Abstract
In sheep's fescue, Festuca ovina, genes coding for the cytosolic enzyme phosphoglucose isomerase, PGIC, are not only found at the standard locus, PgiC1, but also at a segregating second locus, PgiC2. We have used PCR-based sequencing to characterize the molecular structure and evolution of five PgiC1 and three PgiC2 alleles in F. ovina. The three PgiC2 alleles were complex in that they carried two gene copies: either two active genes or one active and one pseudogene. All the PgiC2 sequences were very similar to each other but highly diverged from the five PgiC1 sequences. We also sequenced PgiC genes from several other grass species. Phylogenetic analysis of these sequences indicates that PgiC2 has introgressed into F. ovina from the distant genus Poa. Such an introgression may, for example, follow from a non-standard fertilization with more than one pollen grain, or a direct horizontal gene transfer mediated by a plant virus.
Keywords: Festuca ovina, gene duplication, Pgi, introgression, horizontal gene transfer
1. Introduction
In higher eukaryotes, polyploidy and large duplicated chromosome segments play an important role in determining gene number and function, as shown by many genome sequencing studies (Rubin et al. 2000; Eichinger et al. 2005). A change in the copy number of a single gene is, however, of no less interest, since this phenomenon results from a combination of a rare molecular event and a unique selective process that illustrates the smallest step whereby genome evolution occurs. A new, additional gene is particularly informative when it is not yet fixed but segregates in the species, since both the molecular origin and the selective forces acting on the duplication are directly available for study (Lootens et al. 1993; Long et al. 2003).
Extra gene copies for cytosolic phosphoglucose isomerase (PGIC) have been reported to segregate in sheep's fescue, Festuca ovina L. (Bengtsson et al. 1995; Prentice et al. 1995). Previous studies showed that this variation is due to a polymorphic second locus, PgiC2 (Ghatnekar 1999; Ghatnekar & Bengtsson 2000). The two loci, PgiC1 and PgiC2, assort independently and functioning hybrid enzymes are readily formed by all possible intergenic heterodimers (Ghatnekar 1999). In southern Sweden, up to 10% of plants have at least one active gene at the second locus. The allelic variation at PgiC1 is extensive (Bengtsson et al. 1995). At the second locus, PgiC2, an investigation based on crosses demonstrated the presence of two alleles, PgiC2b and PgiC2c, that code for different allozymes; an additional complex PgiC2 variant codes for both alleles b and c in close linkage (Ghatnekar 1999). The possibility that PgiC2 originated via a retrotransposon-mediated duplication was refuted when we showed that the genes at this locus have introns (Ghatnekar & Bengtsson 2000). The same investigation also indicated that plants lacking active PgiC2 genes do not have any related but inactivated genes at this locus. This result suggests that here evolution is promoting the spread of new additional active genes at a chromosomal site normally devoid of PgiC genes.
Here we report on the genetic diversity within and between the two PgiC loci in F. ovina as revealed by PCR-based DNA sequencing. Our results show that the two loci are highly diverged, and we suggest that PgiC2 has entered F. ovina from the genus Poa via an introgression event.
2. Material and methods
(a) Plant material
The F. ovina plants analysed belonged to the diploid subspecies F. ovina ssp. vulgaris (Koch) Sch. & Kell (2n=14). Plants were collected in southern Sweden (Bengtsson et al. 1995) or were derived from such plants by controlled crosses (Ghatnekar 1999). For the study of PgiC1, five plants known to be homozygous for the alleles a1, a2, b, c and d were used. Except for PgiC1 d/d, a natural homozygote for a very rare allele, the plants were produced in crossing experiments and were homozygous by descent with respect to PgiC1. Three F. ovina plants hemizygous for PgiC2 were also studied. These plants were the result of crossing experiments (Ghatnekar 1999), and their genotypes with respect to the two PgiC loci were PgiC1 d/d PgiC2 b/0, PgiC1 d/d PgiC2 c/0 and PgiC1 d/d PgiC2 bc/0.
