 2002), but have not been demonstrated in root-parasitic Loranthaceae, although all of the latter lack root hairs like many mycorrhizal plants (Kuijt 1963, 1969). If there was a fungal bridge between a terrestrial loranth and B. virginianum, the HGT postulated here may have been mediated by the fungus.
If fungi are functioning as a conduit for gene transfer, that may help to explain some of the many reported HGTs for which no mechanism has been obvious (Bergthorsson et al. 2003; Won & Renner 2003). Mycorrhizal fungi are notoriously non-specific in their host selection and connect many distantly related plants in the same community (Smith & Read 1997). This ‘wood-wide web’ (Simard et al. 1997) may be facilitating rapid and widespread exchange of DNA across phylogenetic distances spanning all green plants. If so, such a finding would affect our thinking about the long-term evolution of the terrestrial biota, and may also have commercial implications. Fungus-mediated HGT could make it very difficult to restrict transgenes to their genetically modified organisms—if this is happening, the transgenie would be out of the bottle.
Acknowledgments
We thank B. Carstens, J. Hall, W. Hauk, J. Johnson, M. Latvis, K. Lewis, L. Knowles, D. Mindell, R. Moran, J. Palmer and P. Tucker. C.D was supported by NSF AtoL EF 04-31242 and by the Michigan Society of Fellows. The curators of the following herbaria kindly permitted us to remove leaf material from specimens: A, GH, M, MICH, NY, US, Z. This paper is dedicated to W. H. Wagner and S. Wonder.
Supplementary Material
References
- APG An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG II. Bot. J. Linn. Soc. 2003;141:399–436. doi:10.1046/j.1095-8339.2003.t01-1-00158.x [Google Scholar]
- Barrington D.S. Ecological and historical factors in fern biogeography. J. Biogeogr. 1993;20:275–280. [Google Scholar]
- Berggren W.A, Kent D.V, Swisher C.C, II, Aubry M.P. A revised Cenozoic geochronology and chronostratigraphy. In: Berggren W.A, Kent D.V, Aubry M.P, Hardenbol J, editors. Geochronology, time scales and global stratigraphic correlation. SEPM (Society for Sedimentary Geology); Tulsa: 1995. pp. 129–212. [Google Scholar]
- Bergthorsson U, Adams K.L, Thomason B, Palmer J.D. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature. 2003;424:197–201. doi: 10.1038/nature01743. doi:10.1038/nature01743 [DOI] [PubMed] [Google Scholar]
- Bergthorsson U, Richardson A.O, Young G.J, Goertzen L.R, Palmer J.D. Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc. Natl Acad. Sci. USA. 2004;101:17 747–17 752. doi: 10.1073/pnas.0408336102. doi:10.1073/pnas.0408336102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen R.T. A monograph of the Ophioglossaceae. Mem. Torrey Bot. Club. 1938;19:1–177. [Google Scholar]
- Davis C.C, Wurdack K.J. Host-to-parasite gene transfer in flowering plants: phylogenetic evidence from Malpighiales. Science. 2004;305:676–678. doi: 10.1126/science.1100671. doi:10.1126/science.1100671 [DOI] [PubMed] [Google Scholar]
- Davis C.C, Bell C.D, Mathews S, Donoghue M.J. Laurasian migration explains Gondwanan disjunctions: evidence from Malpighiaceae. Proc. Natl Acad. Sci. USA. 2002;99:6833–6837. doi: 10.1073/pnas.102175899. doi:10.1073/pnas.102175899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrovska O, Qiu Y.L. Distribution of introns in the mitochondrial gene nad1 in land plants: phylogenetic and molecular evolutionary implications. Mol. Phylogenet. Evol. 2004;32:246–263. doi: 10.1016/j.ympev.2003.12.013. doi:10.1016/j.ympev.2003.12.013 [DOI] [PubMed] [Google Scholar]
