Abstract
Wolbachia are maternally inherited endosymbiotic bacteria that infect many arthropod species and may induce cytoplasmic incompatibility (CI) resulting in abortive embryonic development. Among all the described host species, mosquitoes of the Culex pipiens complex display the highest variability of CI crossing types. Paradoxically, searches for polymorphism in Wolbachia infecting strains and field populations hitherto failed or produced very few markers. Here, we show that an abundant source of the long-sought polymorphism lies in WO prophage sequences present in multiple copies dispersed in the genome of Wolbachia infecting C. pipiens (wPip). We identified up to 66 different Wolbachia variants in C. pipiens strains and field populations and no occurrence of superinfection was observed. At least 49 different Wolbachia occurred in Southern Europe C. pipiens populations, and up to 10 different Wolbachia were even detected in a single population. This is in sharp contrast with North African and Cretan samples, which exhibited only six variants. The WO polymorphism appeared stable over time, and was exclusively transferred maternally. Interestingly, we found that the CI pattern previously described correlates with the variability of Gp15, a prophage protein similar to a bacterial virulence protein. WO prophage sequences thus represent variable markers that now open routes for approaching the molecular basis of CI, the host effects, the structure and dynamics of Wolbachia populations.
Keywords: Wolbachia, Culex pipiens, cytoplasmic incompatibility, prophage, WO
1. Introduction
Wolbachia are maternally inherited endocellular bacteria widespread among arthropods and filarial parasitic nematodes. In several species like the mosquito Culex pipiens, Wolbachia induce cytoplasmic incompatibility (CI) leading to embryonic mortality that occurs when infected males mate either with uninfected females or with females infected by incompatible Wolbachia strain(s) (Yen & Barr 1973). Thus, in a mixed population, Wolbachia that induce CI get a selective advantage and are predicted to spread up to fixation (Rousset & Raymond 1991; Turelli & Hoffmann 1999). In support of this hypothesis, the prevalence of Wolbachia infecting C. pipiens (called wPip) was investigated worldwide, and appeared fixed in 67 populations studied (Duron et al. 2005). Crosses between mosquitoes from various origins revealed a high level of incompatibilities (Laven 1951, 1967; Barr 1966; Subbarao 1982; Magnin et al. 1987; Guillemaud et al. 1997) contrasting with other insect situations (Werren 1998). However, the higher CI level was observed between mosquitoes from Europe, where it exhibits an extreme pattern (Laven 1967; Magnin et al. 1987; Guillemaud et al. 1997).
A straightforward hypothesis to explain the complex CI pattern in C. pipiens is the presence of different Wolbachia strains. However, no polymorphism was observed in strains displaying incompatibilities using the ftsZ gene (Guillemaud et al. 1997), driving the authors to conclude that factors other than Wolbachia variants were responsible for incompatibility in this species. Similarly, no polymorphism could be detected either in the 16S rRNA sequences of Wolbachia infecting different C. pipiens subspecies (Rousset et al. 1992) or in the fastest Wolbachia evolving gene wsp of mosquitoes sampled worldwide (Duron et al. 2005). Recent searches for polymorphism in wPip have produced a few markers, all affecting mobile genetic elements. Sanogo & Dobson (2004) delineated three wPip variants among 11 laboratory strains by analysis of the variability of orf7 copy number in the WO prophage. Sinkins et al. (2005) described two wPip variants based on polymorphism of ankyrin-repeat encoding genes (pk genes) associated with a prophage region. We identified previously five distinct wPip strains from 531 mosquitoes by analysing the polymorphism of Tr1, an apparently functional transposable element of the IS5 family (Duron et al. 2005). wPip Tr1 genetic diversity appeared geographically structured and independent of the C. pipiens subspecies status, and affected mostly European populations (four variants detected). Nevertheless, neither orf7 nor pk or Tr1 markers were sensitive enough to explain the 17 cytotypes present in Laven's (1967) crosses.
The purpose of this study was to identify polymorphic markers that could describe the complex CI pattern found in C. pipiens. To this end, we examined the variability of prophage sequences, described as major contributors of genomic flux in bacteria. WO prophage sequences have been shown to be widespread in the genomes of 35 Wolbachia infecting diverse arthropods and to transfer at high rates between divergent lineages (Bordenstein & Wernegreen 2004; Gavotte et al. 2004). This led to the suggestion that bacteriophages could drive significant gene transfer and evolutionary changes in the genomes of Wolbachia and that phage proteins might be linked to CI directly. Here, we report an unprecedented level of Wolbachia polymorphism from the analysis of 15 WO prophage sequences in 12 laboratory strains and 19 natural populations.
2. Material and methods
(a) Mosquito collections
Mosquitoes were collected in breeding sites and raised to the adult stage. They were either stored in liquid nitrogen for further analyses (field samples), or bred in the laboratory (strains). For each sample, references, subspecies (only for strains), years and countries of origin are indicated in tables S1 and S2 in the electronic supplementary material.