Two additional fescues were included in our phylogenetic study of PgiC sequences: Festuca polesica and Festuca altissima. Festuca polesica, like F. ovina, is a diploid outcrossing species belonging to the group of fine-leaved fescues, whereas F. altissima (also diploid and outcrossing) belongs to the broad-leaved fescues. Festuca altissima is, therefore, a more distant relative of F. ovina than F. polesica (Torrecilla et al. 2004). We also examined Bromus sterilis, Poa supina, Poa trivialis, Poa chaixii, all diploid outcrossing species, plus Aira praecox, a diploid self-fertilizing species which according to Torrecilla et al. (2004) falls in the same monophyletic clade as the fine-leaved fescues. All these species have 2n=14 (Lid 1979). One plant from each species was included in the phylogenetic analyses. All grasses were collected in the same geographical area in southernmost Sweden.
(b) DNA isolation, PCR amplification and DNA sequencing
Total genomic DNA was isolated from leaf material using the Qiagen DNeasy Plant Mini Kit. Primers were constructed using the software Oligo 4.0 (MedProbe). The published cDNA sequence for PgiC in maize, Zea mays (GenBank accession no. U 17225; Lal & Sachs 1995), was used to construct primers for the initial amplification and sequencing of allele PgiC1 d from F. ovina. We used information from the PgiC1 genomic sequence from Clarkia lewisii (Thomas et al. 1993) to infer intron/exon boundaries. Additional primers used in later amplifications were constructed from the PgiC1 d sequence.
The large length difference in intron 4 between PgiC1 d and PgiC2 (Ghatnekar & Bengtsson 2000) was used to construct primers specific for PgiC2: a forward primer 1′ (5′-ATCCTTATTATTCCTTCAGCTGTTC-3′) and a reverse primer, 2′ (5′-AATCGGTTCCATCCACTCCA-3′). Both these primers could be used in combination with other PgiC primers to generate PgiC2-specific PCR products. In those instances when F. ovina primers were not satisfactory, we constructed additional primers for the amplifications of PgiC genes from other grass species. Information on primers can be obtained from the authors upon request. A PCR programme with 1 min at 95°, followed by 30 cycles of 30 s at 95 °C, 1 min at 2 or 3 °C below the Tm, 45–60 s at 72 °C and a final 7 min extension at 72 °C, was used. When necessary, PCR fragments were excised and purified with Ultrafree-DA (Millipore) for subsequent sequencing.
Taq DNA Polymerase (Roche) was used for most PCR amplifications up to 2.5 kb, while Expand Long Template PCR System (Roche) was used for longer fragments. Primer 1′ and a reverse primer annealing to exon 21 (5′-CCTAGCTCCACTCCCCACTG-3′) amplified about 5 kb of PgiC2. Nested PCRs of this fragment produced fragments not longer than 1.5–2 kb. PCR products were purified using the QIAquick PCR Purification Kit (Qiagen). PCR primers or internal primers were used for sequencing in both directions. Direct sequencing was performed with an ABI 3100 automated DNA sequencer. Sequences were aligned and ambiguous sites resolved using Sequencher v. 3.0 (Genes Codes Corp.).
In order to obtain unambiguous haplotypes for the PgiC2b and PgiC2c alleles, a PCR product of 5.2 kb was cloned from the PgiC2 locus using the pGEMR-T Easy Vector System (Promega). The fragment was amplified with primer 1′ and a reverse primer in exon 22 (5′-CATGCAACTGTTTCCTCACTC-3′). High efficiency competent cells (Epicurian colir XL blue, AHdiagnostics) were used for transformations with subsequent ampicillin selection. Screening was done on S-Gal/LB Agar blend (Sigma-Aldrich). Thirteen colonies were picked from the PgiC2b and PgiC2c plates, respectively, and DNA was amplified with Expand Long Template PCR System (Roche) at 49 °C annealing temperature.