- Feuer S.M, Kuijt J. Fine structure of mistletoe pollen. III. Large-flowered neotropical Loranthaceae and their Australian relatives. Am. J. Bot. 1980;67:34–50. [Google Scholar]
- Gifford E.M, Foster A.S. W. H. Freeman and Co; New York: 1989. Morphology and evolution of vascular plants. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. doi:10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Harpending H.C. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum. Biol. 1994;66:591–600. [PubMed] [Google Scholar]
- Hauk W.D, Parks C.R, Chase M.W. Phylogenetic studies of Ophioglossaceae: evidence from rbcL and trnL-F plastid DNA sequences and morphology. Mol. Phylogenet. Evol. 2003;28:131–151. doi: 10.1016/s1055-7903(03)00032-0. doi:10.1016/S1055-7903(03)00032-0 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Hillis D.M, Jones R. Parametric bootstrapping in molecular phylogenetics: applications and performance. In: Ferris J.D, Palumbi S.R, editors. Molecular zoology: advances, strategies, and protocols. Wiley-Liss; New York: 1996. pp. 19–45. [Google Scholar]
- Johnson-Groh C, Riedel C, Schoessler L, Skogen K. Belowground distribution and abundance of Botrychium gametophytes and juvenile sporophytes. Am. Fern J. 2002;92:80–92. [Google Scholar]
- Kadowaki K.-I, Kubo N, Ozawa K, Hirai A. Targeting presequence acquisition after mitochondrial gene transfer to the nucleus occurs by duplication of existing targeting signals. EMBO J. 1996;15:6652–6661. [PMC free article] [PubMed] [Google Scholar]
- Karol K.G, McCourt R.M, Cimino M.T, Delwiche C.F. The closest living relatives of land plants. Science. 2001;294:2351–2353. doi: 10.1126/science.1065156. doi:10.1126/science.1065156 [DOI] [PubMed] [Google Scholar]
- Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimates of the evolutionary tree topologies from sequence data, and the branching order in Hominoidea. J. Mol. Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- Kovács G.M, Kottke I, Oberwinkler F. Light and electron microscopic study on the mycorrhizae of sporophytes of Botrychium virginianum—arbuscular structures resembling fossil forms. Plant Biol. 2003;5:574–580. doi:10.1055/s-2003-44786 [Google Scholar]
- Kuijt J. On the ecology and parasitism of the Costa Rican tree mistletoe, Gaiadendron punctatum (Ruiz and Pavón) G. Don. Canad. J. Bot. 1963;41:927–938. [Google Scholar]
- Kuijt J. University of California Press; Berkeley: 1969. The biology of parasitic flowering plants. [Google Scholar]
- Landis F, Gargas A, Givnish T. Annual Meeting of the Botanical Society of America 2002, Madison, Wisconsin. 2002. The plant tree, roots and clades: Mycorrhizae and plant phylogeny.See http://www.botany2002.org/section13/abstracts/4.shtml. p. 174. [Google Scholar]
- Malécot V, Nickrent D.L, Baas P, Oever L.v.d, Lobreau-Callen D. A morphological cladistic analysis of Olacaceae. Syst. Bot. 2004;29:569–586. doi:10.1600/0363644041744301 [Google Scholar]
- Mower J.P, Stefanovic S, Young G.J, Palmer J.D. Gene transfer from parasitic to host plants. Nature. 2004;432:165–166. doi: 10.1038/432165b. doi:10.1038/432165b [DOI] [PubMed] [Google Scholar]
- Nickrent D.L. Encyclopedia of life sciences. Macmillan Publishers Ltd; New York: 2001. Santalales (Mistletoe); treatment A3714. [Google Scholar]
- Nickrent D.L. Mistletoe phylogenetics: current relationships gained from analysis of DNA sequences. In: Angwin P, editor. Proc. 48th Annual Western Int. Forest Disease Work Conf. USDA Forest Service; Redding: 2002. pp. 48–57. [Google Scholar]