(b) Prophage transmission
Cytoplasmic transmission of WO prophage markers was investigated by using reciprocal crosses between two fully compatible mosquito strains (Keo-A and LaVar), infected by distinct Wolbachia (50 males crossed with 50 females). F1 larvae (randomly sampled, N=10 for each cross) were screened by PCR for the presence of maternal- or paternal-specific WO prophage markers (Gp1b, Gp3a and Gp7d).
A second inheritance analysis was performed on F6 larvae from backcrosses between Istanbul females and Keo-B males. In the first generation, 100–200 females were crossed with equal number of males. In the next generation, F1 females were crossed with Keo-B males. The same procedure was repeated in each generation. F6 adults (N=15) were screened by PCR for the presence of maternal- or paternal-specific WO prophage markers (Gp1b, Gp3b, Gp3d, Gp7d and Gp15b).
Horizontal phage transfer was tested between Keo-A and LaVar strains by feeding Keo-A larvae with crushed LaVar mosquitoes (larvae and adults) as a unique source of food from the first to the adult stages. Fifteen adults were screened by PCR for the presence of maternal- or paternal-specific WO prophage markers (Gp1b, Gp3a and Gp7d).
(c) Database analysis
The wPip genome was searched for CauB WO prophage homologues using the Sanger's Institute Web facilities (http://www.sanger.ac.uk/Projects/W_pipientis/). The wSim genome (partial assembly) was searched by Blast analysis on a local computer. Homologues to Gp 1 to Gp 13 (2 copies): AAGC01000-020;-246;-599;-315;025;-248;-172;-391;-122;-350;-372;-035;-368;-596;-469;-142. Homologues to Gp14 to 24 (single copy): AAGC01000-104;-360;-424;-355;-322;-531;-494;-139;-449;-541;-363;-529.
(d) PCR and sequencing
Mosquito DNA was extracted using a CTAB protocol (Rogers & Bendich 1988). Assays for WO prophage sequences were performed by PCR amplification using specific primers listed in table S3 of the electronic supplementary material. The main issue was to amplify unique open reading frames (ORFs) from multiple copies. For example, designing new primers was necessary to amplify specifically the recently described Gp3c gene product (termed as orf7c in Sanogo & Dobson 2004), since the published primers turned out not to be specific and amplified Gp3c, Gp3d and Gp3e copies concomitantly. The PCR was run for 30 cycles (94 °C for 30 s, 52 °C for 30 s and 72 °C for 1 min). At least four mosquitoes were assayed for each laboratory strain. To study sequence polymorphism, sequencing was performed directly on PCR products on an ABI prism 310 sequencer using the Big Dye Terminator kit. Two Control DNAs corresponding to positive and negative strains were included in each group of PCR.
Loss of Wolbachia in tetracycline treated strains was assessed by PCR amplification of a fragment of the wsp gene using the specific primers wolpipdir and wolpiprev described by Berticat et al. (2002). In all mosquitoes negative for Wolbachia infection after treatment the quality of their DNA was checked using the acetylcholinesterase ace-2 gene amplification (Weill et al. 2000).
3. Results
(a) Multiple WO prophage open reading frames copies in the wPip genome
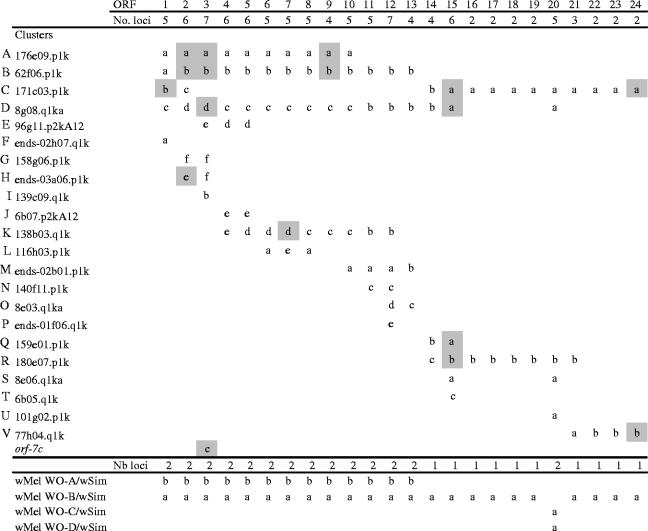
A recent paper reported the nearly complete DNA sequence of a WO phage isolated from Wolbachia infecting the almond moth Cadra cautella (wCauB; Fujii et al. 2004). Twenty-four ORFs were described, including a structural gene module and genes for replication and lysogenic conversion. We took opportunity of the recently available genome of wPip to search for sequences homologous to the 24 gene products (Gp). We identified 22 clusters (labelled A–V) encoding variable numbers of proteins, from one unique partial sequence up to 15 clustered complete ones (table 1). It is noteworthy that the wPip genome assembly is still underway and determination of the final picture of the WO prophage family must await completion of the assembly. For clarity, for each cluster, Gp are numbered as those of the wcauB1 WO phage and for each Gp, variants detected by DNA sequencing are identified by a lower case letter. The number of ORF copies varied from 2 to 6 in the wPip genome, whereas only two WO prophage clusters were found in the wMel genome, one complete and the other partial (Gp1 to Gp13; Wu et al. 2004). Blast analysis of the partially assembled genome of Wolbachia infecting Drosophila simulans (wSim, Salzberg et al. 2005) detected two copies for Gp1 to Gp13 and a single one for Gp14 to 24, suggesting a WO prophage organization identical to wMel (see table 1).