(c) Phylogenetic analyses
Data on nucleotide substitutions, indels and amino acid replacements in 1182 bp of exon sequence were assembled using MacClade v. 4.05 (Maddison & Maddison 2000). Phylogenetic analyses were conducted using maximum parsimony (MP) and neighbour-joining (NJ) algorithms implemented in PAUP* v. 4.0b10 (Swofford 1998). For MP analyses, the branch-and-bound option was used. The NJ analyses were performed using the Kimura two-parameter model (Kimura 1980). Relative stability of phylogenetic trees was assessed with bootstrap analysis using 10 000 replicates.
3. Results
(a) Molecular characterization
All exon sequences have been deposited in GenBank (accession no. DQ225730–DQ225745) as well as the exon–intron sequence for PgiC2c (DQ282377). For a graphic description of the PgiC gene, see figure 1. Five PgiC1 alleles from F. ovina were sequenced for most of their lengths (ranging from 5277 to 6055 bp). The alleles were distinctly different, with 1.3% average nucleotide diversity in exon sequence.
Figure 1.
The PgiC gene as exemplified by the d allele of PgiC1 in Festuca ovina. Exons and introns are drawn to scale. Exon sizes are the same for all genes investigated (except for the pseudogenes that lack 27 bp in exon 13), while intron sizes vary.
When the two alleles b and c from PgiC2 were sequenced over the same region (5742 and 6020 bp), only one nucleotide difference was detected between them (table 1). Allele b had G and allele c had A at position 876 in exon 5. This substitution corresponds to the difference between glutamic acid and lysine and, presumably, caused the amino acid change responsible for the electrophoretic mobility shift. The alleles from PgiC1 and PgiC2 had the same number and length of exons (figure 1), while the length and sequence of their introns differed substantially.
Table 1.
Differences between PgiC2 sequences. (Base pair positions correspond to GenBank accession no. DQ282377.)
| sequence region and position | ||||||
|---|---|---|---|---|---|---|
| exon 5 | intron 12–exon 13 | exon 15 | intron 15 | intron 16 | intron 17 | |
| bp 876 | bp 3258–3286 | bp 3567 | bp 3781 | bp 4140 | bp 4528 | |
| PgiC2b | G | present | G | G | C | G |
| PgiC2c | A | present | G | G | C | G |
| PgiC2ψb | A | deletion | T | A | C | A |
| PgiC2ψc | A | deletion | T | A | T | A |
Cloning of PgiC2b and PgiC2c revealed a molecularly complex structure, in that each allele turned out to consist of one active gene and one pseudogene. The four sequences—PgiC2b and its associated pseudogene PgiC2ψb and PgiC2c and its associated pseudogene PgiC2ψc—were very similar except for a 29 bp deletion covering the junction between intron 12 and exon 13 in the two pseudogenes (table 1). The PgiC2 allelic variant with two expressed genes, PgiC2bc, showed no sign of the deletion characterizing the pseudogenes when sequenced over the intron 12–exon 13 region (590 bp). This variant is, thus, presumed to consist of one active allele of each kind. We could not obtain any PCR products of the region between the two active PgiC2 genes, and the exact molecular organization of this complex locus is therefore still unknown.
From F. polesica, F. altissima, P. supina, P. trivialis, P. chaixii, A. praecox and B. sterilis, we obtained more than 1180 bp PgiC exon sequence, covering exons 5–11 and exons 13–21. In all species, the PgiC gene had the same organization and lengths of exons, but there were large differences in the intron sequences. In none of the investigated species, except F. ovina, did we find evidence for the existence of more than one locus with PgiC-like sequences, active or inactive.
(b) Evolutionary characterization
Phylogenetic analyses of PgiC1 exon sequences from the five F. ovina alleles always resulted in star-like trees (data not shown). The active genes and pseudogenes at PgiC2 exhibited very little sequence variation, as described previously (table 1).