- Nickrent D.L, Malécot V. A molecular phylogeny of Santalales. In: Fer A, Thalouarn P, Joel D.M, Musselman L.J, Parker C, Verkleij J.A.C, editors. Proc. 7th Int. Parasitic Weed Symp. Faculté des Sciences, Université de Nantes; Nantes, France: 2001. pp. 69–74. [Google Scholar]
- Peck J.H, Peck C.J, Farrar D.R. Influence of life history attributes on formation of local and distant fern populations. Am. Fern J. 1990;80:126–142. [Google Scholar]
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Pryer K.M, Schneider H, Smith A.R, Cranfill R, Wolf P.G, Hunt J.S, Sipes S.D. Horsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature. 2001;409:618–622. doi: 10.1038/35054555. doi:10.1038/35054555 [DOI] [PubMed] [Google Scholar]
- Rogers A. Genetic evidence for a Pleistocene population explosion. Evolution. 1995;49:608–615. doi: 10.1111/j.1558-5646.1995.tb02297.x. [DOI] [PubMed] [Google Scholar]
- Rogers A.R, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Ronquist F. Dispersal–vicariance analysis: a new approach to the quantification of historical biogeography. Syst. Biol. 1997;46:195–203. [Google Scholar]
- Rothwell G.W, Stockey R.A. Fossil Ophioglossales in the Paleocene of Western North America. Am. J. Bot. 1989;76:637–644. [Google Scholar]
- Sanderson M.J. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Sanderson M.J. Molecular data from 27 proteins do not support a Precambrian origin of land plants. Am. J. Bot. 2003;90:954–956. doi: 10.3732/ajb.90.6.954. [DOI] [PubMed] [Google Scholar]
- Schmid E, Oberwinkler F. Light and electron microscopy of the host-fungus interaction in the achlorophyllous gametophyte of Botrychium lunaria. Canad. J. Bot. 1994;72:182–188. [Google Scholar]
- Schneider, S., Roessli, D. & Excoffier, L. 2000 Arlequin v2.0: Documentation and program. http://anthro.unige.ch/arlequin
- Schneider H, Schuettpelz E, Pryer K.M, Cranfill R, Magallón S, Lupia R. Ferns diversified in the shadow of angiosperms. Nature. 2004;428:553–557. doi: 10.1038/nature02361. doi:10.1038/nature02361 [DOI] [PubMed] [Google Scholar]
- Simard S.W, Perry D.A, Jones M.D, Myrold D.D, Durall D.M, Molina R. Net transfer of carbon between ectomycorrhizal tree species in the field. Nature. 1997;388:579–582. doi:10.1038/41557 [Google Scholar]
- Smith S.E, Read D.J. Academic Press; London: 1997. Mycorrhizal symbiosis. [Google Scholar]
- Soltis D.E, Soltis P.S. Electrophoretic evidence for inbreeding in the fern Botrychium virginianum (Ophioglossaceae) Am. J. Bot. 1986;73:588–592. [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, MA: 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4.0.610. [Google Scholar]
- Tajima F. The effect of change in population size on DNA polymorphism. Genetics. 1989;123:597–601. doi: 10.1093/genetics/123.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TROPICOS. 2005. Database of the Missouri Botanical Garden; http://mobot.mobot.org/W3T/Search/vast.html
- Vidal-Russell R, Nickrent D. Annual Meeting of the Botanical Society of America 2005 Austin, Texas. 2005. A molecular phylogeny of the mistletoe family Loranthaceae.See http://www.2005.botanyconference.org p. 101. [Google Scholar]
- Wagner W.H, Wagner F.S. Ophioglossaceae Agardh. In: F.O.N.A.E. Committee, editor. Pteridophytes and gymnosperms. vol. 2. Oxford University Press; New York: 1993. pp. 102–105. [Google Scholar]
- Won H, Renner S.S. Horizontal gene transfer from flowering plants to Gnetum. Proc. Natl Acad. Sci. USA. 2003;100:10 824–10 829. doi: 10.1073/pnas.1833775100. doi:10.1073/pnas.1833775100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.