Table 1.
Schematic representation of WO phage ORFs in the wPip, wMel and wSim genomes. (For each cluster, the accession number (http://www.sanger.ac.uk/Projects/W_pipientis/), the identifier used in this study (A–V) and the occurrence of WOcauB1 ORF homologues is indicated. ORF numbering refers to Fujii et al. (2004). Variant ORFs copies are identified by a small case letter. Shading indicates clusters and ORFs analysed in the study. Gp3c corresponds to an additional variant referred to as orf7 (Sanogo & Dobson 2004), absent from the wPip genome (Pel strain; Amin & Pereis 1990). At the bottom are shown wCauB WO homologues encoded in the wMel genome (cluster (A, B) and single loci (C, D), Wu et al. 2004) and the in the wSim genome (Salzberg et al. 2005), without taking into account ORF organization. wPip genome description corresponds to the current situation and will certainly change in term of clusters and copies number when the assembling is achieved.)
(b) Polymorphism of WO prophage ORF copies
(i) Presence/absence of polymorphism
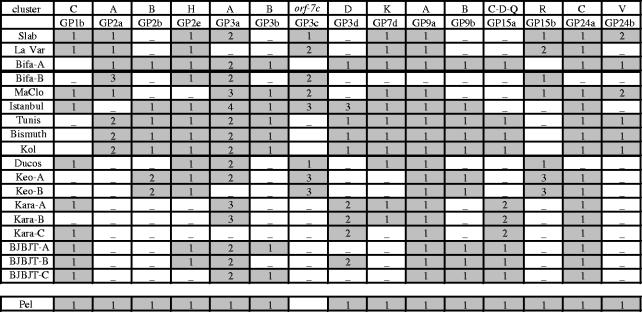
We addressed WO prophage variability by analysis of the presence or absence of specific Gp copies (markers) on individual mosquitoes from 12 C. pipiens strains derived from populations sampled worldwide (table 2 and S1). All markers were absent from tetracycline-treated Wolbachia-free mosquitoes strains, which confirmed the bacterial origin of the WO phage clusters (not shown). Unexpectedly, all markers were polymorphic in the 12 laboratory strains (table 2). Apart from Tunis, Bismuth, Bifa-A and Kol which displayed the same pattern, all strains were different, from two copies (e.g. Slab and LaVar) up to 11 copies (e.g. Tunis and Ducos; table 2). This analysis also showed that 4 of the 12 mosquito strains (Bifa, Keo, Kara and BJBJT) displayed a mixture of Wolbachia variants that can differ by up to 12 markers (Bifa-A and Bifa-B). Interestingly, Gp15a and Gp15b fragments were found to be mutually exclusive in all examined strains, whereas they both are present in the wPip genomic sequences (table 1).
Table 2.
Patterns of presence or absence of WO phage PCR markers in C. pipiens strains. (For each strain, PCR was performed using the amplimer sets listed in table S3. Shaded boxes indicate positive PCR reactions. Allelic sequences of PCR products are identified by numbers. The Pel strain pattern (deduced from the wPip genome analysis) was chosen as a reference. When several Wolbachia variants were identified in the same mosquito strain, distinction was made using a suffix-letter added to the mosquito strain name. References, subspecies, years and countries of collection are reported in table S1.)
(ii) Single nucleotide polymorphism
To get a more accurate view of WO phage polymorphism, we determined and compared the sequences of PCR products for the 15 markers. A high level of single nucleotide polymorphism was found in eight markers (table 2). This suggests that the absence of some PCR products is most likely a consequence of mutations in the priming regions. This was demonstrated for the Istanbul Gp15, which was amplified with Gp14 and Gp16 primers and whose DNA sequence showed divergent Gp15 priming sites (not shown). Ninety per cent of the mutations found in ORFs were conservative, indicating that the encoded proteins are subject to selective constraints and are probably functional (data not shown). Pairwise analysis of cluster A and B markers revealed no significant linkage desequilibrium (Genepop software; Raymond & Rousset 1995), suggesting a high rate of mutations and/or recombination events. For each individual, products amplified and sequenced were all monomorphic, showing the absence of multiple infections in our samples.