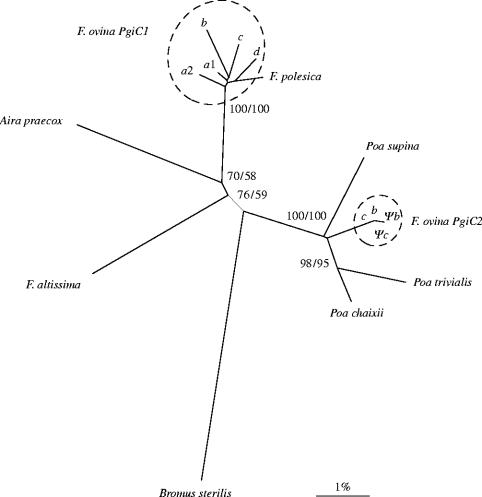
To compare the sequences from the two PgiC loci with each other and with sequences from the other investigated species, phylogenetic analyses based on exons and their corresponding parts in the non-coding pseudogenes were performed. NJ and MP methods gave identical tree topologies. Figure 2 shows the NJ tree based on 1182 bp of exon sequence from the two F. ovina loci and the single PgiC sequences from F. polesica, F. altissima, A. praecox, P. supina, P. trivialis and P. chaixii, with B. sterilis as outgroup. The tree shows that all PgiC1 sequences from F. ovina cluster together. The F. polesica sequence occurs in the same lineage with 100% bootstrap support. The tree also shows the close phylogenetic relationship between Aira and the fine-leaved fescues, represented by F. ovina and F. polesica, as already reported by Torrecilla et al. (2004) based on ITS and trnL-F sequence data.
Figure 2.
Neighbour-joining (NJ) tree based on Kimura two-parameter distances of 11 Festuca, one Aira praecox and three Poa PgiC exon sequences (1182 bp), using Bromus sterilis as outgroup. Sequences from PgiC1 and PgiC2 of F. ovina are circled with dotted lines. Nodal support is given by bootstrap values based on 10 000 replications (neighbour-joining/maximum parsimony).
The surprise in the PgiC tree topology comes from the large divergence between the sequences from the two PgiC loci in F. ovina. Their exon sequences differ by a net divergence of 5.2%, suggesting that they split a long time ago. The PgiC2 sequences do not group with the alleles from the F. ovina PgiC1 locus or with any of the other Festuca sequences, but with the Poa species, in a branch that is separated from the rest of the analysed grasses by a bootstrap value of 100%.
4. Discussion
Our study shows that PgiC1 and PgiC2 sequences in F. ovina are so diverged that they appear to come from different species. The differences are evenly distributed along the length of the sequences (data not shown), implying a long-term accumulation of nucleotide substitutions in introns and exons, as well as of indels in introns. This pattern indicates that a substantial period of isolation, rather than a few dramatic events, due to, for example, transposing elements has caused the PgiC1–PgiC2 divergence.
The phylogenetic analyses reveal that the PgiC2 sequences are not closely related to any of the PgiC1 alleles. Instead, the PgiC2 sequences form a well-supported lineage together with the PgiC sequences from three diploid Poa sequences (P. chaixii, P. supina and P. trivialis; figure 2). According to Catalán et al. (2004), molecular phylogenetic analyses based on ITS and trnL-F sequences recognize an early split between the lineage leading to Poa and related grass genera, and the lineage leading to festucoid grasses. Later, the second lineage divided into the two lineages of broad-leaved and fine-leaved types, with genera additional to Festuca present in both. With the exception of the PgiC2 sequences, our data are in perfect agreement with these results: the fine-leaved species F. ovina, F. polesica and A. praecox cluster together; the broad-leaved type F. altissima falls outside this group; and the combined festucoid lineage is clearly distinct from the Poa lineage (figure 2). The unexpected result obtained by us is that the two genes PgiC1 and PgiC2—today found within F. ovina—must have started to diverge well before the split between the fine-leaved and broad-leaved festucoids.
The most likely interpretation of our data is that PgiC2 entered the F. ovina lineage relatively recently from a Poa species or a closely related genus. Poa is a large genus with many allopolyploid species that may harbour genomes of different origins (cf. Patterson et al. 2005). The close similarity between PgiC2 and the PgiC genes of the three diploid Poa species excludes, however, the possibility that PgiC2 is derived from a genome not characteristic for Poa. In future studies of different Poa species, we hope to find sequences even more closely related to PgiC2, which will give us the possibility of identifying the specific donor genome. From the degree of divergence between PgiC2 and this sequence, we should also be able to obtain an estimate of when the gene transfer occurred.
An alternative interpretation of our data would be that the PgiC locus duplicated a long time ago in an early Festuca lineage and that PgiC2 has continued to exist within the lineage leading up to present day F. ovina, while being lost from other lineages and present-day species. According to Gottlieb & Ford (1997), such a model applies to Clarkia, where extant species have either one or two active loci for PGIC. In the F. ovina case, this interpretation is unlikely, since PgiC2 is not fixed but occurs in up to 10% of southern Swedish F. ovina plants. This fact makes it more difficult to understand how PgiC2 could have existed in the lineage for such an extended period of time. In addition, we have been unable to find indications of PgiC2-like sequences in any of the analysed Festuca plants lacking active PgiC2 genes. Most important, however, is the fact that the model assuming an old duplication does not explain today's close similarity between PgiC2 and the three PgiC sequences from Poa species. Thus, we find that the explanation based on an early duplication is much less likely than an explanation based on a recent introgression event.
As far as known, hybrids between Poa and Festuca are not spontaneously formed in nature today (Knobloch 1968; Hegi 1996). The presence of a Poa-like PgiC sequence in F. ovina must therefore be due to an unusual and rare event. Such an introgression may, for example, follow from a non-standard fertilization involving more than one pollen grain; alternatively, some kind of more direct horizontal gene transfer may have been mediated by a plant virus (cf. Bergthorsson et al. 2003, 2004; Martin 2005). The complex molecular structure of PgiC2, with a chromosomal arrangement of two closely linked gene copies, can be taken as a weak indication that the process that brought PgiC2 into its present chromosomal position included at least one step involving transposing elements.
Our results are based on sequences of PgiC2 from three well-separated collection sites. The active sequences and the pseudogenes were all very similar to each other (table 1); so were also the sequences from the complex bc allele that presumably has arisen via non-homologous recombination. Similarly, in a previous population survey we found no variation among 18 alleles of PgiC2 with respect to the length of intron 4, whereas much diversity was observed for PgiC1 (Ghatnekar & Bengtsson 2000). Thus, our data indicate that PgiC2 sequences in southern Sweden are all very similar to each other. This lack of variation implies that a recent spread of PgiC2 must have occurred in F. ovina. More extensive population sampling will be performed to determine whether the observed pattern is due to random events during population expansion after the last glaciation or to natural selection favouring the new chromosome segment.
Acknowledgments
We wish to express our gratitude to Leslie Gottlieb for inspiration. We also want to thank Alf Ceplitis, Deborah Charlesworth and Torbjörn Säll for valuable discussions, Bengt Jacobsson for his tending of the plants, Pernilla Vallenback for help with the Poa sequencing, and Per Lassen, Clive Stace and Peder Weibull for helpful information on systematics and hybridizations in Poaceae. The research was supported by grants from the Swedish Research Council, the Nilsson-Ehle Fund and the Jörgen Lindström Fund.