(c) WO prophage transmission
Phage inheritance was investigated using reciprocal mass crosses between two fully compatible mosquito strains (Keo-A and LaVar), infected by distinct Wolbachia (table 2). All F1 larvae (randomly sampled, N=10 for each cross) displayed a maternally inherited pattern. A second inheritance analysis was performed on F6 larvae from backcrosses between Istanbul females and Keo-B males. All 13 F6 adults tested displayed the Istanbul pattern, confirming maternal phage transmission.
We next investigated horizontal phage transfer through feeding Keo-A larvae with crushed LaVar mosquitoes (larvae and adults) as unique source of food from the first to the adult stages. All 15 adults tested showed a Keo-A pattern. Paternal or horizontal transfers, thus seem to occur at extremely low levels if any.
(d) WO prophage polymorphism in field populations
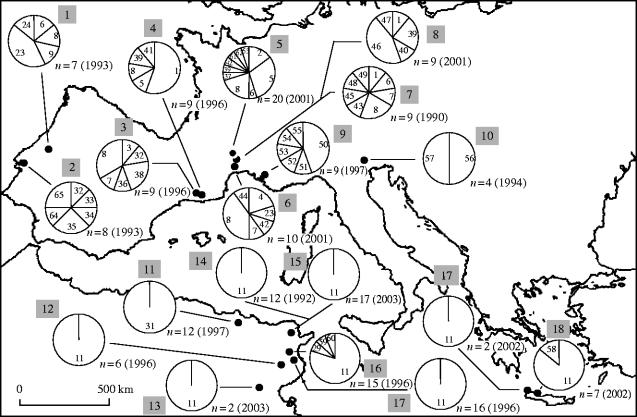
Inter-individual variability within strains suggested that WO polymorphism exists in natural populations. To address this issue, we examined field populations from southern Europe and North Africa. For simplification, we restricted the analysis to 10 phage markers that discriminate all our strains (see legend of table S4 for details). Analysis of 183 mosquitoes from 19 locations (figure 1, table S2) revealed extreme Wolbachia variability, identifying up to 36 additional variants (table S4). The same analysis using the Tr1 transposase (Duron et al. 2005) generated 7 additional variants.
Figure 1.
Distribution of Wolbachia variants in C. pipiens field populations. For each population, identified variants are indicated within circles by numbers referring to table S4. Below circles are indicated the sizes (n) and the years of sampling. Populations are numbered (shaded box) as reported in table S2.
Altogether with those already found in the laboratory strains, the combination of WO prophages and Tr1 PCR assays thus discriminated up to 61 variants in 207 insects from strains and field populations. This represents an underestimation considering the high single nucleotide polymorphism found in strains.
In southern Europe, WO prophage variability was detected in all populations despite low sample sizes. Forty-nine different variants were observed in 103 mosquitoes tested. For most locations, 2–10 variants differing by 1 to 6 markers were observed, a value within the range of the observed inter-strain variations (table S4, electronic supplementary material). Moreover, inter-population polymorphism was unexpectedly high between neighbouring populations sampled during the same period, as illustrated by the Portuguese populations 1 and 2, or the Spanish populations 3 and 4 (figure 1). French populations 7 and 8 were sampled at the same place with an interval of 11 years but display distinct variants.
A dramatically contrasting picture emerged from the analysis of Algerian, Cretan and Tunisian C. pipiens populations, in which only six variants were identified. All variants were distinct from southern European ones. In Tunisia variant 11 appeared nearly at fixation, identified in 65 of 68 mosquitoes analysed from six locations spreading over 500 km. The three remaining variants were found only once in population 16, sampled in 1996 near Monastir. In contrast to southern European populations, North African polymorphism appeared stable over time: we found a strictly identical pattern for the Tunis populations 14 and 15, sampled in 1992 and 2003, respectively, and for the Tunis strain, maintained in the laboratory since 1992. This strengthens the notion that WO prophage transmission is stable across several generations.
(e) WO prophage and cytoplasmic incompatibilities
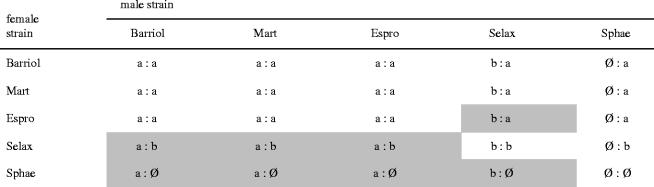
Previous work by Guillemaud et al. (1997) identified four incompatibility types among five C. pipiens strains (Barriol, Mart, Espro, Selax and Sphae). Barriol, Mart, and Espro strains were compatible, whereas Espro and Selax showed bi-directional incompatibility. All other possible crosses were unidirectionally incompatible. Since no sequence polymorphism of the ftsZ gene had been found by the authors, we examined WO phage polymorphism in the Wolbachia infecting the same strains at the time of the crossing experiment (kept in liquid nitrogen). All strains produced distinct presence/absence patterns, generating seven variants (only two were previously found; table S4). Two strains (Selax and Sphae) were infected by a mixture of distinct Wolbachia. Barriol and Mart, which displayed identical compatibility towards the other strains, differed from six markers indicating that distinct Wolbachia strains thus do not necessarily generate CI. Matching WO polymorphism to the CI pattern reported in Guillemaud et al. (1997) failed to show a correlation, except for Gp15. All crosses between Gp15a strains (Barriol, Espro and Mart, table 3) were fully compatible bi-directionnally. Crosses between Gp15a males and Gp15b (Selax) females were all incompatible. Inter-strain crosses with Gp15-negative (Sphae) females were all incompatible but Gp15-negative males did not induce CI in all cases (table 3). However, the correlation failed for crosses between Gp15b males and Gp15a females that were either compatible or incompatible.
Table 3.
Correlation between Gp15 gene product and CI pattern. (Incompatibilities between strains from Guillemaud et al. (1997) were marked by grey area, all other crosses being compatible. The first letter corresponds to the Gp15 status in the male strain, and the second one to the Gp15 status in the female strain. Ø indicates lack of Gp15 PCR product. All crosses with the same parental combination display the same CI pattern, excepted b : a crosses.)
4. Discussion
Availability of genetic markers is crucial to understand Wolbachia evolution and the highly complex CI pattern that affects C. pipiens populations throughout the world. The purpose of this study was to find Wolbachia polymorphic DNA regions and to combine them with those already described in order to genotype precisely Wolbachia strains, to evaluate their role in CI and to approach Wolbachia dynamics in field populations.
Several recent studies revealed that prophage WO is widespread in Wolbachia genomes (Bordenstein & Wernegreen 2004; Gavotte et al. 2004; Wu et al. 2004) and the detection of particles has lent support to the idea that WO prophage genes are activated and mobile (Masui et al. 2000; Masui et al. 2001). This notion was strengthened by the lack of congruence between Wolbachia and phage phylogenies established with the orf7 sequence (ORF Gp3 in our study), suggesting horizontal phage exchange between bacteria co-infecting the same intracellular environment (Bordenstein & Wernegreen 2004; Gavotte et al. 2004; Sanogo & Dobson 2004). Since all polymorphic regions found so far in wPip DNA are mobile genetic elements (Sanogo & Dobson 2004; Duron et al. 2005), the ‘real’ Wolbachia genes being monomorphic (Rousset et al. 1992; Guillemaud et al. 1997; Duron et al. 2005), it was proposed that these mobile genetic elements could favour genome fluidity and enhance rapid adaptation, especially in the highly diversified CI system of C. pipiens. For all these reasons we decided to study WO prophage organization and variability in the wPip genome.
(a) WO putative proteins multiple copies or multiple infections?
For each of the 24 WO ORFs studied, 2–6 copies were found dispersed in the wPip genome (table 1). The situation is clearly different in the wMel and wSim genomes (Wu et al. 2004; Salzberg et al. 2005), which contain only two WO prophage clusters, one complete and the other partial (table 1). The higher copy number in wPip may be overestimated due to a mixture in the Wolbachia used for the wPip genome project. Indeed, the Gp15a and Gp15b WO variants (this study) and the wPip1 and wPip3 Tr1 transposase variants (Duron et al. 2005), each mutually exclusive in all examined strains and field mosquitoes, are both present in the wPip genome sequences (table 1). This strongly suggests that the Pel strain used for the sequenced library was infected by different Wolbachia variants or contaminated by other strains (a contamination with the Bei strain is now mentioned on the wPip genome web page). Several short unassembled WO prophage clusters presented in table 1 might thus derive from different genomes.
Among the six different copies of Gp3 protein, four copies (Gp3a, b, c and d) were found concomitantly in individual mosquitoes from the MaClo strain (table 2). These four copies could derive from a single Wolbachia genome or from different Wolbachia genomes co-infecting the same mosquito. Co-infection has indeed been reported in other insect species, in which doubly infected males turned out to be incompatible with single infected females of either type (Rousset & Solignac 1995; Sinkins et al. 1995; Perrot-Minnot et al. 1996; Vavre et al. 1999). However, two issues do not support the co-infection hypothesis: PCR products amplified from single MaClo individuals were all monomorphic, and a strict mutually exclusive presence of Gp15 and Tr1 is observed. The presence of multiple WO ORFs copies thus probably results from multi-insertion in a single Wolbachia genome. Along this line, we previously proposed that the concomitant presence of the Tr1 transposase variants wPip1 and wPip4 in Slab and several North American samples resulted either from superinfection or from a duplication event (Duron et al. 2005). WO analysis of the same strain and samples showed a unique pattern, which strongly favours the duplication hypothesis.