References
- Bengtsson B.O, Weibull P, Ghatnekar L. The loss of alleles by sampling: a study of the common outbreeding grass Festuca ovina over three geographic scales. Hereditas. 1995;122:221–238. doi:10.1111/j.1601-5223.1995.00221.x [Google Scholar]
- Bergthorsson U, Adams K.L, Thomason B, Palmer J.D. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature. 2003;424:197–201. doi: 10.1038/nature01743. doi:10.1038/nature01743 [DOI] [PubMed] [Google Scholar]
- Bergthorsson U, Richardson A.O, Young G.J, Goertzen L.R, Palmer J.D. Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc. Natl Acad. Sci. USA. 2004;101:17 747–17 752. doi: 10.1073/pnas.0408336102. doi:10.1073/pnas.0408336102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalán P, Torrecilla P, López Rodrígues J.Á, Olmstead G. Phylogeny of the festucoid grasses of subtribe Loliinae and allies (Poeae, Pooideae) inferred from ITS and trnL-F sequences. Mol. Phylogenet. Evol. 2004;31:517–541. doi: 10.1016/j.ympev.2003.08.025. doi:10.1016/j.ympev.2003.08.025 [DOI] [PubMed] [Google Scholar]
- Eichinger L, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. doi:10.1038/nature03481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghatnekar L. A polymorphic duplicated locus for cytosolic PGI segregating in sheep's fescue (Festuca ovina L.) Heredity. 1999;83:451–459. doi: 10.1038/sj.hdy.6885750. doi:10.1038/sj.hdy.6885750 [DOI] [PubMed] [Google Scholar]
- Ghatnekar L, Bengtsson B.O. A DNA marker for the duplicated cytosolic PGI genes in sheep's fescue (Festuca ovina L.) Genet. Res. 2000;7:319–322. doi: 10.1017/s0016672300004705. doi:10.1017/S0016672300004705 [DOI] [PubMed] [Google Scholar]
- Gottlieb L.D, Ford V.S. A recently silenced, duplicate PgiC locus in Clarkia. Mol. Biol. Evol. 1997;14:125–132. doi: 10.1093/oxfordjournals.molbev.a025745. [DOI] [PubMed] [Google Scholar]
- Hegi G. Parey; Berlin: 1996. Illustrierte Flora von Mittel-Europa. Bd 1.T.3. Lief 8/9. [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. doi:10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Knobloch I.W. Michigan State University; East Lansing: 1968. A check list of crosses in the Gramineae. [Google Scholar]
- Lal S.K, Sachs M.M. Cloning and characterization of an anaerobically induced cDNA encoding glucose-6-phosphate isomerase from maize. Plant Physiol. 1995;108:1295–1296. doi: 10.1104/pp.108.3.1295. doi:10.1104/pp.108.3.1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lid J. Det Norske Samlaget; Oslo: 1979. Norsk og svensk flora. [Google Scholar]
- Long M, Betrán E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat. Rev. Genet. 2003;4:865–875. doi: 10.1038/nrg1204. doi:10.1038/nrg1204 [DOI] [PubMed] [Google Scholar]
- Lootens S, Burnett J, Friedman T.B. An intraspecific gene duplication polymorphism of the urate oxidase gene of Drosophila virilis: a genetic and molecular analysis. Mol. Biol. Evol. 1993;10:635–646. doi: 10.1093/oxfordjournals.molbev.a040028. [DOI] [PubMed] [Google Scholar]
- Maddison D.R, Maddison W.P. Sinauer Associates; Sunderland, MA: 2000. MacClade 4: analysis of phylogeny and character evolution. Version 4.0. [Google Scholar]
- Martin W. Lateral gene transfer and other possibilities. Heredity. 2005;94:565–566. doi: 10.1038/sj.hdy.6800659. doi:10.1038/sj.hdy.6800659 [DOI] [PubMed] [Google Scholar]
- Patterson J.T, Larson S.R, Johnson P.G. Genome relationships in polyploid Poa pratensis and other Poa species inferred from phylogenetic analysis of nuclear and chloroplast DNA sequences. Genome. 2005;48:76–87. doi: 10.1139/g04-102. doi:10.1139/g04-102 [DOI] [PubMed] [Google Scholar]
- Prentice H.C, Lönn M, Lefkovitch L.P, Runyeon H. Associations between allele frequencies in Festuca ovina and habitat variation in the alvar grasslands on the Baltic island of Öland. J. Ecol. 1995;83:391–402. [Google Scholar]
- Rubin G.M, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. doi:10.1126/science.287.5461.2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, MA: 1998. PAUP* phylogenetic analysis using parsimony and other methods. [Google Scholar]
- Thomas V.S, Ford B.R, Pichersky E, Gottlieb L.D. Molecular characterization of duplicate cytosolic phosphoglucose isomerase genes in Clarkia and comparison to the single gene in Arabidopsis. Genetics. 1993;135:895–905. doi: 10.1093/genetics/135.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrecilla P, López-Rodríguez J.-A, Catalán P. Phylogenetic relationships of Vulpia and related genera (Poeae, Poaceae) based on analysis of ITS and trnL-F sequences. Ann. Missouri Bot. Gard. 2004;91:124–158. [Google Scholar]