Taken together, our data call for an extremely low level of super-infection in C. pipiens. This does not fit the prediction that doubly or multi-infected females should have a reproductive advantage over single or uninfected females, which would facilitate their spreading and fixation (Frank 1998). This suggests that multi-infection in C. pipiens is sharply limited by selective constraints that remain to be identified.
(b) Contrasted WO prophage polymorphism levels in southern Europe and North Africa
The combination of 10 WO and Tr1 markers allowed the detection of an unexpected level of polymorphism, discriminating up to 66 variants in a sample of moderate size (N=208) and even not including allelic polymorphism from sequence data. Variability mostly affects southern European populations, where 51 Wolbachia strains were identified, up to 10 variants being found in population 5. Availability of these markers now opens access to population dynamics to examine whether WO polymorphism is neutral or maintained by selection. However, since the coexistence of multiple bacterial variants that generate CI is predicted to be unstable within a population (Rousset et al. 1991), it is likely that many of the Wolbachia variants identified in these regions will not generate CI. The CI patterns observed in southern Europe (Laven 1967; Magnin et al. 1987; Guillemaud et al. 1997) would thus be restricted to contact areas between particular cytotypes, a possibility that could be evaluated by the frequency of incompatible egg-rafts in natural populations.
In contrast with southern European populations, North African populations showed a very low level of WO prophage polymorphism. This situation appears stable over time, as exemplified by Tunis samples which have kept the same WO pattern over at least the last 10 years. The reasons why a single WO variant is present all over Tunisia remain speculative. First, the Wolbachia variant 11 might generate CI that has facilitated its spread. Alternatively, the narrow C. pipiens habitat between the Mediterranean sea and the Sahara desert might have reduced migration and favoured genetic drift up to the fixation of a single Wolbachia variant. Lastly, a strong population bottleneck generated by the constant and massive use of insecticides may have occurred. All Tunisian populations indeed carry the G119S resistant allele at the ace-1 resistance locus, except Menzel wherein the additional variants were found (not shown; Weill et al. 2003).
(c) WO prophage and cytoplasmic incompatibility
The discovery of so many variants in southern Europe might now help explain the complex CI situation found in C. pipiens. Although the WO and Tr1 marker patterns do not correlate globally with the CI patterns already described in Guillemaud et al. (1997), we found an interesting but not fully consistent linkage between Gp15 variability and CI. This putative secretory protein was reported to share sequence homology with a virulence-related protein of a pathogenic bacteria and proposed to be responsible for sexual alteration (Fujii et al. 2004). Analysis of a much larger number of crosses will be necessary to unambiguously establish the correlation between CI and the Gp15 protein. Besides, the implication of Wolbachia variants in the establishment of CI does not preclude the influence of others factors like nuclear genes, in particular restorer genes (Rousset et al. 1991; Sinkins et al. 2005). Construction of strains harbouring distinct Wolbachia variants in a same nuclear genetic background is currently underway to address this latter issue.
In conclusion, our data demonstrating the high variability of WO prophage sequences give new insights into the C. pipiens and Wolbachia relationship: (i) different Wolbachia variants do not necessarily generate CI. (ii) Wolbachia genotyping is an obligatory step before addressing the CI status between mosquito strains. Indeed, the mixture of variants found in several strains may induce a wrong CI interpretation and hinder the finding of markers correlated with CI. (iii) The extreme and apparently stable over time level of polymorphism of Wolbachia infecting southern European mosquitoes, in contrast with the quasi-monomorphism found in North African Wolbachia populations, raises novel issues on the dynamics of Wolbachia infection and warrants the use of the WO markers to analyse the genetic structure of Wolbachia infecting field populations. This may open routes for the control of vector-borne diseases since Wolbachia are considered as a promising driving force for manipulating gene pools of mosquito populations (Sinkins 2004).
Acknowledgments
The authors thank C. Berticat and A. Berthomieu for technical help, N. Pasteur and M. Raymond for critical reading of the manuscript. This work was financed by APR ‘Evaluation et réduction des risques liés à l'utilisation des pesticides’ (Ministère de l'Ecologie et du Développement Durable). Contribution 2005-090 of the Institut des Sciences de l'Evolution de Montpellier (UMR CNRS 5554). The authors declare they have no competing financial interests.
Supplementary Material
References
- Amin A.M, Pereis H.T.R. Detection and selection of organophosphorate and carbamate resistance in Culex quinquefasciatus from Saudi Arabia. Med. Vet. Entomol. 1990;4:269–273. doi: 10.1111/j.1365-2915.1990.tb00439.x. [DOI] [PubMed] [Google Scholar]
- Barr, A. R. 1966 Cytoplasmic incompatibility as a means of eradication of Culex pipiens L. In The 33th Annual Conf. of the Californian Mosquito Control Association, Inc., pp. 32–35. Monterey. [PubMed]
- Berticat C, Rousset F, Raymond M, Berthomieu A, Weill M. High Wolbachia density in insecticide-resistant mosquitoes. Proc. R. Soc. B. 2002;269:1413–1416. doi: 10.1098/rspb.2002.2022. doi:10.1098/rspb.2002.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein S.R, Wernegreen J.J. Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol. Biol. Evol. 2004;21:1981–1991. doi: 10.1093/molbev/msh211. doi:10.1093/molbev/msh211 [DOI] [PubMed] [Google Scholar]
- Duron O, Lagnel J, Raymond M, Bourtzis K, Fort P, Weill M. Transposable element polymorphism of Wolbachia in the mosquito Culex pipiens: evidence of genetic diversity, super-infection and recombination. Mol. Ecol. 2005;14:1561–1573. doi: 10.1111/j.1365-294X.2005.02495.x. doi:10.1111/j.1365-294X.2005.02495.x [DOI] [PubMed] [Google Scholar]
- Frank S.A. Dynamics of cytoplasmic incompatibility with multiple Wolbachia infections. J. Theor. Biol. 1998;192:213–218. doi: 10.1006/jtbi.1998.0652. doi:10.1006/jtbi.1998.0652 [DOI] [PubMed] [Google Scholar]
- Fujii Y, Kubo T, Ishikawa H, Sasaki T. Isolation and characterization of the bacteriophage WO from Wolbachia, an arthropod endosymbiont. Biochem. Biophys. Res. Commun. 2004;317:1183–1188. doi: 10.1016/j.bbrc.2004.03.164. doi:10.1016/j.bbrc.2004.03.164 [DOI] [PubMed] [Google Scholar]
- Gavotte L, Vavre F, Henri H, Ravallec M, Stouthamer R, Boulétreau M. Diversity, distribution and specificity of WO phage infection in Wolbachia of four insect species. Insect Mol. Biol. 2004;13:147–153. doi: 10.1111/j.0962-1075.2004.00471.x. doi:10.1111/j.0962-1075.2004.00471.x [DOI] [PubMed] [Google Scholar]
- Guillemaud T, Pasteur N, Rousset F. Contrasting levels of variability between cytoplasmic genomes and incompatibility types in the mosquito Culex pipiens. Proc. R. Soc. B. 1997;264:245–251. doi: 10.1098/rspb.1997.0035. doi:10.1098/rspb.1997.0035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laven H. Crossing experiments with Culex strains. Evolution. 1951;5:370–375. [Google Scholar]
- Laven H. Speciation and evolution in Culex pipiens. In: Wright J, Pal R, editors. Genetics of insect vectors of disease. vol. 7. Elsevier; Amsterdam: 1967. pp. 251–275. [Google Scholar]
- Magnin M, Pasteur N, Raymond M. Multiple incompatibilities within populations of Culex pipiens L. in southern France. Genetica. 1987;74:125–130. doi: 10.1007/BF00055223. doi:10.1007/BF00055223 [DOI] [PubMed] [Google Scholar]
- Masui S, Kamoda S, Sasaki T, Ishikawa H. Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J. Mol. Evol. 2000;51:491–497. doi: 10.1007/s002390010112. [DOI] [PubMed] [Google Scholar]
- Masui S, Kuroiwa H, Sasaki T, Inui M, Kuroiwa T, Ishikawa H. Bacteriophage WO and virus-like particles in Wolbachia, an endosymbiont of arthropods. Biochem. Biophys. Res. Commun. 2001;283:1099–1104. doi: 10.1006/bbrc.2001.4906. doi:10.1006/bbrc.2001.4906 [DOI] [PubMed] [Google Scholar]
- Perrot-Minnot M, Guo L.R, Werren J.H. Single and double infections with Wolbachia in the parasitic wasp Nasonia vitripennis: effects on compatibility. Genetics. 1996;143:961–972. doi: 10.1093/genetics/143.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. Genepop (version 1.2), a population genetics software for exact tests and ecumenicism. J. Hered. 1995;86:248–249. [Google Scholar]
- Rogers S.O, Bendich A.J. Extraction of DNA from plant tissues. In: Gelvin S.B, Schilperoort R.A, editors. Plant molecular biology manual. vol. A6. Kluwer Academic Publishers; Boston, MA: 1988. pp. 1–10. [Google Scholar]
- Rousset F, Raymond M. Cytoplasmic incompatibility in insects: why sterilize females? Trends Ecol. Evol. 1991;6:54–57. doi: 10.1016/0169-5347(91)90123-F. doi:10.1016/0169-5347(91)90123-F [DOI] [PubMed] [Google Scholar]
- Rousset F, Solignac M. Evolution of single and double Wolbachia symbioses during speciation in the Drosophila simulans complex. Proc. Natl Acad. Sci. USA. 1995;92:6389–6393. doi: 10.1073/pnas.92.14.6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F, Raymond M, Kjellberg F. Cytoplasmic incompatibilities in the mosquito Culex pipiens: how to explain a cytotype polymorphism? J. Evol. Biol. 1991;4:69–81. doi:10.1046/j.1420-9101.1991.4010069.x [Google Scholar]
- Rousset F, Bouchon D, Pintureau B, Juchault P, Solignac M. Wolbachia endosymbionts responsible for various alterations of sexuality in arthropods. Proc. R. Soc. B. 1992;250:91–98. doi: 10.1098/rspb.1992.0135. [DOI] [PubMed] [Google Scholar]
- Salzberg S.L, Dunning Hotopp J.C, Delcher A.L, Pop M, Smith D.R, Eisen M.B, Nelson W.C. Serendipitous discovery of Wolbachia genomes in multiple Drosophila species. Genome Biol. 2005;6:R23. doi: 10.1186/gb-2005-6-3-r23. doi:10.1186/gb-2005-6-3-r23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanogo Y.O, Dobson S.L. Molecular discrimination of Wolbachia in the Culex pipiens complex: evidence for variable bacteriophage hyperparasitism. Insect Mol. Biol. 2004;13:365–369. doi: 10.1111/j.0962-1075.2004.00498.x. doi:10.1111/j.0962-1075.2004.00498.x [DOI] [PubMed] [Google Scholar]
- Sinkins S.P. Wolbachia and cytoplasmic incompatibility in mosquitoes. Insect Biochem. Mol. Biol. 2004;34:723–729. doi: 10.1016/j.ibmb.2004.03.025. doi:10.1016/j.ibmb.2004.03.025 [DOI] [PubMed] [Google Scholar]
- Sinkins S.P, Braig H.R, Oneill S.L. Wolbachia superinfections and the expression of cytoplasmic incompatibility. Proc. R. Soc. B. 1995;261:325–330. doi: 10.1098/rspb.1995.0154. [DOI] [PubMed] [Google Scholar]
- Sinkins S, Walker T, Lynd A.R, Steven A.R, Makepeace B.L, Godfray H.C.J, Parkhill J. Wolbachia variablity and host effects on crossing type in Culex mosquitoes. Nature. 2005;436:257–260. doi: 10.1038/nature03629. doi:10.1038/nature03629 [DOI] [PubMed] [Google Scholar]
- Subbarao S.K. Cytoplasmic incompatibility in mosquitoes. In: Steiner W.W.M, Tabachnick W.J, Rai K.S, Narang S, editors. Recent developments in the genetics of insect disease vectors. Stipes; Champaign, IL: 1982. pp. 313–342. [Google Scholar]
- Turelli M, Hoffmann A.A. Microbe-induced cytoplasmic incompatibility as a mechanism for introducing transgenes into arthropod populations. Insect Mol. Biol. 1999;8:243–255. doi: 10.1046/j.1365-2583.1999.820243.x. doi:10.1046/j.1365-2583.1999.820243.x [DOI] [PubMed] [Google Scholar]
- Vavre F, Fleury F, Lepetit D, Fouillet P, Bouletreau M. Phylogenetic evidence for horizontal transmission of Wolbachia in host–parasitoid associations. Mol. Biol. Evol. 1999;16:1711–1723. doi: 10.1093/oxfordjournals.molbev.a026084. [DOI] [PubMed] [Google Scholar]
- Weill M, Berticat C, Raymond M, Chevillon C. Quantitative polymerase chain reaction to estimate the number of amplified esterase genes in insecticide-resistant mosquitoes. Anal. Biochem. 2000;285:267–270. doi: 10.1006/abio.2000.4781. doi:10.1006/abio.2000.4781 [DOI] [PubMed] [Google Scholar]
- Weill M, et al. Insecticide resistance in mosquito vectors. Nature. 2003;423:136–137. doi: 10.1038/423136b. doi:10.1038/423136b [DOI] [PubMed] [Google Scholar]
- Werren J.H. Wolbachia and speciation. In: Howard D, Berlocher S, editors. Endless forms: species and speciation. Oxford University Press; 1998. pp. 245–260. [Google Scholar]
- Wu M, et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004;2:327–341. doi: 10.1371/journal.pbio.0020069. doi:10.1371/journal.pbio.0020327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen J.H, Barr A.R. The etiological agent of cytoplasmic incompatibility in Culex pipiens. J. Invertebr. Pathol. 1973;22:242–250. doi: 10.1016/0022-2011(73)90141-9. doi:10.1016/0022-2011(73)90141-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.